Abstract
Recombination hot spots (RHP), caused by meiosis, are considered to play crucial roles in improvement and domestication of crop. Cultivated peanut is one of the most important rich-source of oil and protein crops. However, no direct scale of recombination events and RHP have been estimated for peanut. To examine the scale of recombination events and RHP in peanut, a RIL population with 200 lines and a natural population with 49 cultivars were evaluated. The precise integrated map comprises 4837 SLAF markers with genetic length of 2915.46 cM and density of 1.66 markers per cM in whole genome. An average of 30.0 crossover (2.06 cMMb−1) events was detected per RIL plant. The crossover events (CE) showed uneven distribution among B sub-genome (2.32) and A sub-genome (1.85). There were 4.34% and 7.86% of the genome contained large numbers of CE (> 50 cMMb−1) along chromosomes in F6 and natural population, respectively. High density of CE regions called RHP, showed negative relationship to marker haplotypes conservative region but positive to heatmap of recombination. The genes located within the RHP regions by GO categories showed the responding of environmental stimuli, which suggested that recombination plays a crucial role in peanut adaptation to changing environments
Similar content being viewed by others
Introduction
Peanut, also known as groundnut (Arachis hypogaea L.; AABB, 2n = 4x = 40), is a major source of edible oils worldwide, of which the annual production is about ~ 46 million tons (FAOSTAT 2015, https://faostat3.fao.org/home/). As a domesticated allotetraploid, the A subgenome and B subgenome of peanut are from the diploid species A. duranensis and A. ipaensis respectively, and it arised from natural hybridization of the two diploid species over 9400 years ago1. Cultivated peanut clusters the beneficial traits from the elites of germplasm. Limited by allotetraploidy and large genome (~ 2.7 Gb), it has been proven difficult to identify the required markers and to assemble a genome sequence in cultivated peanut (A. hypogaea)1.
In previous studies, quantitative trait loci (QTL) mapping has been a useful tool for dissecting the genetic architecture of complex traits in a great number of species. Many of those have focused on variation in important economic traits, such as oil content a polygenic, quantitative trait resulting from interactions between the environment and multiple genes2,3. For instance, a mass of QTLs that associated with important agronomic traits of Arachis hypogaea have been identified based on bi-parental linkage mapping. Nearly, 31 epistatic QTLs associated with five component traits of bruchid resistance throughout the total developmental period were screened in 2 years4. A total of 54 and 23 QTLs were identified for spotted wilt virus and leaf spot of tomato in the F2 and F5 peanut populations, respectively5. A total of 6 and 9 QTLs associated with oil content of peanut were identified in two different recombinant inbred line (RIL) populations respectively6. Recent improvements in sequencing technologies have reduced the cost of genotyping large numbers of accessions and increased the feasibility of performing pre-breeding at genome level as an alternative to QTL mapping or genome-wide associated studies.
The frequencies of recombination during crossover of homologous chromosomes could be detected by molecular markers, based on which a genetic map is constructed. Meiotic recombination which evolved into homologous chromosomes and the two sister chromatids formed after chromosomal replication could create genetic variation in gametes and new allele combinations7,8. As a fundamental biological process, meiotic recombination generates a new genetic diversity through alleles shuffling via crossover (reciprocal exchange of large chromosomal segments) or gene conversion (non-reciprocal exchange of small chromosomal segments), which impacts the genetic evolution of organisms9. So, recombination events is of great significance for the genomic evolution, especially for the crop domestication and improvement. For a offspring population, the allele distribution and haplotype structure is largely determined by the rate and distribution of recombination events happened in the whole genome of a species. The recombination crossover rates between species were different by the detection, for example, 20–50 crossover events per meiosis in human genome2,10,11; an average of 90 crossover events per meiosis w in budding yeast genome12; 81 crossover events per meiosis honey bee genome13; 10–37 crossover events per meiosis in Arabidopsis genome14 and 28–33 crossover events per meiosis in rice15,16. There were some regions showed uneven distributions of crossovers in the genomes. Some regions is entirely devoid the crossovers (0 cMMb−1) (cold spot crossover regions) and others showed the concentration of a large number of crossovers within a few kilobases (> 50 cMMb−1) (hot spot crossover regions)7,8,14,15,16. Many studies have shown that the hot spot crossover regions may contribute largely to evolution by generating novel heritable variations due to adaptive mutation or recombination16.
