Abstract
Cadmium (Cd) accumulation in rice is a serious threat to food safety and human health. Breeding rice varieties with low Cd accumulation is one of the most effective approaches to reducing health risks from Cd-polluted rice. However, the genetic basis of Cd accumulation in grains, especially in indica rice varieties, has not been fully elucidated. The evaluation of Cd-accumulation capacity was conducted among 338 diverse rice accessions grown in Cd-contaminated soils with different Cd contents. Thirteen rice lines with relatively low Cd accumulation, including six indica rice lines, were identified. Then, 35 QTLs significantly associated with Cd accumulation were identified through sequencing-based SNP discovery and a genome-wide association study (GWAS) in the two experimental years, and only qCd8-1 was detected in both years. Among of them, nine QTLs were co-localized with identified genes or QTLs. A novel QTL, qCd1-3, with the lowest P value was selected for further LD decay analysis and candidate gene prediction. We found differential expression of OsABCB24 between high-Cd-accumulative and low-Cd-accumulative accessions, suggesting it may be a candidate gene for qCd1-3 associated with low Cd accumulation. These results may be helpful for further exploiting novel functional genes related to Cd accumulation and developing rice variety with low Cd accumulation through marker-assisted breeding.
Similar content being viewed by others
Introduction
Rice (Oryza sativa L.) is one of the world’s staple food crops. Due to rapid industrialization and urbanization in recent years, heavy metal contamination in arable soils has become an increasingly severe problem in China. Among the heavy metals, cadmium (Cd) is the most serious contaminant. Reportedly, average Cd concentrations in paddy fields of China reached 0.23 mg/kg, and the highest concentration of 0.73 mg/kg was recorded from Hunan province1. Recently, Cd-polluted rice from Hunan, the largest rice-producing province in China, has become a particular concern in food safety. It has been reported that rice tends to accumulate more Cd than other cereals2. Moreover, the acidification of paddy soil caused by frequent applications of nitrogen fertilizer increases Cd solubility and results in more Cd absorption by rice plants3. In rice, Cd inhibits growth and development by causing misfolding of functional proteins and interfering the homeostasis of other essential metal ions4. For people, excessive accumulation of Cd may result in diseases such as cancer, anemia, heart failure, hypertension, as well as other chronic disorders5,6. A range of measures have been adopted to reduce Cd bioaccumulation, including soil remediation, Cd immobilization and transgenic techniques. The most promising strategy is to screen and breed rice varieties with low Cd accumulation, especially crops grown on slightly to moderately Cd-contaminated soils2.
Genetic variation naturally occurs among rice varieties in their abilities to accumulate Cd. Thus, extensive screening has been carried out to identify rice genetic resources with low Cd accumulation7,8,9. In general, indica rice varieties accumulate higher amounts of Cd in the grains than do japonica rice varieties9,10. Indica rice varieties are grown predominantly in southern China, and their planting areas coincide with the high-Cd-contaminated region, which further increases the risk of greater Cd accumulation in rice. Using populations derived from crosses between indica and japonica varieties, many quantitative trait loci (QTLs) for Cd accumulation have been identified11,12,13,14,15. Ueno et al.7 detected a major QTL (qCd11) controlling the Cd concentration in shoots using a F2 population. Liu et al.16 identified seven QTLs and validated qCd2 associated with grain Cd content using a recombinant inbred line (RIL) population. Another major QTL (qCd7) was repeatedly detected using different genetic populations and finally cloned17,18,19,20. The candidate gene of qCd7, OsHMA3, encodes a tonoplast-localized P1B-type ATPase. OsHMA3 is involved in Cd sequestration from the cytoplasm into the vacuoles of root cells and its dysfunction promotes root-to-shoot Cd translocation and consequently increases Cd accumulation in the shoots and grains in some varieties18,19. Several dysfunctional alleles of OsHMA3 have been identified in different japonica rice accessions21,22. However, one SNP in the promoter of OsHMA3, which alters the normal expression of OsHMA3, is responsible for the differential Cd accumulation between indica and japonica rice varieties23. Like the function of OsHMA3, ATP-binding cassette (ABC) proteins mediate Cd sequestration and confer Cd tolerance in Arabidopsis thaliana24,25.
