Abstract
Tuft (or brush) cells are solitary chemosensory cells scattered throughout the epithelia of the respiratory and alimentary tract. The actin-binding protein villin (Vil1) is used as a marker of tuft cells and the villin promoter is frequently used to drive expression of the Cre recombinase in tuft cells. While there is widespread agreement about the expression of villin in tuft cells there are several disagreements related to tuft cell lineage commitment and function. We now show that many of these inconsistencies could be resolved by our surprising finding that intestinal tuft cells, in fact, do not express villin protein. Furthermore, we show that a related actin-binding protein, advillin which shares 75% homology with villin, has a tuft cell restricted expression in the gastrointestinal epithelium. Our study identifies advillin as a marker of tuft cells and provides a mechanism for driving gene expression in tuft cells but not in other epithelial cells of the gastrointestinal tract. Our findings fundamentally change the way we identify and study intestinal tuft cells.
Similar content being viewed by others
Introduction
Tuft (also called brush, multivesicular, fibrovesicular, caveolated) cells are unusual epithelial cells that have been identified in numerous epithelial tissue including the salivary glands, stomach, gall bladder, bile duct, pancreatic duct, small intestine, cecum, colon, nasal cavity, auditory tube, trachea, urethra, and the thymus of multiple species, all with remarkably similar ultrastructure1,2. Compared to other mucosal epithelial cells in the alimentary and respiratory tract, tuft cells contain numerous intermediate filaments, microtubules and a dense network of actin and actin-binding proteins suggesting a major role for the cytoskeleton in their morphology and function3. Additionally, unlike enterocytes, tuft cells are characterized by an apical candle-like “tuft” or “brush” of longer and thicker microvilli that extend into the perinuclear region and like enterocytes the tuft/brush is composed of actin filaments cross-linked by the actin bundling proteins villin-1 (henceforth referred to as villin) and fimbrin4. Consistent with that finding, villin was the first marker used to identify tuft/brush cells, despite the fact that villin is also expressed in differentiated epithelial cells of the intestine5,6. The villin promoter has also been used extensively to drive gene expression or gene deletion from tuft/brush cells. One consequence of that is that data obtained using the Vil1-Cre or Vil1-Cre/ERT2 mice to study tuft cells, have been difficult to recapitulate when compared to similar studies done with other non-villin recombinase drivers such as the Rosa26-Cre/ERT2, doublecortin like kinase 1, Dclk1-Cre, or the leucine rich repeat containing G protein-coupled receptor 5, Lgr5+-Cre7,8,9,10,11,12. Similarly, the microtubule associated DCLK1, the transient receptor potential cation channel (TRPM5), prostaglandin-endoperoxide synthase 1 (PTGS1) are used to identify tuft cells, although all these proteins are also expressed in other cells within the gastrointestinal and respiratory tract and at levels comparable to those seen in tuft/brush cells11,13,14,15,16,17. Consequently, pairing these markers with each other, together with the unique candle-like “tufted” morphology are frequently employed to identify tuft/brush cells. Nonetheless, we note that within the gastrointestinal tissue, there is no definitive marker that is restricted to tuft cells. Consequently, the lineage of tuft cells as well as their functions have remained poorly understood largely because neither tuft cell deficient mice have been generated nor is it possible to modulate gene expression and activity only in intestinal tuft cells.