With lots of studies in diploid species, recombination crossover events in polyploid species are still unknown. To understand the crossing over of peanut, a RIL population (with 200 F6 lines) and a small natural panel (with 49 peanut cultivars) were used in our study. In total, 605,631 accurate markers, which were developed by specific-locus amplified fragment sequencing (SLAF-seq) approach17, were adopted to detect the recombination activity in the whole genome of F6 genetic map. A total of 61,942 InDels detected using transcriptome were adopted to detect the recombination activity in the whole genome of natural population. Together, present research aims to address the following objectives: (1) to construct a genetic linkage map of high-density SNP; (2) to detect the recombination crossover events in genetic map and natural population; (3) to identify the vicinity of genes located within the recombination hot spot regions by gene ontology (GO) categories.
Results
Quality control of specific-locus amplified fragment (SLAF) sequencing
DNA fragments of 314–414 bp digested by Rsal and EcoRV Enzymes were defined as SLAF tags. A total of 392,911 (with average of 19,646) SLAF tags located in a physical map of 2438.09 Mb were obtained by electronic digestion (Fig. 1d). The quality score of SLAF base calling were showed in Fig. 1b. The score of most base calling of SLAF showed equal to 40 (when Q ≥ 30, P value ≤ 0.001). To test whether the sequencing and libraries bring AT or GC base separation were good, the base distribution along reads were detected. The average of AT and CG equaled to 30 and 20, respectively (Fig. 1a), suggesting that the sequencing and libraries were of good quality. Because small insert size of SLAF fragments can lead to spurious mapping, the SLAF fragments with size from 264 to 464 bp (with average of 364 bp) were selected for preparation of libraries (Fig. 1c).
A total of 605,631 SLAF markers were developed by the SLAF-seq. The average individuals’ integrity was nearly 0.85 (Fig. 2a), suggesting that the markers were developed successfully. A total of 21,133 showed polymorphisms between cultivars 950527 and Huayu22 (Fig. 2c). Limited with the high similarity between A and B sub-genomes, the hemi-SNP type (hk × hk; lm × ll; nn × np) were selected and used for linkage map construction (Fig. 2b).
SLAF-markers selection and the construction of genetic linkage maps of RIL (F6) population. (a) The integrity of individuals in F6 population; (b) genotype distribution of SLAF markers; (c) polymorphic SLAF markers distribution on the physical map of peanut; (d) co-linearity of genetic linkage map and physical map of RIL (F6) population.
Construction of a high-density SLAF based genetic linkage map.
A total of 11,076 poly-SLAF markers (normal and hemi-SNP) were developed by the SLAF-seq. Of these, 4837 SLAF markers with ≤ 5% missing data were successfully assigned to 20 linkage groups representing the A01-A10 and B01-B10 linkage groups of A. hypogaea, with total genetic distances of 1533.47 cM and 1381.99 cM for the A and B sub-genomes, respectively (Fig. 3; Table 1). The number of markers mapped on A and B sub-genome were 2383 and 2454, respectively (Table 1). The physical distance of A and B sub-genome were 1026.62 Mb and 1326.03 Mb. The marker number and density varied considerably with different chromosomes. The marker number ranged from 77 on chromosome A08 to 694 on chromosome B04 (with average of 241.85 per chromosome). The marker density ranged from 0.48 markers per cM on A08 to 5.80 markers per cM on B04 (with average density of 1.66 markers per cM). The quality of the linkage map could be detected by max gap and ratio of gap < 5 cM of each chromosome (Table 1). The collinear analysis of linkage map and physical map were showed in Fig. 2d.
Identification of recombination events and recombination hot spots
The linear analysis was performed between the number of recombination events and the physical length of the chromosomes. The mean number of recombination events per chromosome pair was positively correlated with the physical length of the chromosome in the peanut F6 RIL population (Fig. 4; r = 0.99, P = 0.00062) and 49 peanut accessions (Table 2; r = 0.72, P = 0.00039), indicating that longer chromosomes have more recombination events. The recombination rate of each linkage group ranged from 1.16 cM per Mb on B05 chromosome to 5.36 cM per Mb on B04 (with an average of 2.06 cM per Mb) in the F6 RIL population (Table 1). The recombination rate of each chromosome ranged from 6.10 n per Mb on A04 to 15.71 n per Mb on A08 chromosome (with an average of 8.40 n per Mb) (Table 2).