Most of the Cd-related QTLs were identified using bi-parental populations. However, the progress of QTL mapping is hindered due to limited allele diversity and less recombination in bi-parental populations26. Genome-wide association study (GWAS) can overcome these two limitations and is a powerful tool to identify genome regions associating with complex traits27,28. Through a GWAS of 276 diverse accessions, 60 QTLs were detected for accumulation of arsenic, cadmium, and lead in rice29. Zhao et al.30 identified 14 QTLs associated with Cd accumulation in rice by GWAS and predicted OsNRAMP2 as the candidate gene of qCd3–2. Using a composite method combining GWAS and other analyses, Yan et al.10 found that a missense mutation in OsCd1 resulted in the indica-japonica differentiations of Cd accumulation in rice grain. These reports clearly demonstrate that GWAS is an effective approach to elucidate the genetic mechanism underlying Cd accumulation. Unfortunately, for HMA3 and most of these Cd-related QTLs, the favorable alleles for reducing Cd accumulation were basically derived from japonica rice varieties and therefore limited their breeding application in indica rice.
To identify rice germplasms with low Cd accumulation and the responsible loci for Cd accumulation, we selected 338 accessions mainly composed of indica rice to evaluate their Cd accumulation in different Cd-polluted paddy fields. 13 rice lines, including six indica rice lines, were identified as low Cd-accumulative germplasms. Based on the specific-locus amplified fragment sequencing (SLAF-seq) method31, genome-wide SNP discovery and a GWAS strategy were used to identify QTLs associated with Cd concentration in polished rice. 35 QTLs significantly associated with Cd accumulation were identified in two experimental years, but only qCd8-1 was detected in both years. Through a combined analysis of LD decay and gene expression, we predicted OsABCB24 as the candidate gene of qCd1-3. These results will be helpful to elucidate the genetic mechanism of Cd accumulation and provide a good basis for breeding low Cd-accumulative indica rice varieties.
Results
SLAF-based SNPs discovery among rice accessions
After sequencing and quality control, a total of 688,782 SLAF tags were obtained for each of the 338 accessions, and 515,447 polymorphic SLAFs were identified by conducting sequence alignment with the 93-11 reference genome (Supplementary Table 1). The average sequence depth was 14.3× , ranging from 8.7 to 25.5 × among different accessions. Using the GATK and Samtools software packages, 3,960,919 SNPs were called from the SLAFs for the 338 genotypes. Based on the criterion of having MAF larger than 0.05 and missing genotype rate less than 0.2, only 123,865 SNPs (3.11%) passed filters from the SNP dataset and were used for subsequent analysis. These high-quality SNPs were evenly distributed on 12 chromosomes with average density of approximately one SNP per 3.02 Kb (Supplementary Table 2). The highest maker density was detected on chromosome 7 (one SNP per 2.63 Kb), while the lowest density was detected on chromosome 8 (one SNP per 3.55 Kb).
Population structure and relative kinship
Population structure analysis can provide information on the origin and composition of individuals. Based on the filtered high-quality SNPs, the optimal number of ancestors (K) was estimated using the STRUCTURE software. The ∆K value was lowest when K was set to 2, suggesting that the whole group of rice lines could be divided into two subgroups (Fig. 1a). Consistently, the Principal Component Analysis (PCA) results showed that two clusters clearly separated along the eigenvector of PC1, which accounted for 32.9% of the genetic variation (Fig. 1b). Based on the neighbor-joining algorithm, a phylogenetic tree for each sample was constructed with two subgroups (1 and 2) (Fig. 1c). Subgroup_1 contained 268 rice lines, which were in accordance with the indica subspecies. Subgroup_2 contained 70 lines, including a portion of the foreign germplasm and landraces, which were in accordance with the japonica subspecies. Most of the breeding accessions and landraces belonged to subgroup_1. Interestingly, glutinous lines of landraces genetically diverged significantly, with 15 and 20 lines classified to subgroup_1 and subgroup_2, respectively. Moreover, there was differentiation within the indica rice subgroup. Most of the bred varieties had close relationships with foreign indica germplasms but were distantly related to the landraces, indicating that these landraces were rarely used in modern rice breeding in China.