Advillin is a member of the villin/gelsolin superfamily that shares the highest structural homology with villin with a shared six domain structure and a carboxyl-terminal headpiece domain18. Interestingly, villin was identified in several chemosensory and mechanosensory cells that share many of the structural and functional characteristics of tuft cells. This includes Merkel cells in the skin, taste receptor cells, and exocrine glands with an endodermic lineage such as the thymus19,20,21,22,23,24. Villin was also frequently used as a marker for Merkel cells and taste receptor cells. However, more recently using immunohistochemistry and two advillin reporter mouse strains it has been shown that Merkel cells express advillin; while single cell RNA-Seq studies have demonstrated that Merkel cells contain advillin not villin mRNA20,25. Similarly, advillin protein has been identified in the taste receptor cells and advillin not villin mRNA has been identified using single-cell RNA-Seq on taste bud cells; likewise thymic brush cells have also been shown to express high levels of advillin but not villin mRNA26,27,28. More importantly, one of these studies has also identified advillin protein in solitary enteroendocrine-like epithelial cells in the mouse duodenal epithelium20. Although, we note that no markers of enteroendocrine cells were used in this study and that these cells appear to have the unique morphology of candle-like “tufted” cells rather than enteroendocrine cells20. In light of these discoveries, we reasoned that a careful evaluation of the villin family protein expression in intestinal tuft cells (and by extension in brush cells) was necessitated. Here, we report our surprising finding that intestinal tuft cells, in fact, do not express villin protein and that like other chemosensory and mechanosensory cells described above, they express advillin protein. To our surprise, a careful evaluation of more recently published studies that have employed single cell RNA-Seq to identify genes expressed in tuft/brush cells also identify advillin mRNA but do not describe villin mRNA in tuft/brush cells9,29,30,31,32,33. However, none of these studies describe the significance of these data. In light of our findings, we suggest that a thorough assessment of tuft cell lineage commitment and function is warranted. Furthermore, while it has been suggested that like trigeminal ganglia neurons, enteric neurons may also express advillin, immunohistochemistry and RNA-Seq studies reveal the absence of advillin from enteric neurons or enteric glia cells20,34,35. Based on that we propose that advillin expression in the gastrointestinal tract is restricted to tuft cells.
Results and Discussion
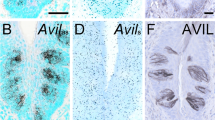
Based on a recent report, we noted that both villin and advillin expressing cells are present in the gastrointestinal epithelium20. Since villin and advillin share very significant (~75%) structural homology, our goal was to first identify antibodies that can distinguish between these two proteins18. Using recombinant human villin and advillin proteins we determined that most villin and advillin antibodies cross-react (Fig. 1a). We have previously shown that these antibodies do not cross-react with GST36,37. Careful characterization allowed us to identify two antibodies raised against the amino-terminus of villin (N-Villin and N20-Villin) that do not cross react with advillin (Fig. 1a). The region of human villin used to generate these antibodies shows some divergence when compared to human advillin gene. Furthermore, we noted that multiple advillin antibodies identify advillin much better than they cross-react with villin, this includes an advillin antibody raised against its amino-terminus (a.a. 440-526; N-advillin) and one raised against the carboxyl-terminus of advillin (a.a. 750-813; C-advillin). Using H & E (Hematoxylin and Eosin) staining of mouse distal ileum we identified less than 0.5% of the cells with a unique candle-like “tufted” morphology associated with intestinal tuft cells (Fig. 1b). Consistent with previous reports, these cells were determined to be approximately 10 μm long and approximately 5 μm in thickness. Based on the thickness of these cells, and the paucity of these cells in the normal mouse intestine, several attempts to use paraffin-embedded serial sections to correlate the morphological features of the same tuft cells with the expression of villin, advillin, and other known tuft cell markers, were unsuccessful. This may also be, one of the reasons, why such studies have not been performed previously. With that knowledge, we identified advillin expressing cells in the terminal ileum of C57/BL6 mice that resembled histomorphologically tuft cells, rather than enteroendocrine cells with a candle-like “tufted” morphology (Fig. 1c)20. Using paraffin-embedded ileal tissue, we noted that cells with this tufted morphology do not express villin protein (Fig. 1c). Cryopreserved tissue sections (which preserve F-actin better than paraffin embedded tissue) from distal ileum also showed solitary tuft cell-like advillin expressing cells that do not express villin protein (Fig. 1d). The advillin expressing cells show protein localization at the apical tufts and at the basolateral surface in what appears to be vesicular structures (Fig. 1c,d). It may be noted that previous studies have identified villin in the apical tufts and along the basolateral membrane of tuft cells38. This is different from the apical brush border restricted distribution of villin in enterocytes. Using the villin knockout (VKO) mice we confirmed the absence of villin (as shown here and as reported previously; Fig. 1e) and the presence of advillin expressing cells that resembled morphologically, tuft cells (Fig. 1f)39. Please note the absence of advillin staining in the gastrointestinal epithelium of the VKO mice, demonstrating that in the absence of villin protein the advillin antibodies do not detect any other cells in the epithelium (Fig. 1f). Tuft cells are marked by DCLK1 and are enriched in PTGS19,40. We confirmed our hypothesis that these advillin expressing cells are tuft cells by double labeling with antibodies against these commonly used markers of tuft/brush cells (Fig. 2a,b). Not all DCLK1 positive cells were positive for advillin and likely represent insulinoma-associated 1 positive enteroendocrine cells as has been suggested before11,13. Cryopreserved tissue sections show F-actin enriched in candle-like apical “tufts” of epithelial cells that are also positive for advillin expression (Fig. 2c). As described before, a unique morphological feature of tuft cells is the presence of axial bundles of actin filaments with a candle-like tuft or brush morphology41. This unique morphology identifies tuft cells and we find advillin but not villin enriched in these cells of the intestinal epithelium.