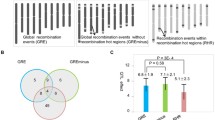
The SLAF marker haplotypes of each individual in the F6 linkage map was shown in Fig. 4a, and this suggests the recombination frequently regions mapped on genome. The relative recombination of each two markers was valued in the RIL (F6) linkage maps (Fig. 4c). Within linkage group, there were 4837 recombination crossover events detected (Fig. 5a). The number of recombination crossover events in the natural population was 2073 (Figs. 5b, 6, S1). The recombination frequency regions (recombination hot spots) on chromosomes are shown in Figs. 4 and S1. A total of 210 (4.34% of total recombination crossover events) of the loci were recombination hot spots in RIL population (Figure S1). Those recombination hot spots were located in 120 loci (2.35% of total recombination crossover events of total recombination crossover events) in A genome and 90 loci (1.86%) in B genome (Fig. 4). There were 163 (was 7.86% of whole recombination crossover events) regions which were recombination hot spots in the peanut natural population. The number of recombination hot spots was 49 (2.36% of all recombination crossover events regions) and 114 (5.50% of all recombination crossover events regions) located A and B genome (Figs. 6, S1), respectively.
Genotyping and identification of crossover events in RIL (F6) population and 49 peanut accessions. (a) Identification of crossover events and recombination hot spots detection in RIL (F6) population; (b) identification of crossover events and recombination hot spots detection in 49 peanut accessions.
Functional characterization of genes with a high recombination rate
A total of 3865 genes were located in the interval (100 kb) of recombination hot spot loci. Nearly 841 types of functions could be classified by using peanut Gene Ontology annotation (https://geneontology.org/).
The top 60 functions (54.30% of total genes function) were shown in Fig. 6. The genes could be divided into three functions including biological process (9.49% of total genes), cellular component (22.41% of total genes) and molecular function (22.41% of total genes) (Table S1). Most genes in cellular component and molecular function were involved into synthesis of ATP, DNA, RNA, and protein binding. Most of these genes in biological process have functions responding to the environmental stimulus (Fig. 7), for example, response to cadmium ion, stress, auxin stimulus, fructose stimulus, wax biosynthetic process, fungus, , etc., suggesting that the genes located in the region of high recombination rates on chromosome tend to be involved in the response to environmental stimuli and biotic stress . The frequent recombination in peanut RIL population and natural population may be beneficial to the adaptation of peanut in changing environments.
Discussion
High density SNP-based genetic linkage map construction
Cultivated peanut is an allopolyploid crop of which the genome structure iscomplex. A large number of homologous sequences exist between the peanut A- and B-subgenome, and simultaneously both the homologous and non-homologous exchanges are extensively observed between them1. This complexity leads to the difficulty of developing high-quality molecular markers used in association studies. Recent advances in sequencing technologies have reduced the cost of genotyping large numbers of accessions and increased the feasibility of constructing a high density, SNP-based genetic linkage map as an alternative to more traditional genetic mapping.
Genetic linkage maps could provide valuable references for studying genome structure and evolution, analyzing comparative genomes and localizing genes of interest18. In our study, a genetic linkage map of peanut with high density SNP markers was constructed (Fig. 3; Table 1). The number of markers mapped onto A- subgenome (2383 markers) was roughly same as those mapped onto B-subgenome (2454 markers) (Table 1). The physical and genetic distances between markers on the A genome were the same as on the B genome. The length of the genetic map in our studies was 2915.46 cM, which bigger than the lengths of 1935.4 cM6 and 1446.7cM19 published. The total number of markers mapped onto the genetic linkage map was 4837, which is larger number than 1267 bin marker19 and 418 markers3 mapped to genetic linkage maps previously. According to Zhou et al.19 and our results, the recombination distance in the A and B genome showed balance which is different from that observed in other polyploid species, such as Brassica napus (The recombination distance on A genome is greater than on C genome), indicating similar evolutionary history of Arachis duranensis and Arachis ipaensis20,21. Moreover, there appear to have been no further interspecific hybridization events during cultivated peanut breeding1.
