Abstract
Staphylococcus aureus strains that are susceptible to the β-lactam antibiotic oxacillin despite carrying mecA (OS-MRSA) cause serious clinical problems globally because of their ability to easily acquire β-lactam resistance. Understanding the genetic mechanism(s) of acquisition of the resistance is therefore crucial for infection control management. For this purpose, a whole-genome sequencing-based analysis was performed using 43 clinical OS-MRSA strains and 100 mutants with reduced susceptibility to oxacillin (MICs 1.0–256 µg/mL) generated from 26 representative OS-MRSA strains. Genome comparison between the mutants and their respective parent strains identified a total of 141 mutations in 46 genes and 8 intergenic regions. Among them, the mutations are frequently found in genes related to RNA polymerase (rpoBC), purine biosynthesis (guaA, prs, hprT), (p)ppGpp synthesis (relSau), glycolysis (pykA, fbaA, fruB), protein quality control (clpXP, ftsH), and tRNA synthase (lysS, gltX), whereas no mutations existed in mec and bla operons. Whole-genome transcriptional profile of the resistant mutants demonstrated that expression of genes associated with purine biosynthesis, protein quality control, and tRNA synthesis were significantly inhibited similar to the massive transcription downregulation seen in S. aureus during the stringent response, while the levels of mecA expression and PBP2a production were varied. We conclude that a combination effect of mecA upregulation and stringent-like response may play an important role in acquisition of β-lactam resistance in OS-MRSA.
Similar content being viewed by others
Introduction
Staphylococcus aureus is an important bacterial pathogen that can cause life-threatening infections in both humans and animals1,2. A known feature of S. aureus is its evolutionary potential to develop antibiotic resistance under selection pressure via antibiotic treatment. Methicillin-resistant S. aureus (MRSA) is resistant to the entire class of β-lactam antibiotics, including penicillin, methicillin, and cefazolin3. It was first recognized as a problematic pathogen in hospital settings, but it has subsequently emerged in community settings and livestock3,4,5. MRSA infections remain a major concern in the clinical setting because they are more difficult to treat than infections caused by other β-lactam-susceptible strains of S. aureus. The β-lactam resistance in MRSA is primarily mediated by a non-native mecA gene encoding modified penicillin-binding protein 2a (PBP2a), which has an extremely low affinity for β-lactams. The expression of PBP2a is dependent on the presence of functional MecI/MecR1/MecR2 and BlaI/BlaR1 regulators in the mec and bla operons, respectively6,7. However, the level of β-lactam resistance does not always correlate with that of PBP2a expression6,7,8.
Recently, oxacillin-susceptible mecA-positive S. aureus (OS-MRSA) strains have been increasingly reported worldwide in clinical isolates as well as in animals and food9,10,11,12,13,14,15. In clinical microbiology laboratories, an oxacillin minimum inhibitory concentration (MIC) ≥ 4 µg/mL or cefoxitin MIC ≥ 8 µg/mL is routinely used as a breakpoint for diagnosing MRSA, whereas the presence of mecA has been used as a genetic marker for identification of MRSA16. Owing to its susceptibility to oxacillin, OS-MRSA might be misidentified as methicillin-susceptible S. aureus (MSSA) in routine clinical laboratories in which mecA detection is unavailable17. In addition, despite being susceptible to β-lactam antibiotics, OS-MRSA is prone to develop β-lactam resistance following antibiotic therapy due to its carriage of mecA10,11,12,14,15, ultimately leading to β-lactam treatment failure18,19,20. Recent findings in study of mechanism of the β-lactam resistance in OS-MRSA include either (1) restoration of frameshift mutation in mecA or (2) mutations in other genes that are not directly relevant to function of mecA21,22,23. In the present study, a collection of 43 clinical OS-MRSA strains and 100 mutants with reduced susceptibility to β-lactam resistance selected by exposing the OS-MRSA to oxacillin were analyzed to identify genome mutations responsible for β-lactam resistance.
Results
Characterization of clinical OS-MRSA isolates
A total of 43 OS-MRSA isolates recovered from various clinical specimens collected from Japan and Taiwan were included in this study (Table S1). The characteristics of OS-MRSA were re-confirmed via determination of their oxacillin susceptibility and the presence of mecA (Table 1). Our results revealed that all strains maintained the typical characteristics of OS-MRSA, including mecA positivity and susceptibility to oxacillin, but they were susceptible to oxacillin with MICs ranging from 0.125 to 2 µg/mL. According to CLSI, cefoxitin can also be used to detect MRSA. Thus, cefoxitin susceptibility testing was conducted to investigate whether there was discrepancy between oxacillin and cefoxitin susceptibility among the OS-MRSA isolates. The cefoxitin MICs for all OS-MRSA isolates ranged 1.5–12 µg/mL, with 24 of the 43 OS-MRSA isolates (56%) exhibiting susceptibility to cefoxitin.
Genomic analysis of the clinical OS-MRSA isolates
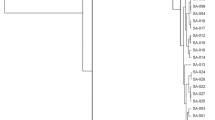
To determine the genetic background of the strains used in this study, the whole-genome sequences of the 43 clinical OS-MRSA isolates were determined, and their phylogenetic relationships were analyzed by constructing a phylogenetic tree using kSNP3 (Fig. 1). The phylogenetic tree revealed extensive genomic diversity among the isolates, which could be classified into seven main phylogenetic clades. In addition, these isolates could also be grouped into 11 MLST types (ST1, ST5, ST8, ST59, ST89, ST91, ST121, ST338, ST772, ST1516, and ST6217), and they carried four different SCCmec types (II, IVa, IVc, and V). The majority of OS-MRSA isolates were belonged to ST121-SCCmec type V (16 strains, 37%), followed by ST59-SCCmec type V (seven strains, 16%), ST89-SCCmec type V (six strains, 14%), ST89-SCCmec type IVa (three strains, 7.0%), ST8-SCCmec type IVa (two strains, 4.7%), and ST6217-SCCmec type V (two strains, 4.7%). In addition, seven singletons (ST1-SCCmec type IVa, ST5-SCCmec type II, ST59-SCCmec type IVa, ST91-SCCmec type IVa, ST338-SCCmec type V, ST772-SCCmec type V, and ST1516-SCCmec type IVc), each of which comprised 2.3% of all strains, were identified. SCCmec types V (33 strains, 77%) and IVa (eight strains, 19%) were predominant among the OS-MRSA isolates, whereas only one isolate harbored each of SCCmec type II and IVc, respectively. Single nucleotide polymorphisms (SNPs) found in promoter and coding region of mecA are listed in Table 2. The type of SNPs located on mecA promoter region, consisting of MecI/BlaI-binding site (− 19 to − 50) and ribosome-binding site (− 7 to − 11)24, and coding region were closely correlated with SCCmec types. For the SNPs in promotor region, SCCmec type II strain JMUB1293 carried an C-30A mutation (A replace C of 30th bases upstream of mecA CDS), all 8 strains belong to SCCmec type IVa had G-7 T mutations, and all strains belong to SCCmec type IVc (1 strain) and V strains (34 strains) carried C-33 T mutations. For the SNPs in mecA coding region, all OS-MRSA strains had a synonymous mutation of C75A, and all SCCmec type V strains carried nonsynonymous mutations of T675A (Ser225Arg), when compared to a prototypic pre-MRSA strain N315, which carries intact mec operon (composed of mecA, mecI, and mecR1) and its mecA gene expression is strongly repressed by mecI25.
Phylogenetic relationships among clinical isolates of oxacillin-susceptible mecA-positive S. aureus (OS-MRSA). A maximum parsimony tree of 43 OS-MRSA isolates was generated with the majority of single nucleotide polymorphisms in the core genome using FigTree ver.1.4.3. The sequence type (ST) of MLST, SCCmec type, and blaI genotypes of each strain were appended to this figure. OS-MRSA isolates were classified into seven main clades (clades 1–7).
Moreover, whole-genome sequencing demonstrated that 34 of the 43 (79%) OS-MRSA isolates carried a complete bla operon (Table 1), which could be classified into two genotypes, namely bla operon-1 and bla operon-2, based on the nucleotide sequences. These two operons shared nucleotide identities of 94% for blaZ, 92% for blaR1, and 94% for blaI. Twelve strains (28%) carried bla operon-1, and all but one (JMUB217) bla operon-1 was located on plasmids. Meanwhile, 23 isolates (53%) carried intact bla operon-2 in their chromosomes. JMUB217 carried both bla operons on its chromosome. An incomplete bla operon-2 lacking blaZ but having intact blaR1 and blaI was present in isolates JMUB1301 and JMUB1313. The absence of blaZ in the bla operons of these isolates was confirmed by PCR (data not shown). Lastly, seven isolates (16%) lacked a bla operon.