Mouse intestinal cells that express advillin do not express villin. (a) Specificity of villin and advillin antibodies was confirmed by Western analysis using recombinant full-length human villin and human advillin proteins. Two antibodies raised against the amino-terminus of villin (N-villin and N20-villin) did not cross-react with advillin compared to antibodies raised against the carboxyl-terminus of villin (C-villin) or full-length villin (FL-villin) protein. Antibodies raised against the amino-terminus of advillin (N-advillin) and those raised against the carboxyl-terminus of advillin (C-advillin) detect advillin better than they cross-react with villin. Original full length data are available in supplementary Fig S1. (b) H & E staining of mouse distal ileum identifies solitary cells with candle-like “tufted morphology” (identified by arrowhead). Right panel shows higher magnification of the boxed area. (c) Immunohistochemistry of paraffin embedded tissue from distal ileum of C57BL/6 J mice using N-villin (red) and advillin (green) antibodies. Nuclei are counter stained with DAPI. N-villin antibody does not cross-react with advillin in cells that express advillin in apical tufts and along the basolateral surface. Right panel shows higher magnification of boxed area. (d) Immunohistochemistry of cryopreserved tissue from distal ileum of C57BL/6 J mice using N-villin (red) and advillin (green) antibodies. Nuclei are counter stained with DAPI. N-villin does not co-localize with advillin in cells that express advillin in apical tufts and along the basolateral surface. Right panel shows higher magnification of boxed area. (e) Western analysis of distal ileal tissue from villin knockout (VKO) mice and their wild type (WT) littermates show the absence of villin protein in VKO mice. Original full length data are available in supplementary Fig S2. (f) Immunohistochemistry of paraffin embedded tissue from distal ileum of villin knockout mice identify advillin but not villin expressing cells. Nuclei are counter stained with DAPI. Right panel shows higher magnification of boxed area. Data shown in Western blots are representative of three independent experiments and in immunohistochemistry of n = 5 animals. Scale bars represent 10 μm.
Intestinal tuft cells express advillin. (a) Immunohistochemistry of paraffin-embedded tissue from distal ileum of C57BL/6 J mice using advillin (green) and DCLK1 (red) antibodies. Nuclei are counter stained with DAPI. Both proteins co-localize in the same cell. Note that not all DCLK1 positive cells are positive for advillin expression. Right panel shows higher magnification of boxed area and shows co-localization of DCLK1 and advillin in a tuft cell. (b) Immunohistochemistry of paraffin embedded tissue from distal ileum of C57BL/6 J mice using advillin (green) and PTGS1 (red) antibodies. Nuclei are counter stained with DAPI. Right panel shows higher magnification of boxed area and co-localization of PTGS1 and advillin in a tuft cell. (c) Immunohistochemistry of cryopreserved tissue from distal ileum of C57BL/6 J mice using Alexa Fluor 568 Phalloidin (red) and advillin (green). Nuclei are counter stained with DAPI. Right panel shows higher magnification of boxed area and co-localization of advillin and F-actin at the apical surface of a tuft cell. Data shown are representative of n = 5 animals. Scale bar represents 10 μm.