Recombination detected in peanut RIL and natural population
Meiotic recombination as an important, fundamental biological process can produce crossover and gene conversions, which greatly influences the genomic evolution, particularly the process of crop improvement or breeding. There were 24,185 crossover events per plant detected in the RIL population (Table 1), which is lower than that in rice16; that in budding yeast12; that in honey bee13 and that in Arabidopsis15. However, crossover events per plant species may not be suitable scales for detecting recombination crossover events across the genome. Because population crossover events depend on the population size, the density of recombination rate was a better scale for valuing genome crossover events. In our study, the average number of crossover events was 2.06 cM/Mb (Table 1), smaller than that in rice 4.53 cM/Mb16, suggesting a negative trend between genome size and recombination crossover events rates. Similar recombination crossover rates were observed in yeast and honey bee14,15. There was a weak correlation between genetic distance and recombination crossover rate in the RIL population (r = 0.1549, P = 1.08 × 10–32). There was also a weak correlation between genetic distance of each linkage group and recombination crossover rate (r = 0.1265, P = 1.35 × 10–14) too. The number of recombination showed a weak correlation between density of recombination crossover events rate and physical distance in 49 peanut cultivars (r = 0.3033, P = 7.71 × 10–15) (Table 2), suggesting that the recombination crossover rates are too complex to be controlled by one element. There was a strong correlation between recombination crossover events and physical distance in RIL and natural populations (Table 1).
Recombination hot spots were defined as the regions which showed high distributions of crossovers in the genomes7,8. In our studies, nearly 4.34% and 7.86% of recombination events on the genome were recombination hot spots (Figs. 4, 6), which are larger than those in rice (0.72% recombination hot spots of recombination crossover events)16. Compared with the RIL population, the natural population had higher recombination hot spot rates. The recombination hot spot rates on the A genome are larger than on the B genome in RIL population, suggesting more diversity within the A genome has been selected in the breeding process. However, the recombination hot spots rates were larger in the B genome than the A genome in the natural population, suggesting that there has been more balanced selection between the two genomes in nature.
Higher recombination rates in the RIL and natural population and in the vicinity of environment-related genes
As an important polyploid edible oil crop, the recombination crossover events in the peanut genome appear to have played a crucial role in crop environment adaptation, breeding and improvements. Previous studies have shown that the genes in the hot spot crossover regions tended to be involved in responses to environmental stimuli8,15. In our work, a total of 3865 genes were located in the recombination hot spots regions. Most genes (accounting for 22.41% of total genes) were involved in cellular components and 22.41% of total genes in molecular function (Fig. 7; Table S1). Most genes in cellular components and molecular functions were involved in ATP, DNA, RNA, and protein binding, suggesting that the genes within hot spot regions have played an important role in maintaining normal regulation mechanisms of peanut. The genes involved in biological processes within recombination hot spot regions primarily tended to be involved in responses to environmental stimuli. This suggests that frequent recombination plays an important role in adaptive evolution in changing environment of peanut breeding or improvement (Fig. 7). Compared with the GO analysis in rice16, the GO analysis likely provides a pool of highly dynamic targets for selection, which is potentially a result of the elevated recombination rates in peanut.
Experimental procedures
Plant materials and DNA isolation
The RIL (F6) population including 200 peanut (Arachis hypogaea L.) lines was obtained from the crossing between female parent 950527 and male parent Huayu22. A total of 49 peanut accessions made up a natural population, which were collected from peanut breeding or improvement programs. All of the 249 genotypes were grown in a field screening nursery at Qingdao, China (120.41°E, 36.39°N) in May of 2017. Each accession was planted in a single-row with 12 individual plants within each row. The trial management followed standard breeding field protocols. Young leaves from one individual plant of each accession were collected and kept at − 80 °C freezer, and genomic DNA was isolated using CTAB method22.
Genotyping of the RIL population
SNP genotyping of the association panel was performed using a SLAF-seq approach17. Construction of the peanut DNA libraries and Illumina sequencing of the plants were performed at Biomarker Technologies Corporation in Beijing, China. Through restriction enzymes HaeIII and Hpy166II (New England Biolabs, NEB, USA) that digest peanut genomic DNA into DNA fragments of 364–464 bp23, the sequencing libraries of 202 peanut accession were constructed. The physical position of the markers were identified by aligning the sequence of a 125 bp paired-end reads attached to each marker with the ‘pseudomolecules’ genome sequences of diploid peanut (Arachis duranensis-AA and Arachis ipaensis-BB, https://www.peanutbase.org) using local BLASTn (BLAST: Basic Local Alignment Search Tool, https://blast.ncbi.nlm.nih.gov/Blast.cgi). If the reads matched two or more locations in the reference genome of peanut, the markers were regarded as non-specific markers and discarded. Accurate markers were selected throught three steps. First, all candidate markers must be called in the mixed reads from parents and all the F6 samples using GATK (https://software.broadinstitute.org/gatk/). Second, all candidate markers must be called less than 20%. Third, some hemi-SNPs which showed polymorphism in sub-genomes were used for polymorphism markers in genetic linkage map in RIL population.