PBP2a production and mecA expression in OS-MRSA
To assess the correlation between oxacillin susceptibility and level of PBP2a production in OS-MRSA, PBP2a agglutination assay (semi-quantitation) was performed using representative OS-MRSA strains of seven different main phylogenetic clades (Fig. 1). Oxacillin-resistant (OR) MRSA strains such as USA300_C02 (ST8, IVa, mecI−, blaI-1+, OXA = 48 μg/mL), JMUB5028 (ST89, II, mecI+, blaI-2+, OXA = 128 µg/mL), JMUB611 (ST5, II, mecI+, blaI-1+, OXA = 256 µg/mL), and COL (ST256, I, mecI−, blaI−, OXA > 256 µg/mL) and pre-MRSA strain N315 (ST5, II, mecI+, blaI-1+, OXA = 6 µg/mL) were also analyzed as controls. Results showed that all OS-MRSA strains tested produced detectable level of PBP2a (Fig. 2A). JMUB1315 showed weak agglutination reaction similar to pre-MRSA N315, while other strains showed moderate or strong agglutination reactions. In parallel, mecA expression level of these strains was also determined by qRT-PCR, and the results were very similar to that of PBP2a semi-quantitation (Fig. 2B). There was no clear correlation seen between the levels of oxacillin MIC and mecA expression as well as PBP2a production in OS-MRSA.
The levels of PBP2a production and mecA expression in oxacillin-resistant MRSA (OR-MRSA), oxacillin-susceptible MRSA (OS-MRSA), and pre-MRSA. (A) PBP2a agglutination assay results of (1) OR-MRSA, (2) OS-MRSA, (3) pre-MRSA and (4) OS-MRSA JMUB217 ∆mecA strain. PBP2a production was detected by the MRSA-screen test (Denka Seiken) based on the agglutination of latex particles sensitized with monoclonal antibodies against PBP2a. (B) Quantitation of mecA mRNA in OR-, OS- and pre-MRSA by qRT-PCR. The qRT-PCR data are shown as means ± SD of three biological triplicates. *, **, *** and ns indicate P < 0.05, 0.01, 0.001 and not significance, respectively by Student’s t-test.
Influence of bla operon on reduced susceptibility to oxacillin in OS-MRSA
A previous study suggested that blaI expression levels were associated with reduced oxacillin susceptibility in OS-MRSA isolates26. To understand how the bla operons affect oxacillin susceptibility in the tested OS-MRSA strains, mutants with knockout of β-lactamase repressor gene blaI were generated and their effect on the oxacillin susceptibility was analyzed. Our whole-genome sequencing analysis showed that bla operon were carried by 36 of the 43 (84%) OS-MRSA isolates, and the bla operons could be classified into two genotypes, bla-1 and bla-2. The OS-MRSA JMUB217 (ST772, V, mecI−, blaI-1+, blaI-2+, OXA = 0.75 µg/ml) carried both blaI-1 and blaI-2, therefore, we deleted either one or both of their blaI to generate single and double blaI-knockout mutants, and their MICs of penicillin G and oxacillin were determined (Fig. 3A). Knockout of blaI-1 or blaI-2 alone did not significantly affect MICs of the penicillin G and oxacillin, whereas the double knockout could raise MIC of penicillin G significantly from 1.5 to 8 µg/mL, but of oxacillin slightly from 0.75 to 2 µg/mL. Similar to the results of MIC determination, knockout of blaI-1 or blaI-2 alone did not affect the levels of mecA expressions and PBP2a production, whereas the double knockout enhanced mecA expression and PBP2a production but to a lesser extent (Fig. 3B). These results indicated that the influence of blaI on oxacillin susceptibility is limited.
The levels of PBP2a production and mecA expression in mutants of blaI deletion, mecA overexpression and/or mecA deletion generated from OS-MRSA strain JMUB217. (A) PBP2a agglutination assay results of strains (a) JMUB217, (b) JMUB217 ∆blaI-1, (c) JMUB217 ∆blaI-2, (d) JMUB217 ∆blaI-1/∆blaI-2, (e) JMUB217 pKAT (vector), (f) JMUB217 pKATmecA, and (g) JMUB217 ∆mecA. PBP2a production was detected by the MRSA-screen test (Denka Seiken) based on the agglutination of latex particles sensitized with monoclonal antibodies against PBP2a. (B) qRT-PCR quantitation results of mecA mRNA in blaI deletion and mecA overexpression mutants of JMUB217. The data are shown as means ± SD of three biological replicates. *, **, *** and ns indicate P < 0.05, 0.01, 0.001 and not significance, respectively by Student’s t-test.
Identification of mutations associated with reduced susceptibility to oxacillin in OS-MRSA
To elucidate the pathway(s) leading to the acquisition of β-lactam resistance in OS-MRSA, laboratory-derived mutants with reduced susceptibility to oxacillin were obtained from the parent OS-MRSA strains via single-step exposure to oxacillin. Although resistant colonies growing inside the inhibition zone were generated from all 43 parent strains, not all selected colonies displayed increased oxacillin MICs after single-colony purification. This might be due to the hetero-resistance phenotype of OS-MRSA as some OS-MRSA strains were reported to show heterogeneous resistance to oxacillin18,22. Re-determination of the oxacillin susceptibility of the isolated colonies identified 100 mutants with increased oxacillin MICs (ranged from 1 – 256 µg/mL) generated from 26 OS-MRSA strains representing all seven phylogenetic clades (Fig. 1). Among them, 86 mutants exhibited MICs of greater than 4 µg/mL (Table S2). The comparative genomic analysis of the 100 mutants and their respective parent OS-MRSA strains identified a total of 141 mutations, and all mutations were verified via Sanger sequencing (Table S2). It was found that 70 out of 100 mutants carried only one mutation, four mutants carried two mutations in a single gene, and 26 mutants carried multiple mutations in different gene or intergenic region. Moreover, 96 mutants carried at least one nonsynonymous or frameshift mutation, and 4 mutants (JMUB1283-3, JMUB1972-1, JMUB1281-7, and JMUB1310-6) had silent mutations (HP7T450C, HP10C651T, guaAG1158A, and tilSA1287C; Table S2). Of the 129 mutations identified in coding sequences, 98 (76%), 13 (10%), 10 (7.8%), and 8 (6.2%) were missense, nonsense, frameshift, and synonymous mutations, respectively. Among them, 121 nonsynonymous mutations were distributed within 46 genes (Table S2; Fig. 4), but no mutation was found in mec or bla operons. The 46 mutated genes could be classified into 13 functional categories: (1) DNA/RNA polymerase, rpoC (22 mutations), rpoB (20 mutations), and dnaE (one mutation); (2) purine biosynthesis, guaA (nine mutations), and hprT (three mutations); (3) (p)ppGpp synthase, relSau (four mutations), and relQ (one mutation); (4) protein quality control, clpP (six mutations), clpX (three mutations), ftsH (one mutation), and yjbH (one mutation); (5) membrane protein associated with glycopeptide resistance, mprF (four mutations), tcaA (one mutation), and vraT (three mutations); (6) glycolysis, fruB (five mutations), fbaA (two mutations), ptsI (one mutation), and pykA (one mutation); (7) pentose phosphate pathway, rpiA (one mutation) and prs (five mutations); (8) tRNA synthesis, thrS (one mutation), tilS (one mutation), gltX (one mutation), and lysS (one mutation); (9) folate biosynthesis, folC (one mutation); (10) peptidoglycan biosynthesis, sgtB (one mutation); (11) transcriptional regulation, mraZ (one mutation); (12) extracellular matrix protein, ebhA (one mutation); and (13) unknown function, HP1-HP18 (27 mutations). On the other hand, the 12 mutations identified in eight intergenic regions were located between genes of SA0499 and rpoB, sgtB and SA1692, E8M03_00305 and hsdR, SA2092 and ssaA2, SAS044 and SA1196, norB and ebhA, tnp and proP, and SA1447 and alaS, respectively. These results clearly demonstrated that mutations responsible for oxacillin resistance in the OS-MRSA-derived mutants are quite diversified.
Relationship between gene mutation and fold changes of oxacillin MIC in the mutants with reduced oxacillin susceptibility. Each mutant with reduced oxacillin susceptibility is represented by a closed circle, and different color-coded circles indicate different phylogenetic clades. (A) Fold changes of oxacillin MICs for mutants carrying single or double mutations in the same gene. (B) Fold changes of oxacillin MICs for mutants carrying single or double mutations in different genes.