In the normal mouse intestine the number of tuft cells is very low (~0.5%) although this number can be increased 10-fold in the gut following parasitic (enteric metazoan and protozoan) colonization and infection42,43. Alternatively, this can be achieved by exogenous addition of interleukins (IL) IL-4 and IL-13 to isolated mouse enteroids, as reported before and as shown here (Fig. 3a)43. Similar to the intestinal tissue, ex vivo in the enteroids, advillin labeled the tuft cells as noted by co-localization of DCLK1 and PTGS1 (Fig. 3b). Using Virtual Channels to acquire multichannel confocal images, we show that all three proteins, PTGS1, DCLK1 and advillin are localized to the same cells (Fig. 4a). As expected, in mouse enteroids advillin co-localizes with the cytoskeletal proteins F-actin and tubulin (Fig. 4b). F-actin and advillin localized primarily to the eponymous apical tuft consisting of actin microfilaments that terminate at the perinuclear region. In contrast, tubulin localizes to the upper half of the cell and the basolateral surface of tuft cells where advillin co-localizes with the cytoplasmic tubulin. As reported before and as shown here, tubulin expression in the upper half of the cell is also unique to tuft cells and is never seen in other gastrointestinal or respiratory epithelial cells3. Similar to data shown in Figs. 1c,d,f, 2a–c, 3a,b, the expression of advillin in tuft cells appears to be associated with vesicular structures (Fig. 4b,c). More notably, in mouse enteroids like in the mouse intestine, advillin expressing cells do not express villin protein (Fig. 4c).
In intestinal enteroids, tuft cells express advillin. (a) Immuno-histochemistry of enteroids from distal ileum of C57BL/6 J mice shows tuft cell hyperplasia 72 hours post IL-4 and IL-13 treatment. Control refers to untreated enteroids from C57BL/6 J mice. Advillin (green) and DCLK1 (red) co-localization was used to identify tuft cells. Nuclei are counter stained with DAPI. Right panel shows higher magnification of the boxed area. (b) Immunohistochemistry of IL-4 and IL-13 treated enteroids from distal ileum of C57BL/6 J mice, show co-localization in tuft cells of advillin (green) and DCLK1 (red) in the upper panel; and advillin and PTGS1 (red) in the lower panel. Nuclei were counter stained with DAPI. Right panels show higher magnification of boxed area. Data shown are representative of n = 5 animals. Scale bar represents 10 μm.
Tuft cells positive for advillin are negative for villin. (a) Immunohistochemistry and Virtual Channel confocal microscopy of IL-4 and IL-13 treated enteroids from distal ileum of C57BL/6 J mice, show co-localization of advillin (green), DCLK1 (red) and PTGS1 (magenta) within the same cells identified as tuft cells. Nuclei are counter stained with DAPI. Right panel shows higher magnification of boxed area. (b) Immunohistochemistry of IL-4 and IL-13 treated enteroids from distal ileum of C57BL/6 J mice show co-localization in tuft cells of advillin (green) with F-actin (red) in the upper panel; and co-localization of advillin (red) and tubulin (green) in the lower panel. Nuclei were counter stained with DAPI. Right panel shows higher magnification of boxed area. Note the co-localization of advillin to the apical tuft that terminates near the perinuclear region of the cells. Advillin appears in vesiculated structures that co-localize with tubulin near the lateral cell surface and in the cytoplasm. (c) Immunohistochemistry of IL-4 and IL-13 treated enteroids from distal ileum of C57BL/6 J mice show the absence of villin (red) from advillin (green) expressing tuft cells. Nuclei are counter stained with DAPI. Right panel shows higher magnification of boxed area. Data shown are representative of n = 5 animals. Scale bar represents 10 μm.