Genotyping of the natural population
Peanut young seed of three plants of each of the 49 accessions were sampled 30–40 days (pegs stage) after flowering. Samples were cleaned and immediately placed in liquid nitrogen before being stored at − 80 °C24. The sequencing libraries of 147 RNA samples were generated using the Illumina RNA Library Prep Kit (NEB #E7760, San Diego, CA USA) and sequenced on an Illumina Hiseq 2000 platform with 100-bp paired-end reads. The physical position of the markers was identified by aligning the sequence of a 100 bp paired-end reads attached to each marker with the ‘pseudomolecules’ genome sequences of peanut using local BLASTn. The InDels markers with call frequencies > 0.8 and minor allele frequencies (MAF) > 0.0525 were selected for recombination crossover events analysis.
Construction of genetic maps, heat maps and haplotype maps in RIL population
A representative SNP marker of a bin was selected to construct the genetic linkage map using the sofware packages of HighMap13. The map evaluation module provide heat maps and haplotype maps for intuitive displays of map quality. The grouping module uses the single-linkage clustering algorithm to cluster the markers into linkage groups, using a pair-wise modified independence LOD score as distance metric.
The independence test G statistic is given by:
\(o{ }\) is observed number of each genotype; \(e\) is expected number in each cell.
The modified LOD score from an approximate transformation is given by:
where \({\text{d}}\) is the degrees of freedom; \(e\) is the expected number, where \( e\) is total row multiplied by total column division by total grand.
According to Liu et al.13, the mapping algorithm applies an iterative process of marker ordering and error genotype correction to ensure the accuracy of map order and map distances in the presence of missing observations and genotyping errors.
Genotyping and identification of crossover events
Based on the nucleotide at the marker sites in 950527 and Huayu22, all the candidate markers were converted to heterozygous genotypes (950527 and Huayu22) (Fig. 5a) in each individual of F6 and natural population (Fig. 5b). The markers on genetic map could be candidate recombination crossover events in RIL population. The recombination crossover events could be detected by the detailed methods in Yang et al.15. The spans with both sides of the breakpoints of ≥ 10 kb were assumed to be the outcomes of recombination crossover events15,16. Based on the markers’ genotypes, some regions along chromosome pairs were also converted into blocks of heterozygous genotypes or parental genotypes (Fig. 5). Slide windows (100 kb) were used for analysis of the loci recombination crossover events. Based on the heat maps and haplotype maps of each F6 population individual, small recombination regions (block length ≤ 200 kb) were checked to exclude potential false positives. Moreover, the recombination crossover events with ambiguous allelic relationships were excluded. The alignment gaps caused by blocks with an abnormal insert size of the paired-end reads were excluded16.
Identification of gene function within hot spot regions
To identify hot spot regions of crossover events, we used a Poisson distribution to find the threshold value of the number of crossover events in each 100-kb region in 49 peanut accessions. To identify the high recombination rate regions in the peanut genome, we divided the whole peanut genome into 2920 non-overlapping windows (1000 kb for each window) and calculated the recombination rate of each window in 49 peanut accessions16. If the rate was ≥ 25 cMMb−1 (threefold of average recombination rate), we assigned this as a high recombination rate window and collected the genes in this window. A recombination rate > 50 cM per Mb indicated recombination hot spots in the RIL population (F6). The recombination hot spots were defined by three principles, which were less SLAF markers haplotypes region, recombination rate > 50 cM per Mb in F6 population, weak relative recombination of each two markers region and high recombination rate in 49 peanut accessions (> triple of genome average recombination). The gene function frequency of all the collected genes and searched for the high-frequency function compared with the whole genome in hot spot region were checked by Gene Ontology data (downloaded from the RGAP website)16.
References
Bertioli, D. J. et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Na. Genet. 48, 438–446 (2016).
Wang, J. B., Fan, H. C., Behr, B. & Quake, S. R. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150, 402–412 (2012).
Pandey, M. K. et al. Genetic dissection of novel QTLs for resistance to leaf spots and tomato spotted wilt virus in peanut (Arachis hypogaea L.). Front. Plant Sci. 8, 25. https://doi.org/10.3389/fpls.2017.00025 (2017).
Mondal, S., Hadapad, A. B., Hande, P. A. & Badigannavar, A. M. Identification of quantitative trait loci for bruchid (Caryedon serratus Olivier) resistance components in cultivated groundnut (Arachis hypogaea L.). Mol. Breed. 33(4), 961–973 (2014).