Contribution of increased mecA expression to reduced oxacillin susceptibility in OS-MRSA
To understand the role of the identified mutations in reduced oxacillin susceptibility, OS-MRSA strain JMUB217 and its oxacillin-resistant mutants were used as representative strains for further study because 24 mutants carrying 26 variants in 11 genes and an intergenic region were derived from the JMUB217. In addition, oxacillin MICs for the 24 mutants ranged widely (1.5–256 µg/mL), and were highly elevated compared to their parent strain JMUB217 (0.75 µg/mL). A sequential experiment was carried out. First, a mecA-overexpressing mutant was created to investigate whether changes in mecA expression affects oxacillin susceptibility in OS-MRSA. A vector pKAT containing mecA and its native promoter was introduced into JMUB217 to generate the mecA-overexpressing mutant JMUB217 (pmecA), and MIC determination found that the generated mutant exhibited increment of oxacillin MIC from 0.75 to 12 µg/mL (Fig. 3; Table 3). Next, mecA-knockout mutant JMUB217 (∆mecA) was generated and its oxacillin MIC was measured. The oxacillin MIC decreased slightly from 0.75 to 0.38 µg/mL in this mecA-deleted mutant (Table 3), indicating that the presence of mecA itself confers a low level oxacillin resistance. Moreover, overexpression of mecA in the mecA-deleted mutant JMUB217 (∆mecA) resulted in an increment of the oxacillin MIC to 12 µg/mL, similar to that of the mecA-overexpressing mutant JMUB217 (pmecA). Finally, a set of mecA-knockout strains from three oxacillin-resistant mutants (JMUB217-11, JMUB217-22, and JMUB217-24), carrying mutations of RpoCP358L, RpoBG645H, and RpoCG498D, respectively, were generated and their oxacillin MICs were determined. The results found that their MICs of 4, 32, and 256 µg/mL decreased to 0.38 µg/mL, similar to the level of the mecA-knockout mutant JMUB217 (∆mecA) (Table 3). These results indicated that mecA expression is a key factor for promoting reduced oxacillin susceptibility in OS-MRSA.
Correlation of the levels of mecA expression and PBP2a production with oxacillin MIC in mutants with reduced oxacillin susceptibility
To examine the correlation between PBP2a production and oxacillin susceptibility in laboratory mutants, PBP2a agglutination assay was performed on 11 JMUB217-derived mutants (Fig. 5A). Despite the increase of oxacillin MIC, PBP2a production was not significantly changed in these mutants. Next, mecA expression of JMUB217-derived mutants in the presence and absence of oxacillin was measured (Fig. 5B). The results showed that the natural expression of mecA significantly increased in 9 of 11 mutants with 1.3 to 1.9-fold change. However, the mecA expression levels were still lower than those of OR-MRSA (Fig. 2B). Since bla operons of JMUB217 induced mecA expression (Fig. 3), the mecA expression levels were measured in the presence of low concentration of oxacillin (0.1 µg/mL). Results showed that oxacillin induction significantly upregulated the mecA expression level in wild-type (2.8-fold) as well as the resistant mutants (1.3 to 2.9-fold). When compared with wild-type strain, the mecA expression levels induced by oxacillin were significantly increased in three of 11 mutants (JMUB217-11, -23, -24). Interestingly, the resistant mutant with the highest oxacillin MIC did not display the strongest mecA expression in both presence and absence of oxacillin. As seen in Fig. 5B, JMUB217-24 had the highest oxacillin MIC of 256 µg/mL, but showed a similar mecA expression level compared to the mutants JMUB217-23, JMUB217-22, JMUB217-18, JMUB217-11 and JMUB217-9, which had oxacillin MICs of 64, 32, 24, 4, and 4 µg/mL, respectively (Table S2; Fig. 5), indicating that mecA expression was not the only cause of oxacillin resistance in the mutants.
The levels of PBP2a production and mecA expression in JMUB217-derived mutants with reduced oxacillin susceptibility. (A) PBP2a agglutination assay of mutants with reduced oxacillin susceptibility. PBP2a production was detected by the MRSA-screen test (Denka Seiken) based on the agglutination of latex particles sensitized with monoclonal antibodies against PBP2a. (B) qRT-PCR quantitation of mecA mRNA in mutants with reduced oxacillin susceptibility under the absence (white bars) and the presence (grey bars) of oxacillin induction. The qRT-PCR data are shown as means ± SD from biological triplicates. *, **, *** and ns indicate P < 0.05, 0.01, 0.001 and not significance, respectively by Student’s t-test.
Transcriptome analysis revealed a stringent-like response in the oxacillin-resistant mutants
To figure out the overall gene regulation in the oxacillin-resistant mutants, which alters transcriptional profile and ultimately biases gene regulation toward the expression of oxacillin resistance, the whole-genome expression profiles of five representative mutants of JMUB217, JMUB217-7 carrying RpiAA64E (oxacillin MIC = 4 µg/mL), JMUB217-11 carrying RpoCP358L (oxacillin MIC = 4 µg/mL), JMUB217-18 carrying RpoCV488G,K673N (oxacillin MIC = 24 µg/mL), JMUB217-20 carrying RpoBV1016E,H1042Q (oxacillin MIC = 24 µg/mL), and JMUB217-22 carrying RpoBQ645H (oxacillin MIC = 32 µg/mL), were analyzed and compared with that of the parent strain JMUB217. The transcriptome analyses were performed under both oxacillin-induced and drug-free growth conditions. Differentially expressed genes (DEGs) with log2-fold change of > ±1 (adjusted P value of < 0.01) were identified by pair-wise comparison (Table S3). In concordance with qRT-PCR data (Fig. 5B), the results of transcriptome analysis showed that mecA expression was significantly induced by oxacillin in both the mutants and parent strain, while the differences in the basal mecA expression between the mutants and its wild-type was small (log2-fold change < 1) as shown in Table S3.
A Venn diagram analysis showed that two rpoC mutants JMUB217-11 and JMUB217-18 shared 217 DEGs, of which 64 or 153 genes were commonly upregulated or downregulated, respectively (Fig. 6A). Two rpoB mutants JMUB217-20 and JMUB217-22 shared 297 DEGs with 168 up- and 129 down-regulated genes (Fig. 6B). In case of all five strains carrying mutation of either rpiA or rpoC or rpoB, 13 genes were up- and 15 genes down-regulated commonly (Fig. 6C; Table S4). Among the commonly regulated genes, upregulation of tryptophan biosynthesis genes (trpBDEFG) and downregulation of nucleotide transporter and biosynthesis genes (pyrRP, hisIG). These genes were known to be related with RelSau/RSH-dependent stringent response mediated by amino-acid deprivation in S. aureus27. RelSau is a bifunctional (p)ppGpp synthase/hydrolase and induces the classic stringent response by accumulation of (p)ppGpp28. In addition, downregulation of purine biosynthesis genes such as xprT, purF and guaB, which were usually seen in the stringent response were observed in the four rpoC and rpoB mutants (Table S4; Fig. 7I).
Venn diagram summary of differently expressed genes identified among the five representative mutants from OS-MRSA strain JMUB217. The number of genes co-upregulated and -downregulated with log2-fold change of > 1 (P value of < 0.01) in the mutants with mutation of (A) rpoC, (B) rpoB , and (C) rpoC, rpoB, and rpiA , respectively, are listed.
Gene expression profiles of OS-MRSA strain JMUB217 and its mutants with reduced susceptibility to oxacillin in the presence and absence of oxacillin induction. Differentially expressed genes between the parent strain JMUB217 and its mutants were classified into 12 different functional categories, including (A) antibiotic resistance, (B) protein quality control, (C) tRNA synthesis, (D) autolysis, (E) RNA polymerase activity, (F) DNA polymerase activity, (G) glycolysis, (H) the pentose phosphate pathway, (I) purine biosynthesis, (J) folate biosynthesis, (K) (p)ppGpp synthesis, and (L) peptidoglycan biosynthesis. The color scales indicate the degree of log2-fold changes of transcriptional expression in the mutants compared with that in wild-type strains without induction. Mutated genes identified in the mutants with reduced oxacillin susceptibility are shown in red font.
In addition to the alteration of expression of genes directly related to the stringent response, the mutants with reduced oxacillin susceptibility also exhibited downregulation of genes involved in protein quality control and tRNA synthesis. Notably, clpP, clpX, and ftsH were significantly downregulated in mutants with reduced oxacillin susceptibility (Fig. 7B). Moreover, 16 out of 25 tRNA genes were also downregulated in at least one of the five mutants (Fig. 7C). These changes in gene expression might contribute to oxacillin resistance, as previous studies described that deficiencies of protein quality control and tRNA synthesis were associated with the stringent response and β-lactam resistance29,30,31.
Transcriptome profiles of the rpoBC and rpiA mutants also showed alteration in gene expression relevant to the peptidoglycan biosynthesis, for example, upregulation of mecA and sgtB, and downregulation of murBJY, femABX, pbp4, ftsW, and rodA (Fig. 7L). Furthermore, changes in the expression of genes involved in autolysis of S. aureus were observed in strains carrying mutations of rpoBC and rpiA, for example, lytM and sle1 were upregulated, whereas lytH, isaA, and ssaA were downregulated (Fig. 7D). All these changes in combination with mecA expression and stringent-like response might direct bacterial metabolism towards acquisition of oxacillin resistance in the resistant mutants.