Although advillin was identified over two decades ago as a new member of the gelsolin/villin family of proteins, the advillin protein has not been studied to determine either its actin binding ability or its ability to modify actin dynamics18. Based on that we elected to study, for the first time, the actin regulatory functions of advillin. All in vitro studies were performed with comparable levels of recombinant villin and advillin proteins. Using a standard in vitro assay for the measurement of actin binding by purified recombinant villin and advillin, we show that like villin, advillin is an actin binding protein (Fig. 5a)44. Advillin contains a villin-like carboxyl-terminal headpiece domain that is associated with villin’s bundling function18. Using a standard sedimentation assay for actin bundling we now show that this domain in advillin is functional and that advillin bundles actin similar to the villin protein (Fig. 5b). Advillin also shares the six domain structure of gelsolin and villin that is responsible for the actin depolymerizing functions of both proteins6. We now show, for the first time, that like its family members villin and gelsolin, advillin can nucleate, cap and sever actin filaments (Fig. 5c–e). These data demonstrate, for the first time, that advillin shares structural but also functional homology with other members of its family. Both villin and advillin are expressed in the gastrointestinal epithelium but are restricted to distinct cell types. While villin expression is restricted to differentiated intestinal epithelial cells, advillin expression is restricted to the chemosensory tuft cells. This lack of villin from tuft cells may also explain the unique ultrastructural features of tuft cells not shared by enterocytes namely, an apical tuft of stiff microvilli with long microvillar actin rootlets and no terminal web45. Furthermore, we hypothesize that unlike the restricted apical brush border localization of villin, the apical and basolateral expression of advillin in tuft cells may also have functions unique to solitary sensory cells. Additionally, these findings suggest that enterocytes and tuft cells may also have distinct lineage.
Regulation of actin dynamics by villin and advillin. (a) Villin and advillin bind F-actin. Recombinant villin and advillin proteins (60 nM) were incubated with 3 μM F-actin and centrifuged at 200,000 x g for 15 min. The supernatant (S) precipitated with 2 volumes of acetone and the pellet (P) were analyzed by SDS-PAGE and gels were stained with GelCode Blue. Control refers to assay in the absence of either protein. (b) Villin and advillin bundle F-actin. Recombinant villin and advillin proteins (1 μM) were incubated with F-actin and centrifuged at 10,000 x g for 15 min. The supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE and gels were stained with GelCode Blue. Control refers to assay in the absence of either protein. (c) Villin and advillin (60 nM each) nucleate actin filaments. Pyrene-labeled G-actin (6 μM) was polymerized in the presence of villin or advillin protein and fluorescence intensity measured over time. (d) Villin and advillin (60 nM each) cap actin filaments. Polymerization of 1.4 μM pyrene-labeled G-actin in the presence of F-actin seeds (290 nM) was measured in the presence of villin or advillin protein. (e) Villin and advillin sever actin filaments. Pyrene-labeled F-actin (1 μM) was diluted to 0.1 μM in actin-polymerizing buffer in the presence of villin or advillin protein (60 nM each) and the decrease in fluorescence intensity was followed over time. All gels are representative of three experiments with similar results. In all actin kinetic assays, values represent the mean of three independent experiments.