Variation of Cd accumulation in polished rice among 323 lines
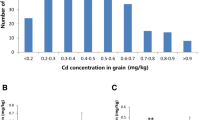
In order to reduce confounding effects from variable growth durations among accessions, only 323 lines with moderate durations of growth were selected for Cd determination and GWAS analysis. The Cd concentrations of polished rice collected from Cd-polluted fields were determined using atomic-absorption spectrometry. The Cd accumulation varied significantly among different rice accessions in both years (Supplementary Table 3). In 2016, Cd concentrations in polished rice ranged from 0.57 mg/kg to 4.03 mg/kg, with an average of 1.61 mg/kg and a median of 1.53 mg/kg (Fig. 2a). All the rice lines showed higher Cd accumulation over 0.2 mg/kg, which is the allowable concentration for human consumption as stipulated by the National Food Hygiene Standard of China. In 2017, Cd concentrations ranged from 0.06 mg/kg to 2.24 mg/kg, with an average of 0.43 mg/kg and a median of 0.33 mg/kg (Fig. 2b). Only 58 lines displayed Cd concentrations less than 0.2 mg/kg, accounting for 18.0% of all rice lines. Overall, rice Cd accumulation in 2017 was significantly lower than that in 2016, indicating that Cd concentrations of soil was a key factor determining Cd accumulation in grains (Fig. 2c). Despite large differences between two years, thirteen rice lines were identified with relatively low Cd accumulation (Cd2016 < 0.8 mg/kg and Cd2017 < 0.2 mg/kg) in both years (Table 1). For example, breeding material BS114 showed the lowest Cd accumulation (0.57 mg/kg) in 2016 and also extremely low Cd (0.07 mg/kg) in 2017, respectively. Additionally, many landraces from Hunan province, such as “Shenshuinuo” and “Daganzaogu,” were found in this low-Cd-accumulative group and could be used as potential donors in future low-Cd rice breeding.
Distribution of Cd concentration in polished rice. (a) Cd concentration of polished rice in 2016; (b) Cd concentration of polished rice in 2017; (c) Boxplot of Cd accumulation in 2016 and 2017; (d) Boxplot of Cd accumulation in different subgroups in 2016 and 2017, G1 and G2 indicate group_1 and group_2 respectively.
Considering the presence of distinct population structure, Cd accumulations were compared between different subgroups in both years. As shown in Fig. 2d, Cd accumulation in the indica subgroup was significantly higher than that in the japonica subgroup (P < 0.001). In 2016 and 2017, the mean Cd accumulations in the indica subgroup were 1.75 mg/kg and 0.47 mg/kg respectively, while in the japonica subgroup were 1.04 mg/kg and 0.25 mg/kg, respectively. These results clearly indicated that population structure had effect on the Cd accumulation in these rice lines.
GWAS for Cd accumulation
To investigate the genotypic basis underlying Cd accumulation in polished rice, we performed GWAS to identify the associated SNP loci in the selected 323 rice lines. Considering the effect of population structure on Cd accumulation, the mixed linear model (MLM) model was adopted with kinship matrix and PC matrix as covariates. According to Lv et al.32, a region was considered as one QTL when more than two significant SNPs (P < 0.001) were detected within a 200-Kb window. In total, 35 QTLs with 203 SNPs significantly associated with Cd accumulation were identified in the two experimental years with a well-fitted quantile–quantile (Q–Q) plots (Table 2, Supplementary Table 4, Fig. 3a,b). These QTLs were distributed on all chromosomes except chromosome 10. The comparison of QTLs identified in two years indicated that only qCd8-1 was detected in both years, suggesting that environmental factors might have great influence on the GWAS results of Cd accumulation. To verify the accuracy of the GWAS results, the identified QTLs in this study were further compared with previous reports. We found 9 QTLs were co-localized with previous mapped QTLs, associated markers and characterized genes (Table 2), indicating that GWAS results were reliable in this study. Among the co-localized QTLs, qCd6-2 on chromosome 6 was located close to OsLCT133, and qCd7-1 on chromosome 7 was identified in the genome interval of the well-characterized gene HMA3, which is involved in Cd transport into rice grains. The remaining QTLs have not been reported previously and were considered as novel QTLs.
Manhattan plots and quantile–quantile (Q–Q) plot of GWAS for Cd accumulation. (a) Manhattan plots and Q-Q plot of GWAS for Cd accumulation in 2016 using the whole group; (b) Manhattan plots and Q-Q plot of GWAS for Cd accumulation in 2017 using the whole group; (c) Manhattan plots and Q-Q plot of GWAS for Cd accumulation in 2016 using the Indica subgroup; (d) Manhattan plots and Q–Q plot of GWAS for Cd accumulation in 2017 using the Indica subgroup. The horizontal line in Manhattan plots indicates threshold of P value.
To further exclude interference from population structure, GWAS was conducted using the indica subgroup of 259 accessions and compared with the identified QTLs using the whole group (Fig. 3c,d, Supplementary Table 5). The japonica subgroup was not analyzed separately in this study due to a limited number of rice lines. For the indica subgroup, GWAS results were basically consistent with those for the whole group. However, 16 QTLs including qCd1-1, qCd1-2, qCd2-2, qCd4-1, qCd4-2, qCd5-1, qCd5-2, qCd6-2, qCd7-2, qCd7-4, qCd8-1, qCd8-3, qCd8-4, qCd9-2, qCd11-2 and qCd11-3 were not detected in this subgroup mainly because only one significant SNP was identified in most of these loci. In total, 19 QTLs for Cd accumulation were identified in both the whole group and indica subgroup. Among these QTLs, qCd1-3 showed the lowest P value on chromosome 1 near position 43.3 Mb and was chosen for subsequent analysis.