The mutants with reduced oxacillin susceptibility tend to slow growth
Mutations in genes involved in the stringent response were reported to be associated with slower growth rate32. In addition, some β-lactam–resistant mutants generated in vitro were also found to have slow growth rates or a phenotype of persistent infection31,32,33,34. To investigate whether the mutations identified in the oxacillin-resistant mutants affect cell growth, the doubling time of JMUB217-derived mutants was measured. Seven of eleven mutants showed significantly slower growth speed (doubling time 36.5 ± 0.7 min to 57.7 ± 1.1 min) compared to their parent strain (32.7 ± 1.4 min). In contrast, three of them had similar doubling time (JMUB217-11, 34.1 ± 0.3 min; JMUB217-24, 34.8 ± 0.4 min; JMUB217-9, 35.3 ± 0.9 min) and JMUB217-7 grew faster than wild type (29.0 ± 0.3 min). Surprisingly, growth of the JMUB217-21 carrying GuaAI249fs mutation was very slow (347.1 ± 9.3 min).
Intracellular ATP level in the mutants with reduced oxacillin susceptibility
Some reports examining intracellular ATP levels of S. aureus in relation to stress responses found that lower cellular ATP production was associated with bacterial tolerance to several environmental stresses such as salt, cold, and antibiotics, and it could also induce the conversion of bacterial cells into persistent forms, including small colony variants35,36. Our transcriptomics study with the resistant mutants illustrated that several genes involved in purine biosynthesis and folate biosynthesis were clearly downregulated (Fig. 7I,J), which was similar to the findings of Cassels et al.37. However, some genes involved in ATP biosynthesis were significantly upregulated (Fig. 7J). To analyze whether the mutations of oxacillin-resistant mutants affect ATP biosynthesis, the intracellular ATP levels of 23 mutants and their parent strain JMUB217 were measured. The results showed that the intracellular ATP was increased in the mutants compared to the parent strain, and there was good correlation between the levels of intracellular ATP and oxacillin MIC with correlation coefficient of 0.6047 (p = 0.0017) (Fig. 8).
Correlation between the levels of intracellular ATP and oxacillin MIC among the OS-MRSA strain JMUB217 and its mutants with reduced susceptibility to oxacillin. The levels of intracellular ATP are shown as means ± SD of three biological replicates. The correlation coefficient was evaluated using Spearman’s correlation coefficient test. Open circle denotes parent strain and closed circle does the mutants.
Discussion
Since its first description in 1991, OS-MRSA15,38, which is related to borderline methicillin-resistant MRSA39, has been frequently isolated in hospital and community settings with a prevalence rate of 0.62 – 33.7%12,15,18,20,40,41. The presence of OS-MRSA is currently a challenge in the clinical management of staphylococcal infections and requires great attention because it is prone to be misidentified as MSSA via routine β-lactam susceptibility testing10,12,14. Indeed, the majority of the OS-MRSA isolates used in this study were initially identified as MSSA according to the oxacillin susceptibility profile provided by the original laboratory despite carrying mecA. Similarly, susceptibility testing using cefoxitin, a stronger inducer of the mecA regulatory system than oxacillin that is used to detect methicillin resistance42, failed to accurately identify OS-MRSA (Table 1). These observations suggest that a combination of oxacillin and cefoxitin susceptibility tests, as recommended elsewhere43, or detection of mecA will be more reliable for the identification of MRSA.
Despite being phenotypically susceptible to oxacillin, β-lactam resistance can easily be induced in OS-MRSA20,21,44. The mechanisms regulating oxacillin susceptibility in S. aureus appear to differ depending on the types of mutations and genetic basis of the individual isolates. Chen et al. reported that mutations in the MecI-binding site of the mecA promoter downregulated the expression of PBP2a and increased the susceptibility of ST59-SCCmec type V strains to oxacillin45. Meanwhile, they demonstrated that mutation of the ribosome-binding site of mecA in an ST59-SCCmec type IV strain attenuated its oxacillin resistance. Nevertheless, these mutations affect only oxacillin resistance in the strains of ST59 background, whereas mutations in the same locus barely affected the β-lactam resistance levels of isolates with different genetic backgrounds, such as COL (ST250) and CH482 (ST45)24. Conversely, mutations in the mecA coding region conferred oxacillin resistance to OS-MRSA strains isolated in the US21. These studies suggested that the inclusion of OS-MRSA strains with a diverse genetic backgrounds is crucial for providing a comprehensive insight into understanding the mechanism of oxacillin resistance in OS-MRSA.
Although oxacillin resistance in OS-MRSA might be caused by increased mecA expression, the exact mechanism triggering mecA overexpression is unknown. The structure of bla operon is highly homologous to the mec operon46, and quite a high portion (84%) of OS-MRSA isolates analyzed in this study carried bla operon (Table 1; Fig. 1), which suggest that bla operon might influence oxacillin susceptibility in OS-MRSA. However, deletion of the repressor gene blaI from an OS-MRSA strain JMUB217 resulted in only slight increases of the oxacillin MIC. In addition, mecA levels were not uniformly increased in a set of JUMB217-derived oxacillin-resistant mutants compared to the parent strain JUMB217, as determined by qRT-PCR. Therefore, we postulated that oxacillin resistance in OS-MRSA strains involves a more complex regulatory pathway than simply direct mecA signaling.
The stringent stress response governed by the alarmone (p)ppGpp is involved in the β-lactam resistance of MRSA34,47. Both our whole-genome comparative analysis and RNA-seq analysis demonstrated that many mutations identified in the resistant mutants (Fig. 4) and their altered gene transcriptions (Fig. 7) were associated with depletion of pathway relevant to the purine biosynthesis, protein quality control, and tRNA synthesis, which was very similar to the massive transcription downregulation seen in S. aureus during the stringent response. However, the expression profiles of stringent response elements in oxacillin-resistant mutants derived from this study were not indicative of the classic stringent response elicited by mupirocin treatment48. During the classic stringent response, the cellular stresses resulting from amino acid starvation and mupirocin exposure induce the accumulation of uncharged (deacylated) tRNA49. The uncharged tRNA in turn binds to the A (aminoacyl-tRNA) site of the 70S ribosome and activates RelSau to produce (p)ppGpp50. However, in some Gram-positive bacteria like Bacillus subtilis, (p)ppGpp does not directly regulate RNA polymerase (RNAP). Rather, (p)ppGpp synthesis reduces intracellular GTP levels, subsequently leading to the induction of the stringent response51,52. Hence, mutations in genes involved in glycolysis, pentose phosphate biosynthesis, folate synthesis, and purine biosynthesis might mimic the (p)ppGpp-mediated reduction in intracellular GTP levels and induce “stringent-like response”, as evidenced by the downregulation of genes responsible for GTP biosynthesis (purine and folate biosynthesis, pentose phosphate biosynthesis, and glycolysis) (Fig. 7). In addition, gene mutations identified in this study included many stringent response elements, and most of them were previously reported to be associated with conversion of hetero- to homo-resistance against β-lactam, such as genes associated with RNA polymerase (RNAP; rpoB31,53 and rpoC31,54), purine biosynthesis (guaA31, prs31,33, hprT31,33, (p)ppGpp synthesis (relSau31,33,47), glycolysis (pykA31, fbaA31, and fruB33) protein quality control (ftsH31, clpX30,55, and clpP30,55), and tRNA synthase (lysS31, and gltX31). These results suggest that stringent-like response played an important role in β-lactam resistance in the oxacillin-resistant mutants of OS-MRSA. It remains to be further studied, however, how this response leads to substantial metabolic changes towards acquisition of the resistance.
OS-MRSA is considered problematic in the clinical setting because the strain is prone to develop high-level β-lactam resistance during the course of antibiotic treatment20,44. Because the targets of antibiotics are generally essential proteins in bacteria, the acquisition of antibiotic resistance is usually associated with a fitness cost56. In S. aureus, slow-growth phenotypes, including the formation of small colony variants, are associated with tolerance to antibiotics57,58,59,60. Contrarily, some of the mutations identified in the JMUB217 strain altered its oxacillin susceptibility without affecting its doubling time. This suggested that the mutations conferring reduced oxacillin susceptibility in OS-MRSA may incur only small fitness costs because of the complementary upregulation of ATP synthase genes. The increased expression of ATP biosynthesis genes was supported by the positive correlation between oxacillin MICs and intracellular ATP levels (Fig. 8), which might explain the easy acquisition of oxacillin resistance in OS-MRSA. Nonetheless, chromosomal mutations in rpoBC and other genes involved in purine biosynthesis were identified in slow VISA strains61,62, indicating that the fitness cost of mutations may depend on the genetic background of individual strains.