Since the development of the villin-Cre and villin-Cre/ERT2 mouse lines, more than a dozen studies have relied on them for labeling or knocking out genes in the intestinal tuft cells. While these studies do not claim gene expression or gene deletion restricted to tuft cells, they all identify villin expression in the intestinal tuft cells, and villin expression in tuft cells was important for the interpretations related to the lineage and function of tuft cells. For instance, two independent groups used the atonal bHLH transcription factor 1, Atoh1 flox/flox mice to induce Atoh1 deletion from the intestinal epithelium. One group crossed these mice with the Vil1-Cre/ERT2 mice and reported the complete loss of tuft cells46. Two other groups crossed the Atoh1flox/flox mice to Rosa26-Cre/ERT2 mice and reported the opposite namely, that tuft cells are preserved or that there is a significant increase in the number of tuft cells11,12. However, a subsequent study using the Lgr5-Cre/ERT2 mice crossed to Atoh1flox/flox mice also demonstrated that loss of Atoh1 from Lgr5+ cells (which includes tuft cells) results in an increase in the number of tuft cells in the intestinal epithelium12. Despite that confirmation, the role of Atoh1 in tuft cell development remains controversial, leading some to suggest an Atoh1-dependent and Atoh1-independent lineage for intestinal tuft cells47. Similarly, a role for the regulatory associated protein of MTOR complex 1 (RPTOR) in tuft cell lineage has been reported using the Vil1-Cre/ERT2 and the Raptorflox/flox mice even though ex vivo enteroids derived from these mice have normal tuft cells48. The role of the Wnt target gene Sox9 is likewise debatable. While several groups have demonstrated that DCLK1+ tuft cells express Sox9 the Vil1-Cre Sox9flox/flox mice show no effect of Sox9 loss on tuft cell distribution46,49,50. The role of tuft cells in the regulation of intestinal repair is also discordant. Deleting Dclk1 using the Vil1-Cre Dclk1flox/flox model showed that animals fail to recover normal crypt-villus architecture following genotoxic insult or treatment with dextran sodium sulfate, suggesting a loss of epithelial regeneration accompanied by a dramatic loss of self-renewal pathways regulated by Notch and mammalian target of rapamycin (mTOR)51,52. Consistent with that, two independent studies using the Vil1-Cre Dclk1flox/flox model demonstrated that tuft cells regulate DNA damage response and that the crypts of these mice have significantly higher number of apoptotic cells following radiation-induced injury51,53. In contrast, McKinley et al. demonstrated that when compared to most intestinal epithelial cells, tuft cells are extremely resistant to mucosal atrophy in response to acute fasting33. Additionally, using Dclk1-Cre/ERT2 Rosa26-LacZ reporter mice it was shown that tuft cells do not function as reserve stem cells further supporting the idea that tuft cells play no role in epithelial regeneration, a finding that was supported by studies done by Nakanishi et al.12,54 Our study underscores that the most likely explanation for many of these and similar conflicting findings related to tuft cell lineage commitment and function namely, the absence of villin protein from tuft cells. While the villin-Cre and villin-Cre/ERT2 mice are used to target gene expression or gene loss from a limited number of adult gastrointestinal epithelial cells that endogenously express villin, namely the enterocytes, unexpected expression of Cre recombinase in cells that do not endogenously express villin such as goblet cells is known. This 'ectopic' expression of villin-Cre in goblet cells and potentially tuft cells should be approached with caution. It is also known that a given Cre-expressing mouse can have different recombination efficiencies for different floxed genes in different cell types. Our findings underscore the importance of testing the Cre expression outside the expected cell type and highlight the need for a greater degree of caution that must be used when empolying the Vil1-Cre or villin-Cre/ERT2 mice to study gastrointestinal tuft cells.
Bezencon et al. used the Trpm5 promoter to express enhanced green fluorescent protein (EGFP) in intestinal tuft cells followed by RNA-Seq of the isolated EGFP expressing cells29. Their study reported very high expression of advillin but not villin mRNA in tuft cells and they identified no advillin mRNA in other EGFP null cells of the gastrointestinal epithelium29. Based on that the authors even suggested that it was possible that previously published immunostaining of brush cells with villin antibodies resulted from cross-reactivity with advillin29. We note that multiple single-cell RNA-Seq studies published after that have also identified advillin but none have identified villin mRNA in tuft/brush cells and a few have also shown the absence of advillin mRNA from other intestinal epithelial cells including enterocytes9,30,31,32. Nevertheless, we also note that none of these studies address these