Identification of candidate genes responsible for Cd accumulation
Around the interval of qCd1-3, 14 consecutive SNPs were significantly associated with Cd accumulation in 2017 (Fig. 4a), among which the lead SNP rs1_43287290 (P = 2.78E−06) was selected as the representative of this loci. According to the alleles of the lead SNP, all samples were divided into two groups of a favorable allele G (designated as group G) and an unfavorable allele T (designated as group T) respectively. Cadmium accumulation in group G was significantly lower than that in group T. An average of 0.78 mg/kg and a median of 0.62 mg/kg were observed in group T while an average of 0.36 mg/kg and a median of 0.30 mg/kg were observed in group G. (Fig. 4b). In order to accurately estimate the target interval, LD decay analysis was performed for the region around qCd1-3. With r2 = 0.8 as the threshold, a 136-Kb block containing the lead SNP was identified as the candidate region (Fig. 4c). Based on the annotation of the reference genome, 22 genes were identified in this block including 17 functional protein-coding genes and five lncRNA-encoding genes. One of these genes (OsABCB24) located approximately 71 Kb from the lead SNP was annotated as an ATP-binding cassette (ABC) transporter. Since the ABC transporter had been reported to mediate vacuolar compartmentation of Cd in root tissue, OsABCB24 was regarded as the primary candidate gene of qCd1-3 associated with Cd accumulation. Then we analyzed the expression of OsABCB24 in different tissues at the vegetative stage. Although OsABCB24 showed the highest expression in leaf, its expression in root was higher than those in many other tissues such as stem, leaf sheath and panicle (Supplementary Fig. 1). Based on the genotype of qCd1-3, twelve accessions with contrasting Cd accumulation, including six high-Cd-accumulative accessions and six low-Cd-accumulative accessions, were selected for further expression analysis. As shown in Fig. 4d, OsABCB24 showed lower transcript levels in the high-Cd-accumulative rice accessions than those in the low-Cd-accumulative accessions under normal growth conditions. A similar trend was also observed under Cd-treatment conditions, even though Cd treatment slightly reduced the expression of OsABCB24 in low-Cd-accumulative accessions. These results suggest that OsABCB24 might be a good candidate gene for qCd1-3.
Candidate gene estimation of qCd1-3 on chromosome 1. (a) Local manhattan plots of GWAS for Cd accumulation around qCd1-3; (b) Distribution of Cd concentration in polished rice between the favorable allele G and the unfavorable allele T; (c) LD heatmap of around qCd1-3; (d) Expression of OsABCB24 between high-Cd-accumulative and low-Cd-accumulative rice accessions. Varieties accessions, H1: W261; H2: W233, H3: W32; H4: W357; H5: W333; H6: W278; L1: W49; L2: W30; L3: W132; L4: W18; L5: W135; L6: W95. Different letters indicate significant difference at 1% level (Duncan’s multiple range test).
Discussion
Natural genetic variation is a powerful resource not only for rice breeding but also for investigating the genetic mechanism of complex traits. Cadmium accumulation varied considerably among different rice accessions, suggesting that it is feasible to breed low Cd-accumulative rice varieties. In order to accurately evaluate the Cd-accumulation capacity among different genetic rice lines, we cultivated 338 rice lines in paddy fields that naturally contained different concentrations of cadmium, and the experiment was conducted in two consecutive years. The difference of soil Cd concentration and Cd accumulation of polished rice in 2017 was significantly lower than those in 2016, respectively. These results were consistent with the previous study that the grain Cd accumulation was largely affected by the soil Cd concentration9. Zhao et al.30 planted 312 rice accessions in a field with a pH value of 5.5 and soil Cd level of 1.4 mg/kg (similar to the Cd level in 2016 of this study), while they recorded relatively lower Cd accumulation in the grain. Because pH is probably the most important influencing factor of Cd uptake in rice plants34, it is reasonable to ascribe this difference in grain Cd content to the difference in pH. Consistent with previous studies7,8,9, there are wide variations in Cd accumulation levels among different rice accessions. In addition, our results also indicted that indica rice accessions tended to accumulate more Cd than japonica accessions. Nevertheless, several indica rice lines with relatively low Cd accumulation were identified in both years, most of which are landraces from Hunan province. After long-term evolution under natural and artificial selection, rice landraces show high genetic diversity and outstanding environmental adaptability27. The results showed that most of the landraces were genetically distant from the bred varieties, and they could be ideal donors for breeding low-Cd-accumulative rice varieties, especially for indica rice varieties. Unfortunately, most of the rice lines showed higher Cd accumulation over 0.2 mg/kg in the two-year experiment. Thus, identifying accessions with relatively low Cd accumulation might be an important step towards reducing Cd risks to rice consumers.