This study aimed to understand the genetic pathways associated with oxacillin resistance in OS-MRSA isolates from diverse genetic backgrounds. Our results suggested that OS-MRSA was rendered oxacillin-resistant by a combination effect of stringent-like response (a stress response similar to the classic stringent response) and subsequent expression of antibiotic resistance genes (e.g., mecA, bla operon). The relatively low fitness cost of the mutations may fuel the easy selection of oxacillin-resistant OS-MRSA mutants during the course of antimicrobial treatment.
Materials and methods
Bacterial strains and growth conditions
A total of 43 OS-MRSA isolates from various clinical samples were collected from routine clinical laboratories in hospitals across Japan and Taiwan between 1998 and 2015 (Table S163,64,65). Mueller–Hinton broth (MHB; Becton Dickinson Co., Ltd., Sparks, MD, USA) and tryptic soy broth (TSB; Becton Dickinson) were used to culture S. aureus, whereas Escherichia coli was grown in Luria–Bertani (LB; Becton Dickinson) medium. In some experiments, antibiotics were added to the medium at the following concentrations: ampicillin (Nacalai Tesque, Inc., Kyoto, Japan) at 100 µg/mL for E. coli, tetracycline (Nacalai Tesque) at 5 µg/mL for S. aureus, and chloramphenicol (Nacalai Tesque) at 10 µg/mL for S. aureus and E. coli. For preservation, bacterial cells were cultivated on tryptic soy agar (TSA; Becton Dickinson) and incubated at 37 °C upon receipt. A single colony was then selected and grown overnight in TSB at 37 °C. The overnight culture was aliquoted and stored at − 80 °C in 50% glycerol (Wako Pure Chemical Industries, Ltd., Tokyo, Japan) until use.
Antibiotic susceptibility test
Oxacillin and cefoxitin MICs were determined using the E-test method according to Clinical and Laboratory Standards Institute (CLSI) guidelines16. Briefly, overnight cultures of S. aureus strains grown in 4 mL of MHB at 37 °C were adjusted to 0.5 McFarland turbidity (approximately 1 × 108 to 2 × 108 CFU/mL) and spread on Mueller–Hinton agar (MHA; Becton Dickinson) plates. The E-test gradient strip (bioMérieux SA, Marcy l’Étoile, France) was then placed on the bacterial lawn. The MIC was determined after incubation at 37 °C for 24 h. The isolates with oxacillin MIC ≤ 2 µg/mL or cefoxitin MIC ≤ 4 µg/mL were considered oxacillin- and cefoxitin-susceptible, respectively.
mecA detection via PCR
DNA was extracted from OS-MRSA isolates grown overnight on TSA plates using MightyPrep reagent (Takara Bio Inc., Shiga, Japan) in accordance with the manufacturer’s instructions. PCR was then performed on the extracted DNA using Quick Taq HS DyeMix (Toyobo Co., Ltd., Osaka, Japan). A primer pair (mecA-F and mecA-R, Table S5) was used to amplify a 519-bp region of mecA. The thermal cycling conditions included initial denaturation at 94 °C for 2 min followed by 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 1 min. Finally, the amplified products were electrophoresed on 1% agarose gel, stained with ethidium bromide, and visualized using AE-6933FXES Printgraph (Atto Co., Tokyo, Japan).
PBP2a production
PBP2a was extracted from colonies grow on MHA and was detected using the MRSA-screen latex agglutination test (Denka, Seiken Co. Ltd., Tokyo, Japan) according to the manufacturer’s instructions.
Isolation of mutants with reduced susceptibility to oxacillin from parent OS-MRSA strains
To isolate mutants with reduced oxacillin susceptibility, all 43 OS-MRSA parent strains were exposed to oxacillin according to the E-test method as described for susceptibility testing. Briefly, the oxacillin E-test was performed on OS-MRSA strains inoculated onto MHA plates. A single colony growing inside the inhibition zone after 24–48 h of incubation was randomly picked and sub-cultured in TSB for 24 h at 37 °C. The overnight culture was then serially diluted tenfold with 0.9% NaCl and spread onto a TSA plate. A single colony growing on the TSA plate was again randomly selected and inoculated into TSB for preservation in 50% glycerol at − 80 °C. The oxacillin susceptibility of the stocked cells was determined again using the E-test method to discriminate mutant colonies from persister colonies. The cells exhibiting higher oxacillin MICs were selected as oxacillin-reduced susceptibility mutants, which were then used for subsequent analysis.
Whole-genome sequencing
Genomic DNA was extracted from OS-MRSA and its mutants using the phenol–chloroform method and purified using a QIAamp DNA mini kit (Qiagen, Hilden, Germany) following previously developed methods66. The genomic sequences of parent strains were determined via mate-pair sequencing as previously described66,67. Briefly, a mate-pair library was prepared using a Nextera mate-pair library prep kit (Illumina, Inc., San Diego, CA, USA), and sequencing was performed using an Illumina MiSeq platform with the MiSeq reagent kit version 3 (Illumina). The mate-paired reads of OS-MRSA were trimmed using the FASTQ toolkit version 2.2.0 to generate high-quality reads and assembled using Velvet Assembler version 1.2.10 to construct genome scaffolds. The generated genomic sequences were finally annotated using Microbial Genome Annotation Pipeline (https://www.migap.org/). Meanwhile, the genomic sequences of in vitro-selected mutants with reduced oxacillin susceptibility were determined by sequencing paired-end reads as previously described68. The paired-end library was prepared using a Nextera XT library prep kit and sequenced using the Illumina MiSeq platform with the MiSeq reagent kit version 3. The paired-end reads of each mutant were mapped against the genomic sequences of their corresponding parent OS-MRSA strains, and mutations were detected using CLC Genomics Workbench version 9 (CLCbio, Qiagen, Valencia, CA, USA). Mutations identified in each mutant were verified by Sanger sequencing using the Applied Biosystems 3130xl genetic analyzer (Thermo Fisher Scientific, MA, USA).
Construction of the phylogenetic tree
To construct the OS-MRSA phylogenetic tree, kSNP369, available at https://sourceforge.net/projects/ksnp/, was first used to identify single nucleotide polymorphisms (SNPs) in the whole-genome sequencing data of OS-MRSA strains. The k-mer size was set to an optimum length of 13 as estimated by Kchooser for extracting SNPs from the sequence data. A maximum parsimony tree was then constructed using the majority of the SNPs present in at least 75% of the genomes. The generated phylogenetic tree was visualized using FigTree ver.1.4.3 (tree.bio.ed.ac.uk/software/figtree/).
Growth curve analysis
The bacterial doubling time was determined as described previously70. Briefly, overnight cultures of parent OS-MRSA strains and the laboratory-selected mutants were adjusted to an OD600 of 0.2 in MHB. Then, aliquots of 100 µL were inoculated into 10 mL of MHB (final concentration of 1 × 105 CFU/mL), and the cultures were grown at 37 °C with agitation at 25 rpm in an automatic temperature gradient rocking incubator (model TVS126MB; Advantec, Tokyo, Japan). The cell densities at OD600 were measured every 5 min for 12 h, and the bacterial growth curve was generated by plotting the measured ODs against time. The doubling time was determined by fitting the growth curve to an exponential equation. Bacterial growth was measured from at least three independent experiments.
Determination of intracellular ATP levels
The parent OS-MRSA strains and the laboratory-selected mutants were cultured overnight in MHB at 37 °C with agitation at 150 rpm. The overnight cultures were adjusted to an OD600 of 0.2 in MHB, and 100 µL of the OD-adjusted culture were inoculated into 10 mL of MHB. The cultures were grown at 37 °C with agitation at 25 rpm in an automatic temperature gradient rocking incubator. One mL of each mid-exponential phase culture (OD600 = 0.5) was then transferred to a clean 1.5-mL tube and immediately centrifuged at 15,000 rpm for 1 min at 4 °C to pellet cells. After centrifugation, the cell pellet was stored immediately at − 80 °C until analysis. To determine intracellular ATP levels, a BacTiter-Glo Microbial Cell Viability Assay kit (Promega, WI, USA) was used according to the manufacturer’s instructions. Briefly, the cell pellet was resuspended in 1 mL of MHB, and 25 µL of the cell suspension were mixed with an equal volume of BacTiter-Glo Reagent in a 384-well opaque plate (Iwaki, Tokyo, Japan) and incubated at room temperature for 5 min. The luminescence was then read on an EnVision 2104 Multilabel Reader (Perkin Elmer, Waltham, MA, USA). The ATP concentration was determined with reference to an ATP standard curve prepared from ATP disodium salt hydrate (A2383, Merck KGaA, Darmstadt, Germany). ATP disodium salt was dissolved in distilled water, yielding 1 µM ATP standard solutions. Serial tenfold dilutions of the ATP standard solution were created using MHB to prepare diluted standards that were then used to generate the standard curve. The intracellular ATP concentration of each sample was presented as the mean of three independent experiments performed using three biological replicates.