Wang, H. et al. Genetic mapping and quantitative trait loci analysis for disease resistance using F2 and F5 generation-based genetic maps derived from “Tifrunner” X “GT-C20” in Peanut. Plant Genome 6, 3 (2013).
Pandey, M. K. et al. Identification of QTLs associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L.). BMC Genet. 15, 133 (2014).
Hey, J. What’s so hot about recombination hotspots?. PLoS Biol 2, e190 (2004).
Wahls, W. P. & Davidson, M. K. DNA sequence-mediated, evolutionarily rapid redistribution of meiotic recombination hotspots. Genetics 189, 685–94. https://doi.org/10.1534/genetics.111.134130 (2011).
Baudat, F., Imai, Y. & de Massy, B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet 14, 794–806 (2013).
Cheung, V. G., Burdick, J. T., Hirschmann, D. & Morley, M. Polymorphic variation in human meiotic recombination. Am. J. Hum. Genet. 80, 526–530 (2007).
Coop, G., Wen, X. Q., Ober, C., Pritchard, J. K. & Przeworski, M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319, 1395–1398 (2008).
Mancera, E., Bourgon, R., Brozzi, A., Huber, W. & Steinmetz, L. M. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485 (2008).
Liu, D. et al. Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS ONE 9, e98855. https://doi.org/10.1371/journal.pone.0098855 (2014).
Wu, J. Z. et al. Physical maps and recombination frequency of six rice chromosomes. Plant J. 36, 720–730 (2003).
Yang, S. et al. Great majority of recombination events in Arabidopsis are gene conversion events. Proc. Natl. Acad. Sci. USA 109, 20992–20997 (2012).
Si, W. et al. Widely distributed hot and cold spots in meiotic recombination as shown by the sequencing of rice F2 plants. New Phytol. 206, 1491–1502. https://doi.org/10.1111/nph.13319 (2015).
Sun, X. et al. SLAF-seq: an efficient method of large-scale De novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 8, e58700 (2013).
Zhang, Y. et al. QTL meta-analysis of root traits in Brassica napus under contrasting phosphorus supply in two growth systems. Sci. Rep. 6, 33113. https://doi.org/10.1038/srep33113 (2016).
Zhou, X. et al. Construction of a SNP-based genetic linkage map in cultivated peanut based on large scale marker development using next-generation double-digest restriction-site-associated DNA sequencing (ddRADseq). BMC Genom. 15, 351. https://doi.org/10.1186/1471-2164-15-351 (2014).
Clevenger, J. et al. Genome-wide SNP genotyping resolves signatures of selection and tetrasomic recombination in peanut. Mol. Plant 10, 309–322 (2017).
Leal-Bertioli, S. C. et al. Tetrasomic recombination is surprisingly frequent in allotetraploid Arachis. Genetics 199, 1093–1105 (2015).
Paterson, A. H., Brubaker, C. L. & Wendel, J. F. A rapid method for extraction of cotton (gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 11, 122–127 (1993).
Li, T. et al. Genome-wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 15, 1520–1532 (2017).
Wei, L. et al. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol. J. 14, 1368–1380 (2016).
Wang, X. et al. Genetic variants associated with the root system architecture of oilseed rape (Brassica napus L.) under contrasting phosphate supply. DNA Res. 24, 407–417 (2017).
Acknowledgements
This study was supported by Shandong Provincial Natural Science Foundation (ZR2019PC055; ZR2018PC023), the National Natural Science Foundation of China (31471533, 31701376; 31700553; 31801868), The National Ten Thousand Youth Talents Plan of 2014 (W02070268), China Agriculture Research System (CARS-13), The foundation of Linyi university (40618112; 40618113). Thanks to Thomas D Alcock from the Plant and Crop Sciences Division, School of Biosciences, University of Nottingham, Sutton Bonington Campus, Loughborough, LE12 5RD, United Kingdom, for English proofreading.
Author information
Authors and Affiliations
Contributions
Conceptual and experiment designs by X.W., P.X., L.Y., Y.R. and M.Y.; Experiments were conducted by X.W., P.X.,S.L., Y.W., Y.S. and M.Y.; Data analysis performed by X.W., H.L., X.C., X.C., T.Y., and M.Y.; the report was written by X.W., P.X., M.K.P. and R.K.V. All the authors have commented, read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., Xu, P., Ren, Y. et al. Genome-wide identification of meiotic recombination hot spots detected by SLAF in peanut (Arachis hypogaea L.). Sci Rep 10, 13792 (2020). https://doi.org/10.1038/s41598-020-70354-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70354-x
- Springer Nature Limited