Identifying the loci associated with complex traits in rice is challenging due to high population differentiation26. In this study, the population structure of the rice accessions likely affected Cd accumulation in the polished rice. MLM model has been widely adopted due to its effectiveness in controlling confounding factors and reducing the number of false positives35. Results of the GWAS for the indica subgroup were basically consistent with those for the whole group, demonstrating that the use of relatedness matrixes as covariates in GEMMA could eliminate confounding effects of population structure. The slight difference between the subgroup and whole group might be caused by two reasons: (1) a smaller number of SNPs were used in the GWAS for the indica subgroup; and (2) the criterion of requiring more than two significant SNPs existing within a 200-Kb window to identify a QTL may have been too strict. Another common problem encountered in the GWAS was the large effect of environment, especially when mapping isonomic traits36. Among the identified QTLs, only qCd8-1 was detected under different soil Cd concentration in both years. Except for the inaccurate phenotypic identification caused by environmental factors, it’s quite likely that the genetic mechanisms underlying Cd accumulation might be divergent under different levels of Cd-polluted soils. Since the Cd pollution level of the paddy field in 2017 is more similar to the common level occurring in China1, the QTLs identified in this environment is more valuable for use in rice breeding than those identified in 2016.
The transfer of Cd from soil to grain is controlled by at least four steps: transport of Cd from soil into root cells, sequestration of Cd into the vacuoles, xylem loading, and phloem-mediated Cd transport to grains37. Nramp5 is a major transporter responsible for the Cd uptake from soil, and its knock-out resulted in a significant reduction of Cd accumulation in different genetic backgrounds of rice38. However, no natural allelic variation of OsNramp5 have been reported among different genetic resources. Through GWAS analysis, Zhao et al.30 predicted that another homolog OsNRAMP2 might be involved in Cd uptake in rice. OsHMA3, which specifically sequesters Cd into the vacuole of root cells and prevents its upward transport, was identified as the candidate gene for qCd7-1 in this study. Interestingly, qCd7-1 was also detected in the indica subgroup, implying that there might be natural variations of OsHMA3 among different indica rice varieties. Unlike the function of OsHMA3, another homolog OsHMA2 functions in the transport of both zinc (Zn) and Cd between root and shoot tissues through xylem loading39. The last Cd translocation step from shoot to grain involves a low-affinity cation transporter, OsLCT1, the encoding gene of which was co-localized with qCd6-2 (identified in the present study).
Our bioinformatics and gene expression analyses suggest that OsABCB24 was the candidate gene of qCd1-3. The ABC transport family is one of the largest protein families and conserved in all organisms. There are more than 125 ABC transporters in the rice genome, and their functions have yet to be elucidated40. Several studies have shown that ABC transporters may play important roles in Cd tolerance in plants24,25. Among the genes within the block of qCd1-3, only OsABCB24 was identified to encode a transporter. Reportedly, ABC transporters contribute to detoxifying cadmium by pumping it into vacuoles in yeast41. In Arabidopsis, AtABCC3 functions as a transporter of phytochelatin–Cd complexes into vacuoles42, similar to the function of OsHMA3 in Cd vacuolar sequestration. Our results showed that the expression of OsABCB24 was relatively high in root, and was significantly lower in high-Cd-accumulative rice accessions than that in low Cd-accumulative accessions. Because overexpression of OsHMA3 decreased Cd concentration in shoots, we proposed that the strong expression of OsABCB24 might contribute to enhancing vacuolar compartmentation of Cd in roots, thereby reducing Cd accumulation in rice grains of low Cd-accumulative accessions. Future work will apply functional genomics methodologies such as genetic transformation and CRISPR-cas9 technology to verify the role of OsABCB24 in regulating Cd accumulation.