RNA extraction
Overnight cultures of the parent OS-MRSA strains and the laboratory-selected mutants were adjusted to an OD600 of 0.4. The OD-adjusted cultures were then diluted 1:100 in 1 or 10 mL of MHB for qRT-PCR and RNA-seq analysis, respectively. Each culture was grown to the early log-phase (OD600 = 0.3) before treatment with a final concentration of 0.1 μg/mL oxacillin or equal volume of distilled water (control) for 1 h (qRT-PCR) or until OD600 = 0.6 (RNA-seq). After oxacillin treatment, the bacterial cells were harvested by centrifugation at 15,000 rpm for 1 min at 4 °C (qRT-PCR) or at 8000 rpm for 5 min at 4 °C (RNA-seq). Pelleted cells were resuspended in 600 μL (qRT-PCR) or 6 mL (RNA-seq) of TE buffer (10 mM Tris–HCl and 10 mM EDTA, pH 8.0) and lysed with 25 (qRT-PCR) or 30 µg (RNA-seq) of lysostaphin (Merck KGaA) by incubating the mixtures at 37 °C for less than 5 min. Total RNA was then extracted using acidic-phenol saturated with 20 mM sodium acetate (pH 4.8) and chloroform and enriched via ethanol precipitation. Contaminating DNA was removed from the total RNA preparations by incubating the solutions with 2 (qRT-PCR) or 20 units (RNA-seq) of RNase-free DNase I (F. Hoffmann-La Roche Ltd, Basel, Switzerland) at 37 °C for 30 min. Total RNA was finally purified using acidic-phenol/chloroform and eluted in RNase-free water.
Determination of mecA levels by qRT-PCR
The extracted total RNA (100 ng per sample) was reverse-transcribed into complementary DNA (cDNA) using a PrimeScript 1st Strand cDNA Synthesis Kit (Takara Bio). qRT-PCR was performed using TB Green Premix Ex Taq (Tli RNaseH Plus, Takara Bio) on the Mx3005P Real-Time PCR instrument (Stratagene, La Jolla, CA, USA). A primer set (mecA-F-qRT-PCR and mecA-R-qRT-PCR, Table S5) was used to amplify the 162-bp mecA sequence, whereas the 163-bp housekeeping gene rho was amplified using designated primers (rho-F-qRT-PCR and rho-R-qRT-PCR, Table S5) and used as the reference gene for normalization during gene expression analysis. The thermal cycling conditions included initial denaturation at 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s.
RNA-seq analysis
To perform RNA-seq analysis, ribosomal RNAs (rRNAs) in total RNA preparations of the JMUB217 strain and its mutant derivatives were first depleted using a Ribo-Zero rRNA Removal Kit (Bacteria) from Illumina. Double-stranded cDNA was then synthesized using a PrimeScript Double Strand cDNA Synthesis Kit (Takara Bio). The generated cDNA served as the template for constructing the paired-end library using a Nextera XT library prep kit, and the library was subsequently sequenced using the Illumina MiSeq platform and the MiSeq reagent kit version 3. RNA-seq analysis was performed using CLC Genomics Workbench version 9, and the RNA-seq reads were aligned to the reference genomes of the parent strain JMUB217. Gene expression was normalized by calculating the reads per kilobase per million mapped reads, and differentially expressed genes were identified using Baggerly’s test (β-binomial test) with false discovery rate correction. Genes with adjusted p < 0.01 were considered to be significantly differentially expressed.
Construction of mecA- and blaI-knockout mutants
To construct mecA and blaI-knockout mutants of the JMUB217 strain, the pKFT markerless gene deletion system was used as previously described71. Briefly, to delete mecA, 1-kb upstream and downstream flanking sequences of the target gene were amplified by PCR using the primer sets SacI-mecAKO-UP-2/mecA_fPCR_UP and PstI-mecAKO-UP/mecA_fPCR_DN (Table S5), respectively, with KOD FX Neo (Toyobo). Then, second-round PCR was performed using the first-round PCR products as templates with the primer set SacI-mecAKO-UP-2/PstI-mecAKO-UP. The second-round PCR products and pKFT were digested with the restriction enzymes PstI and SacI (Takara Bio) and ligated using Ligation high ver. 2 (Toyobo), generating the plasmid pmecAKO. pmecAKO was transformed into E. coli DH5α, and the transformed cells were plated on LB agar with 100 µg/mL ampicillin. Regarding the generation of blaI-knockout mutants, DNA fragments containing blaI-1 (locus tag: JMUB217_1395) or blaI-2 (locus tag: JMUB217_2048) were first amplified with the primer sets BlaI-1-1/BlaI-1,2-2 and BlaI-2-1/BlaI-1,2-2 (Table S5), respectively. The PCR fragments and pKFT were then digested using the restriction enzymes BamHI and PstI (Takara Bio) and ligated using Ligation high ver. 2. The ligated DNA fragments were independently transformed into E. coli DH5α, and the transformed cells were plated on LB agar with 100 µg/mL ampicillin. The plasmids were extracted, and second-round PCR was conducted using the primer set BlaI-1,2-3/BlaI-1-4 for blaI-1 knockout and BlaI-1,2-3/BlaI-2-4 for blaI-2 knockout. The self-ligated PCR fragments (pblaI-1KO and pblaI-2KO) were again individually transformed into E. coli DH5α, and transformed cells were plated on LB agar with 100 µg/mL ampicillin. Afterwards, all three plasmids (pmecAKO, pblaI-1KO, and pblaI-2KO) were extracted from the E. coli DH5α transformants and transformed into E. coli BL21. The plasmids extracted from E. coli BL21 were subsequently electroporated into S. aureus JMUB217 and mutants with reduced oxacillin susceptibility as previously described72, and the cells were cultured on TSA with 5 µg/mL tetracycline at 30 °C. An isolated colony was then grown overnight in 4 mL of TSB containing 5 µg/mL tetracycline at 30 °C. Single crossover was performed by growing the overnight culture on TSA with 5 µg/mL tetracycline at 43 °C. Then, double crossover was performed by incubating the single crossover mutant on TSA at 30 °C. The double crossover event was confirmed by PCR and Sanger sequencing.
Complementation of mecA
To generate a mecA-complemented mutant, a DNA fragment containing wild-type mecA from strain JMUB217 was amplified using the primers SmaI-mecAcomp-F-pKAT and SalI-mecAcomp-R-pKAT (Table S5). The PCR fragment and pKAT were digested with SmaI and SalI (Takara Bio) and ligated using Ligation high ver. 2. The ligated DNA fragment was transformed into E. coli DH5α, and the transformed cells were plated on LB agar with 10 µg/mL chloramphenicol. Finally, the complementation plasmid was extracted and electroporated into the JMUB217 strain72.
Statistical analysis
All statistical analyses were performed using Prism 8 (GraphPad Software, San Diego, CA, USA). Statistical comparison was carried out using the Student’s t-test whereas the correlations between variables were calculated using the non-parametric Spearman’s correlation coefficient (rs). Statistical significance was denoted with a P value of < 0.05.
Data availability
The raw sequence data have been deposited in DNA Data Bank of Japan (DDBJ) under accession number DRA009699 and DRA009727.
References
Gordon, R. J. & Lowy, F. D. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46(Suppl 5), S350-359. https://doi.org/10.1086/533591 (2008).
Fluit, A. C. Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 18, 735–744. https://doi.org/10.1111/j.1469-0691.2012.03846.x (2012).
Stapleton, P. D. & Taylor, P. W. Methicillin resistance in Staphylococcus aureus: Mechanisms and modulation. Sci. Prog. 85, 57–72. https://doi.org/10.3184/003685002783238870 (2002).
Boucher, H. W. & Corey, G. R. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46(Suppl 5), S344-349. https://doi.org/10.1086/533590 (2008).
Cuny, C., Wieler, L. H. & Witte, W. Livestock-associated MRSA: The impact on humans. Antibiotics (Basel) 4, 521–543. https://doi.org/10.3390/antibiotics4040521 (2015).
Foster, T. J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol Rev 41, 430–449. https://doi.org/10.1093/femsre/fux007 (2017).
Llarrull, L. I., Fisher, J. F. & Mobashery, S. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new beta-lactams that meet the challenge. Antimicrob. Agents Chemother. 53, 4051–4063. https://doi.org/10.1128/AAC.00084-09 (2009).