discrepancies related to villin/advillin expression in tuft/brush cells. Using the advillin promoter driven EGFP expression and a Human Protein Atlas advillin antibody, Hunter and colleagues reported that advillin is expressed in the duodenum in solitary endocrine cells even though these cells appear to morphologically resemble the candle-like “tufted” cells20. Their study also did not include any specific marker of enteroendocrine cells. Additionally, the authors reported advillin expression in enteric neurons20. We note that while Hunter et al. identify cells in the Meissner’s plexus that are enriched in EGFP-advillin, these cells do not appear to be the same as those identified by the advillin antibody. More significantly, the same antibody does not identify any enteric neurons in the duodenum or any other sections of the gastrointestinal tract but identifies solitary cells in the gastrointestinal epithelium with a candle-like “tufted” morphology (https://www.proteinatlas.org/ENSG00000 135407-AVIL/tissue). Our own studies with these antibodies (N-advillin) identify advillin positive tuft cells. It is known that tuft cells communicate with afferent nerves and that PGP9.5-positive nerves make direct contact with duodenal tuft cells29. We suggest that one possibility is that Hunter et al. have identified enteric neurons (identified by EGFP-advillin) in close proximity of advillin expressing tuft cells20. Alternatively, this discrepancy could be attributed to the ectopic expression of EGFP advillin in enteric neurons. There are a few single cell RNA-Seq studies on enteric neurons that do not identify advillin mRNA in enteric neurons or glia cells (Public databases: PanglaoDB; Single Cell Expression Atlas – EMBL-EBI)34,35,55,56, Based on that we suggest that advillin expression in the gastrointestinal tract may be restricted to the tuft cells. It is generally agreed that only tuft cell specific deletion of genes will provide the most definitive understanding of tuft cell lineage and function in vivo7,31. Unlike the Vil1-Cre or Vil1-Cre/ERT2 mice the availability of the advillin Cre mouse allows, for the first time, the targeting of this subpopulation of gastrointestinal epithelial cells, rather than all epithelial cells. Such approaches are more likely to provide a detailed investigation of the molecular mechanisms and functions of intestinal tuft cells.
The exact function of advillin has not been investigated although its distribution to intestinal tuft cells (and by extension respiratory brush cells) indicates a role in chemosensing and initiation of immune type 2 response. The fact that tuft cells express advillin and not villin suggests that despite the significant structural and functional homology, advillin may be uniquely adapted to regulate chemosensory and mechanosensory functions of tuft cells. Advillin also differs from villin in significant ways such as advillin interacts with the scavenger receptor SREC-1 and contributes to neurite outgrowth in vitro57,58,59. Since tuft cells are found in contact with nerve fibers, advillin may have a unique function in regulating the enteric nervous system. That would also suggest that advillin plays a role in transferring sensory signals to the enteric nervous system.
Methods
Reagents
Murine recombinant epidermal growth factor, Advanced DMEM/F12, penicillin-streptomycin, HEPES buffer, Dulbecco’s Phosphate Buffered Saline and L-glutamine were bought from Gibco. Matrigel was from Corning, Inc. Noggin-conditioned medium and R-spondin conditioned medium (Noggin-producing cells and R-spondin-producing cells) were kindly provided by the Texas Medical Center Digestive Diseases Center (DDC) core. Acrylamide, SDS, Ammonium Persulfate, Tetramethylethylenediamine, N2 supplement, B27 supplement, Tissue-Tek® Optimal Cutting Temperature™ (O.C.T.) compound, and 24 well cell culture treated plate were from Thermo-Fisher Scientific. Glass bottom dishes were from MatTek.
Antibodies
The following antibodies were used in the study: PTGS1 (NB100-867) from Novus Biologicals; N-advillin raised against the amino-terminus of advillin (a.a. 440-526), used in Human Protein Atlas is available from Sigma (HPA058864) and from Novus Biologicals (NBP2-34118); C-advillin antibody raised against the carboxyl-terminus of advillin (a.a. 750-813), from Abcam (ab72210); DCLK1 (H00009201-M03) from Abnova; Tubulin (ab131205) and N-Villin (ab201989) from Abcam; N20-Villin (sc33347), C-Villin (sc7672) from Santa Cruz Biotechnology, Inc.; FL-Villin (610359) from BD Transduction Laboratories; Hematoxylin and Eosin staining kit from VWR (470302-740); Alexa Fluor 568 phalloidin, Alexa Fluor 488, Alexa Fluor 647 and Alexa Fluor 555 from Thermo-Fisher Scientific. N-advillin from Novus Biologicals (NBP2-34118) was used in all immunohistochemical studies. Anti-fade mounting medium with DAPI was from Vector Laboratory. Actin Polymerization Biochem Kit was purchased from Cytoskeleton Inc. Recombinant murine IL-4 and IL-13 were purchased from Peprotech.