Methods
SLAF-seq, sequencing data analysis and SNP calling
A core collection of 338 rice accessions were selected from the genetic resources in the gene bank of Hunan province, China. The whole group was composed of 148 landraces, 92 introduced foreign germplasms, 82 bred varieties and 16 breeding intermediate materials (Supplementary Table 3). Young leaves from each of the 338 accessions were collected, frozen in liquid nitrogen, and used for DNA extraction. Genomic DNA was isolated using the cetyltrimethyl annonium bromide (CTAB) protocol43. The SLAF libraries were constructed for each accession following the method proposed by Sun et al.31, and sequencing was performed on a HiSeq 2500 system (Illumina, CA, USA). The library construction and sequencing were carried out at Biomarker Technologies Corporation (Beijing, China). Because most of the rice lines belong to the subspecies indica, the pair-end reads were aligned to the reference genome of indica rice 93-11 (https://rice.genomics.org.cn/) using the MEM algorithm of Burrows-Wheeler Aligner (BWA) software (version 0.7.10)44. After alignment, SNP calling was conducted by the combined use of GATK (version 3.7)45 and Samtools (version 1.9)46. The identified SNPs were further filtered by the Plink software (version 1.90)47. Only SNPs with minor allele frequencies (MAF) > 0.05 and missing genotype rates < 0.2 were retained for GWAS analysis.
Field and pot experiments
Two years of field experiments were conducted in two separate Cd-polluted paddy fields in Beishan, Hunan province, China. The soil Cd concentration in 2016 was 1.25 mg/kg with a pH value of 5.2, while the Cd concentration in 2017 was 0.69 mg/kg with a pH value of 5.3. To reduce potentially unexpected effects of differential growth duration among accessions, sowing dates were staggered in May based on the days to maturity of each accession to ensure most lines heading at approximately the same time. The 25-d-old seedlings were transplanted in a randomized complete block design with two replications for each line. Each replication contained 16 rice plants grown in two rows with an in-row spacing of 16.7 cm and a between-row distance of 20 cm. Flooded condition was maintained in the field until mid-August. In order to increase bioavailable Cd concentration in the soil, rain-fed irrigation was mainly adopted during the grain filling stage, and flush irrigation was applied when necessary to avoid drought stress. Other field management, including fertilizer application and disease and pest control, was conducted according to standard rice farming practice.
Sampling and Cd determination
Rice grain was harvested 35 days after heading and dried in an oven at 40℃ for three days. The Cd concentration was determined according to the Chinese National Standard (GB 5009.15-2014). Because polished rice is the main edible part of rice, Cd accumulation in polished rice was investigated in this study. Rice grains were polished and then ground into powder. Approximately 0.3–0.5 g samples were digested with a solution of nitric acid and perchloric acid (9:1 v/v). The Cd concentration in the digest solution was measured by atomic absorption spectrometry (Solaar S4; Thermo, USA).
Phylogenetic tree construction, population structure, and principal component analysis
A phylogenetic tree of 338 lines was constructed by MEGA 5.048 using the neighbor-joining method with 1000 bootstrap replicates. The population structure was analyzed using STRUCTURE software49. The following parameters were used for the analysis: K = 2 to 10, burn in 5000, MCMC repeat 50,000 and three replicates for each K. Then we calculated ΔK to determine the optimal K value. The software Clumpp50 and Pophelper51 were used to visualize the population structure. The Q matrix of population structure was analyzed by ADMIXTURE software52. Principal components analysis was carried out using EIGENSOFT53.
Genome-wide association study
A software toolkit of GEMMA was used to perform association mapping according to Zhou and Stephens54. The standard linear mixed model was expressed as y = Wα + xβ + u + e, where y represents the phenotypic observation, W = (w1, … wc) is an n × c matrix of covariates, α is the vector of the corresponding coefficients including the intercept, x is an n-vector of marker genotypes, β is the effect size of marker, u and e represent random effects and errors, respectively. To minimize the effect of population structure, PCA matrix and kinship matrix were used as covariates in this study. P values of ≤ 0.001 were used as the threshold to identify significantly associated SNPs. The SNP with the minimum P value in a locus was considered as the lead SNP. The allele contributing to reduction of cadmium content was regarded as the favorable allele.
Gene prediction and expression analysis
The LD heatmap around the peak SNP in GWAS was constructed using HaploView software55 and the candidate region was estimated using r2 > 0.8. The local Manhattan plot was produced using R package qqman56. The reference sequences of a candidate region were downloaded for gene annotation. Based on the annotations, genes related to transport of metal ion were selected as candidate genes.