Fuda, C. C., Fisher, J. F. & Mobashery, S. Beta-lactam resistance in Staphylococcus aureus: The adaptive resistance of a plastic genome. Cell Mol. Life Sci. 62, 2617–2633. https://doi.org/10.1007/s00018-005-5148-6 (2005).
Mistry, H. et al. Prevalence and characterization of oxacillin susceptible mecA-positive clinical isolates of Staphylococcus aureus causing bovine mastitis in India. PLoS ONE 11, e0162256. https://doi.org/10.1371/journal.pone.0162256 (2016).
Saeed, K. et al. Oxacillin-susceptible methicillin-resistant Staphylococcus aureus (OS-MRSA), a hidden resistant mechanism among clinically significant isolates in the Wessex region/UK. Infection 42, 843–847. https://doi.org/10.1007/s15010-014-0641-1 (2014).
Andrade-Figueiredo, M. & Leal-Balbino, T. C. Clonal diversity and epidemiological characteristics of Staphylococcus aureus: High prevalence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with clinical isolates in Brazil. BMC Microbiol. 16, 115. https://doi.org/10.1186/s12866-016-0733-4 (2016).
Song, Y., Cui, L., Lv, Y., Li, Y. & Xue, F. Characterisation of clinical isolates of oxacillin-susceptible mecA-positive Staphylococcus aureus in China from 2009 to 2014. J. Glob. Antimicrob. Resist. 11, 1–3. https://doi.org/10.1016/j.jgar.2017.05.009 (2017).
Quijada, N. M. et al. Oxacillin-susceptible mecA-positive Staphylococcus aureus associated with processed food in Europe. Food Microbiol. 82, 107–110. https://doi.org/10.1016/j.fm.2019.01.021 (2019).
Conceição, T., Coelho, C., de Lencastre, H. & Aires-de-Sousa, M. Frequent occurrence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) strains in two African countries. J. Antimicrob. Chemother. 70, 3200–3204. https://doi.org/10.1093/jac/dkv261 (2015).
Hososaka, Y. et al. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus: A new type of MRSA. J. Infect. Chemother. 13, 79–86. https://doi.org/10.1007/s10156-006-0502-7 (2007).
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-three Informational Supplement M100-S23. CLSI, Wayne, PA, USA, 2013.
Pu, W. et al. High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China. PLoS ONE 9, e88134. https://doi.org/10.1371/journal.pone.0088134 (2014).
Sakoulas, G. et al. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J. Clin. Microbiol. 39, 3946–3951. https://doi.org/10.1128/JCM.39.11.3946-3951.2001 (2001).
Ikonomidis, A. et al. In vitro and in vivo evaluations of oxacillin efficiency against mecA-positive oxacillin-susceptible Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 3905–3908. https://doi.org/10.1128/AAC.00653-08 (2008).
Duarte, F. C. et al. Fatal sepsis caused by mecA-positive oxacillin-susceptible Staphylococcus aureus: First report in a tertiary hospital of southern Brazil. J. Infect. Chemother. 25, 293–297. https://doi.org/10.1016/j.jiac.2018.09.010 (2019).
Goering, R. V., Swartzendruber, E. A., Obradovich, A. E., Tickler, I. A. & Tenover, F. C. Emergence of oxacillin resistance in stealth methicillin-resistant. Antimicrob. Agents Chemother. 63, e00558-e619. https://doi.org/10.1128/AAC.00558-19 (2019).
Chung, M. et al. Heterogeneous oxacillin-resistant phenotypes and production of PBP2A by oxacillin-susceptible/mecA-positive MRSA strains from Africa. J. Antimicrob. Chemother. 71, 2804–2809. https://doi.org/10.1093/jac/dkw209 (2016).
Gratani, F. L. et al. Regulation of the opposing (p)ppGpp synthetase and hydrolase activities in a bifunctional RelA/SpoT homologue from Staphylococcus aureus. PLoS Genet. 14, e1007514. https://doi.org/10.1371/journal.pgen.1007514 (2018).
Ender, M., McCallum, N. & Berger-Bächi, B. Impact of mecA promoter mutations on mecA expression and beta-lactam resistance levels. Int. J. Med. Microbiol. 298, 607–617. https://doi.org/10.1016/j.ijmm.2008.01.015 (2008).
Kuroda, M. et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357, 1225–1240. https://doi.org/10.1016/s0140-6736(00)04403-2 (2001).
Liu, P., Xue, H., Wu, Z., Ma, J. & Zhao, X. Effect of bla regulators on the susceptible phenotype and phenotypic conversion for oxacillin-susceptible mecA-positive staphylococcal isolates. J. Antimicrob. Chemother. 71, 2105–2112. https://doi.org/10.1093/jac/dkw123 (2016).
Geiger, T. et al. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8, e1003016. https://doi.org/10.1371/journal.ppat.1003016 (2012).
Geiger, T. et al. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 78, 1873–1883. https://doi.org/10.1128/IAI.01439-09 (2010).
Steiner, K. & Malke, H. relA-Independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183, 7354–7364. https://doi.org/10.1128/JB.183.24.7354-7364.2001 (2001).
Bæk, K. T. et al. β-Lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease. Antimicrob. Agents Chemother. 58, 4593–4603. https://doi.org/10.1128/AAC.02802-14 (2014).
Dordel, J. et al. Novel determinants of antibiotic resistance: identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. MBio 5, e01000. https://doi.org/10.1128/mBio.01000-13 (2014).
Kim, C. et al. The mechanism of heterogeneous beta-lactam resistance in MRSA: Key role of the stringent stress response. PLoS ONE 8, e82814. https://doi.org/10.1371/journal.pone.0082814 (2013).
Pardos de la Gandara, M. et al. Genetic determinants of high-level oxacillin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents. Chemother. 62, e00206-18. https://doi.org/10.1128/AAC.00206-18 (2018).
Aedo, S. & Tomasz, A. Role of the stringent stress response in the antibiotic resistance phenotype of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 60, 2311–2317. https://doi.org/10.1128/AAC.02697-15 (2016).
Bui, L. M., Conlon, B. P. & Kidd, S. P. Antibiotic tolerance and the alternative lifestyles of. Essays Biochem. 61, 71–79. https://doi.org/10.1042/EBC20160061 (2017).
Onyango, L. A. & Alreshidi, M. M. Adaptive metabolism in staphylococci: Survival and persistence in environmental and clinical settings. J. Pathog. 2018, 1092632. https://doi.org/10.1155/2018/1092632 (2018).
Cassels, R., Oliva, B. & Knowles, D. Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J. Bacteriol. 177, 5161–5165. https://doi.org/10.1128/jb.177.17.5161-5165.1995 (1995).
Murakami, K. et al. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29, 2240–2244 (1991).
Hiramatsu, K., Kihara, H. & Yokota, T. Analysis of borderline-resistant strains of methicillin-resistant Staphylococcus aureus using polymerase chain reaction. Microbiol. Immunol. 36, 445–453. https://doi.org/10.1111/j.1348-0421.1992.tb02043.x (1992).
Kampf, G., Adena, S., Rüden, H. & Weist, K. Inducibility and potential role of MecA-gene-positive oxacillin-susceptible Staphylococcus aureus from colonized healthcare workers as a source for nosocomial infections. J. Hosp. Infect. 54, 124–129. https://doi.org/10.1016/s0195-6701(03)00119-1 (2003).
Phaku, P. et al. Unveiling the molecular basis of antimicrobial resistance in Staphylococcus aureus from the Democratic Republic of the Congo using whole genome sequencing. Clin. Microbiol. Infect. 22(644), e641-645. https://doi.org/10.1016/j.cmi.2016.04.009 (2016).
Roisin, S., Nonhoff, C., Denis, O. & Struelens, M. J. Evaluation of new Vitek 2 card and disk diffusion method for determining susceptibility of Staphylococcus aureus to oxacillin. J. Clin. Microbiol. 46, 2525–2528. https://doi.org/10.1128/JCM.00291-08 (2008).
Sharma, S., Srivastava, P., Kulshrestha, A. & Abbas, A. Evaluation of different phenotypic methods for the detection of methicillin resistant Staphylococcus aureus and antimicrobial susceptibility pattern of MRSA. Int. J. Community Med. Public Health 4(9), 3297–3301 (2017). https://doi.org/10.18203/2394-6040.ijcmph20173832.
Proulx, M. K. et al. Reversion from methicillin susceptibility to methicillin resistance in Staphylococcus aureus during treatment of bacteremia. J. Infect. Dis. 213, 1041–1048. https://doi.org/10.1093/infdis/jiv512 (2016).
Chen, F. J., Wang, C. H., Chen, C. Y., Hsu, Y. C. & Wang, K. T. Role of the mecA gene in oxacillin resistance in a Staphylococcus aureus clinical strain with a pvl-positive ST59 genetic background. Antimicrob. Agents Chemother. 58, 1047–1054. https://doi.org/10.1128/AAC.02045-13 (2014).