For the expression analysis, the seedlings were grown in quarter-strength Hoagland solution in a growth chamber. Ten-days-old seedlings were then transferred to a nutrient solution with a cadmium concentration of 1.0 mg/kg, while the control group of seedlings continued to grow normally in the same nutrient solution without cadmium. After one week of treatment, roots were sampled and immediately frozen in liquid nitrogen. Total RNAs were isolated using Trizol Reagent (TransGen, Beijing, China) and were used for cDNA synthesis using RT SuperMix (Vazyme, Nanjing, China). Quantitative PCR was performed on a LightCycler 96 system (Roche, Rotkreuz, Switzerland) using SYBR qPCR Master Mix (Vazyme, Nanjing, China). Gene-specific primers for OsABCB24 were 5′- TCTTTACGAGTGACCCTGACC-3′ and 5′- CTCCATACTACCGACCCGTT-3′. Actin was used as an internal control with primers 5′- CATTGGTGCTGAGCGTTTCC-3′ and 5′- AGAAACAAGCAGGAGGACGG-3’.
Data availability
The raw reads of 338 rice accessions generated in this study have been deposited in the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra) under the accession number of PRJNA629658.
References
Liu, X., Tian, G., Jiang, D., Zhang, C. & Kong, L. Cadmium (Cd) distribution and contamination in Chinese paddy soils on national scale. Environ. Sci. Pollut. Res.. 23, 17941–17952 (2016).
Hu, Y., Cheng, H. & Tao, S. The challenges and solutions for cadmium-contaminated rice in China: a critical review. Environ. Int. 92, 515–532 (2016).
Wang, P., Chen, H., Kopittke, P. M. & Zhao, F. Cadmium contamination in agricultural soils of China and the impact on food safety. Environ. Pollut. 249, 1038–1048 (2019).
DalCorso, G., Farinati, S., Maistri, S. & Furini, A. How plants cope with cadmium: staking all on metabolism and gene expression. J. Integr. Plant Biol. 50, 1268–1280 (2008).
Godt, J. et al. The toxicity of cadmium and resulting hazards for human health. J. Occup. Med. Toxicol. 1, 22 (2006).
Satarug, S. et al. A global perspective on cadmium pollution and toxicity in non-occupationally exposed population. Toxicol. Lett. 137, 65–83 (2003).
Ueno, D. et al. A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa). New Phytol. 182, 644–653 (2009).
Yao, W. et al. Additive, dominant parental effects control the inheritance of grain cadmium accumulation in hybrid rice. Mol. Breed. 35, 39 (2015).
Sun, L. et al. Genetic diversity, rather than cultivar type, determines relative grain Cd accumulation in hybrid rice. Front. Plant. Sci. 7, 1407 (2016).
Yan, H. et al. Natural variation OsCd1V449 contributes to reducing cadmium accumulation in rice grain. Plant. Sci. https://doi.org/10.20944/preprints201802.0075.v1 (2018).
Abe, T. et al. Detection of a QTL for accumulating Cd in rice that enables efficient Cd phytoextraction from soil. Breed. Sci. 61, 43–51 (2011).
Hu, D. W. et al. Identification of QTLs associated with cadmium concentration in rice grains. J. Integr. Agric. 17, 1563–1573 (2018).
Norton, G. J. et al. Genetic mapping of the rice ionome in leaves and grain: identification of QTLs for 17 elements including arsenic, cadmium, iron and selenium. Plant Soil 329, 139–153 (2010).
Zhang, X. et al. Identification of quantitative trait loci for Cd and Zn concentrations of brown rice grown in Cd-polluted soils. Euphytica 180, 173–179 (2011).
Zhang, M. et al. Mapping and validation of quantitative trait loci associated with concentrations of 16 elements in unmilled rice grain. Theor. Appl. Genet. 127, 137–165 (2014).
Liu, W. et al. Identification of QTLs and validation of qCd-2 associated with grain cadmium concentrations in rice. Rice Sci. 26, 42–49 (2019).
Ueno, D. et al. Identification of a novel major quantitative trait locus controlling distribution of Cd between roots and shoots in rice. Plant. Cell Physiol. 50, 2223–2233 (2009).
Ueno, D. et al. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. 107, 16500–16505 (2010).
Miyadate, H. et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 189, 190–199 (2011).
Sui, F. et al. Map-based cloning of a new total loss-of-function allele of OsHMA3 causes high cadmium accumulation in rice grain. J. Exp. Bot. 70, 2857–2871 (2019).
Ueno, D., Koyama, E., Yamaji, N. & Ma, J. F. Physiological, genetic, and molecular characterization of a high-Cd-accumulating rice cultivar, Jarjan. J. Exp. Bot. 62, 2265–2272 (2010).
Yan, J. et al. A loss-of-function allele of OsHMA3 associated with high cadmium accumulation in shoots and grain of Japonica rice cultivars. Plant Cell Environ. 39, 1941–1954 (2016).
Liu, C. L. et al. Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between Indica and Japonica rice. J. Integr. Plant Biol. https://doi.org/10.1111/jipb.12794 (2019).
Kim, D. Y., Bovet, L., Maeshima, M., Martinoia, E. & Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant. J. 50, 207–218 (2007).
Park, J. et al. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant. J. 69, 278–288 (2012).
Korte, A. & Farlow, A. The advantages and limitations of trait analysis with GWAS: a review. Plant. Methods 9, 29 (2013).
Huang, X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961 (2010).
Huang, X. et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32 (2012).
Liu, X. et al. Association study reveals genetic loci responsible for arsenic, cadmium and lead accumulation in rice grain in contaminated farmlands. Front. Plant Sci. 10, 61 (2019).
Zhao, J. et al. Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice 11, 61 (2018).
Sun, X. et al. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 8, e58700. https://doi.org/10.1371/journal.pone.0058700 (2013).
Lv, Y. et al. New insights into the genetic basis of natural chilling and cold shock tolerance in rice by genome-wide association analysis. Plant Cell Environ. 39, 556–570 (2016).
Uraguchi, S. et al. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Natl. Acad. Sci. 108, 20959–20964 (2011).
Rafiq, M. T. et al. Cadmium phytoavailability to rice (Oryza sativa L.) grown in representative Chinese soils. A model to improve soil environmental quality guidelines for food safety. Ecotoxicol. Environ. Saf. 103, 101–107 (2014).
Yu, J. et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203 (2006).
Famoso, A. N. et al. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. 7, e1002221. https://doi.org/10.1371/journal.pgen.1002221 (2011).
Zhao, F. J. & McGrath, S. P. Biofortification and phytoremediation. Curr. Opin. Plant Biol. 12, 373–380 (2009).
Sasaki, A., Yamaji, N., Yokosho, K. & Ma, J. F. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24, 2155–2167 (2012).
Satoh-Nagasawa, N. et al. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 53, 213–224 (2011).
Moon, S. & Jung, K. Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J. Plant Physiol. 171, 1276–1288 (2014).
Mendoza-Cózatl, D. G. et al. Tonoplast-localized Abc2 transporter mediates phytochelatin accumulation in vacuoles and confers cadmium tolerance. J. Biol. Chem. 285, 40416–40426 (2010).
Brunetti, P. et al. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 66, 3815–3829 (2015).
Murray, M. & Thompson, W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8, 4321–4326 (1980).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Jakobsson, M. & Rosenberg, N. A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007).
Francis, R. M. pophelper: an R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17, 27–32 (2017).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904 (2006).
Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821 (2012).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2004).
Turner, S. D. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. J. Open Source Softw. 3, 731 (2014).
Acknowledgements
The research was supported by grants from the Ministry of Science and Technology in China (2016YFD0100101-12), Finance project of Hunan province-The breeding of rice varieties with low cadmium accumulation, Training Program for Excellent Young Innovators of Changsha (kq1802034), and Department of Science and Technology in Hunan province (2019RS2047).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. X.L. designed the study. X.P. and Y.L. performed most of the experimental work and manuscript writing. Y.D., Y.Y. and J.M. prepared seeds of rice accessions. X.P., W.L., S.L., H.X. and Z.D. conducted data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pan, X., Li, Y., Liu, W. et al. QTL mapping and candidate gene analysis of cadmium accumulation in polished rice by genome-wide association study. Sci Rep 10, 11791 (2020). https://doi.org/10.1038/s41598-020-68742-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-68742-4
- Springer Nature Limited
This article is cited by
-
Characterisation of a low methane emission rice cultivar suitable for cultivation in high latitude light and temperature conditions
Environmental Science and Pollution Research (2023)
-
Understanding the physiological and molecular mechanisms of grain cadmium accumulation conduces to produce low cadmium grain crops: a review
Plant Growth Regulation (2023)
-
Cadmium toxicity impacts plant growth and plant remediation strategies
Plant Growth Regulation (2023)
-
Combined linkage analysis and association mapping identifies genomic regions associated with yield-related and drought-tolerance traits in wheat (Triticum aestivum L.)
Theoretical and Applied Genetics (2023)
-
Phytotoxic Responses and Plant Tolerance Mechanisms to Cadmium Toxicity
Journal of Soil Science and Plant Nutrition (2023)