McKinney, T. K., Sharma, V. K., Craig, W. A. & Archer, G. L. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and beta-lactamase regulators. J. Bacteriol. 183, 6862–6868. https://doi.org/10.1128/JB.183.23.6862-6868.2001 (2001).
Mwangi, M. M. et al. Whole-genome sequencing reveals a link between β-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb. Drug Resist. 19, 153–159. https://doi.org/10.1089/mdr.2013.0053 (2013).
Anderson, K. L. et al. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188, 6739–6756. https://doi.org/10.1128/JB.00609-06 (2006).
Hughes, J. & Mellows, G. On the mode of action of pseudomonic acid: Inhibition of protein synthesis in Staphylococcus aureus. J. Antibiot. (Tokyo) 31, 330–335. https://doi.org/10.7164/antibiotics.31.330 (1978).
Haseltine, W. A. & Block, R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. USA 70, 1564–1568. https://doi.org/10.1073/pnas.70.5.1564 (1973).
Liu, K., Bittner, A. N. & Wang, J. D. Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 24, 72–79. https://doi.org/10.1016/j.mib.2015.01.012 (2015).
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. https://doi.org/10.1038/nrmicro3448 (2015).
Aiba, Y. et al. Mutation of RNA polymerase β-subunit gene promotes heterogeneous-to-homogeneous conversion of β-lactam resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 4861–4871. https://doi.org/10.1128/AAC.00720-13 (2013).
Matsuo, M., Yamamoto, N., Hishinuma, T. & Hiramatsu, K. Identification of a novel gene associated with high-level β-Lactam resistance in heterogeneous vancomycin-intermediate Staphylococcus aureus strain Mu3 and methicillin-resistant S. aureus Strain N315. Antimicrob. Agents Chemother.63, e00712-18, https://doi.org/10.1128/AAC.00712-18 (2019).
Thalsø-Madsen, I. et al. The Sle1 Cell wall amidase is essential for β-Lactam resistance in community acquired methicillin resistant. Antimicrob. Agents Chemother. 64, e01931-e2019. https://doi.org/10.1128/AAC.01931-19 (2019).
Andersson, D. I. & Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance?. Nat. Rev. Microbiol. 8, 260–271. https://doi.org/10.1038/nrmicro2319 (2010).
Baumert, N. et al. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8, 253–260. https://doi.org/10.1089/10766290260469507 (2002).
Chuard, C., Vaudaux, P. E., Proctor, R. A. & Lew, D. P. Decreased susceptibility to antibiotic killing of a stable small colony variant of Staphylococcus aureus in fluid phase and on fibronectin-coated surfaces. J. Antimicrob. Chemother. 39, 603–608. https://doi.org/10.1093/jac/39.5.603 (1997).
Garcia, L. G. et al. Antibiotic activity against small-colony variants of Staphylococcus aureus: Review of in vitro, animal and clinical data. J. Antimicrob. Chemother. 68, 1455–1464. https://doi.org/10.1093/jac/dkt072 (2013).
Cui, L., Neoh, H. M., Iwamoto, A. & Hiramatsu, K. Coordinated phenotype switching with large-scale chromosome flip-flop inversion observed in bacteria. Proc. Natl. Acad. Sci. USA 109, E1647-1656. https://doi.org/10.1073/pnas.1204307109 (2012).
Saito, M. et al. “Slow VISA,” a novel phenotype of vancomycin resistance, found in vitro in heterogeneous vancomycin-intermediate Staphylococcus aureus strain Mu3. Antimicrob. Agents Chemother. 58, 5024–5035. https://doi.org/10.1128/AAC.02470-13 (2014).
Katayama, Y. et al. Prevalence of slow-growth vancomycin nonsusceptibility in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 61, e00452-e517. https://doi.org/10.1128/AAC.00452-17 (2017).
Kanesaka, I. et al. Characterization of compensatory mutations associated with restoration of daptomycin-susceptibility in daptomycin non-susceptible methicillin-resistant Staphylococcus aureus and the role mprF mutations. J. Infect. Chemother. 25, 1–5. https://doi.org/10.1016/j.jiac.2018.09.009 (2019).
Wada, A. et al. Ratio of mecA gene in oxacillin-insusceptible and susceptible Staphylococcus aureus. Jpn. J. Chemother. 55(5), 374–377 (2007).
Chen, F. J. et al. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J. Clin. Microbiol. 50, 1679–1683. https://doi.org/10.1128/JCM.06711-11 (2012).
Watanabe, S. et al. Complete genome sequencing of three human clinical isolates of Staphylococcus caprae reveals virulence factors similar to those of S. epidermidis and S. capitis. BMC Genomics 19, 810. https://doi.org/10.1186/s12864-018-5185-9 (2018).
Watanabe, S. et al. Complete genome sequence of streptococcus pyogenes Strain JMUB1235 isolated from an acute phlegmonous gastritis patient. Genome Announc. 4, e01133-e1216. https://doi.org/10.1128/genomeA.01133-16 (2016).
Watanabe, S. et al. Composition and diversity of CRISPR-Cas13a systems in the genus Leptotrichia. Front. Microbiol. 10, 2838. https://doi.org/10.3389/fmicb.2019.02838 (2019).
Gardner, S. N., Slezak, T. & Hall, B. G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. https://doi.org/10.1093/bioinformatics/btv271 (2015).
Neoh, H. M. et al. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 52, 45–53. https://doi.org/10.1128/AAC.00534-07 (2008).
Kato, F. & Sugai, M. A simple method of markerless gene deletion in Staphylococcus aureus. J. Microbiol. Methods 87, 76–81. https://doi.org/10.1016/j.mimet.2011.07.010 (2011).
Sato’o, Y. et al. Optimized universal protocol for electroporation of both coagulase-positive and -negative Staphylococci. J. Microbiol. Methods 146, 25–32. https://doi.org/10.1016/j.mimet.2018.01.006 (2018).
Acknowledgements
We thank Dr. Tetsuro Muratani at Kyurin Medical Laboratory, Dr. Tetsu Mizutani and Ms. Kana Sawa at Osaka Police Hospital, Dr. Tsai-Ling Yang Lauderdale at the National Health Research Institutes of Taiwan, and Dr. Intetsu Kobayashi at Toho University for kindly providing OS-MRSA strains. We thank Dr. Motoyuki Sugai for kindly gifting plasmids (pKFT and pKAT).
Funding
This work was supported by JMU Graduate Student Start-up Award to TB, JSPS KAKENHI (Grant Nos. 15H05654 and 19K08960 to SW, 18K15149 to KK, 17K15691 to YS, and 17K19570 to LC), the Takeda Science Foundation (SW, LC), and the Japan Agency for Medical Research and Development (Grant Nos. JP17fm0208028, JP18fm0208028, and JP19fm0208028, JP20fk0108134 to LC). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
T.B., S.W., and L.C. designed the study, analysed the data, and wrote the manuscript. T.B., S.W., R.N., K.S., and R.T. performed the experiments. X.T. contributed to data interpretation and assisted with the preparation of the manuscript. K.T., R.N., K.S., R.T., Y.S., Y.A., K.K., T.S.,Y.T., F.L., Y.Z., A.H.A., and T.K. contributed to data collection and interpretation. All authors reviewed and revised the manuscript. The final version of the manuscript was approved and agreed by all authors for all aspects of the work in ensuring that questions related to the integrity or accuracy of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Boonsiri, T., Watanabe, S., Tan, XE. et al. Identification and characterization of mutations responsible for the β-lactam resistance in oxacillin-susceptible mecA-positive Staphylococcus aureus. Sci Rep 10, 16907 (2020). https://doi.org/10.1038/s41598-020-73796-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-73796-5
- Springer Nature Limited
This article is cited by
-
Phenotypic and genotypic characteristics of mecA - positive oxacillin-sensitive Staphylococcus aureus isolated from patients with bloodstream infection in a tertiary hospital in Southern Brazil
Brazilian Journal of Microbiology (2024)
-
Efficient synthesis of CRISPR-Cas13a-antimicrobial capsids against MRSA facilitated by silent mutation incorporation
Scientific Reports (2024)
-
Phenotypic and genomic characteristics of oxacillin-susceptible mecA-positive Staphylococcus aureus, rapid selection of high-level resistance to beta-lactams
European Journal of Clinical Microbiology & Infectious Diseases (2023)
-
An inhibitory effect of schisandrone on α-hemolysin expression to combat methicillin-resistant staphylococcus aureus infections
World Journal of Microbiology and Biotechnology (2023)
-
Molecular Typing and Global Epidemiology of Staphylococcus aureus
Current Pharmacology Reports (2021)