Mice
All experimental protocols were approved by Institutional Animal Care and Use Committee (IACUC) of the University of Houston. Villin-1 knock-out (VKO) mice and their wild-type (WT) littermates have been described by us previously60. C57BL/6 J mice were purchased from the Jackson laboratory.
Enteroid 3D culture and cytokines treatment
All methods were performed in accordance with the relevant guidelines and regulations. Crypt isolation and enteroid 3D cultures were derived from distal ileum of mice as previously described61. Organoid lines were passaged up to 10 times before experiments to ensure pure epithelial cultures. Enteroid 3D cultures were treated with a mixture of recombinant murine IL-4 (400 ng ml−1) and recombinant murine IL-13 (400 ng ml−1)43. Organoids were fixed and permeabilized for 1 hour using 3.7% paraformaldehyde and 0.2% Triton X-100. All images were acquired on Olympus Fluoview FV1200 Laser Scanning confocal microscope with a 60X NA 1.35 objective. The FV1200 Confocal microscope has three photodetectors, to image four fluorophores a Virtual Channel was created and imaging was performed under sequential mode.
Immunohistochemistry using paraffin and Optimal Cutting Temperature (O.C.T.) embedded tissue
Tissue from distal ileum of mice were fixed and paraffin-embedded as described previously61. Hematoxylin and eosin (H & E) staining was performed as described previously62. To preserve the F-actin, some tissues were embedded in O.C.T. compound and stored at -80 °C. O.C.T. frozen blocks were sectioned 5 μm thick using Leica CM3050 Cryostat. The immunofluorescence studies were performed as described previously61. All fixed tissues were embedded and sectioned by the Texas Medical Center Digestive Diseases Center (DDC) core.
Measurement of actin dynamics regulated by villin and advillin proteins
Recombinant full-length human villin and advillin proteins were cloned, expressed and purified as described previously36,63. Actin capping, nucleating and severing assays were performed as described by us64. Actin binding and actin bundling assays were performed as described by us previously65. Fluorescence measurements were performed using the FluoroMax-4 spectrofluorometer (Horiba Scientific).
References
Von Moltke, J. Intestinal tuft cells. Physiology of the Gastrointestinal Tract Chapter 31, 721–733 (2018).
Isomaki, A. M. A new cell type (tuft cell) in the gastrointestinal mucosa of the rat. A transmission and scanning electron microscopic study. Acta Pathol Microbiol Scand A Suppl 240, 241–235 (1973).
Hofer, D. & Drenckhahn, D. Cytoskeletal markers allowing discrimination between brush cells and other epithelial cells of the gut including enteroendocrine cells. Histochem Cell Biol 105, 405–412 (1996).
Allan, E. M. The ultrastructure of the brush cell in bovine lung. Res Vet Sci 25, 314–317 (1978).
Hofer, D. & Drenckhahn, D. Cytoskeletal markers allowing discrimination between brush cells and other epithelial cells of the gut including enteroendocrine cells. Histochem Cell Biol 105, 405–412 (1996).
Khurana, S. Structure and function of villin. Chapter 5, 89-117. In: Advances in Molecular and Cell Biology, 37. Aspects of the Cytoskeleton. Series Editor: E. Edward Bittar. Volume Editor: Seema Khurana., Vol. 37 (Elsevier, 2006).
Banerjee, A., McKinley, E. T., von Moltke, J., Coffey, R. J. & Lau, K. S. Interpreting heterogeneity in intestinal tuft cell structure and function. J Clin Invest 128, 1711–1719, https://doi.org/10.1172/JCI120330 (2018).
Middelhoff, M. et al. Dclk1-expressing tuft cells: critical modulators of the intestinal niche? Am J Physiol Gastrointest Liver Physiol 313, G285–G299, https://doi.org/10.1152/ajpgi.00073.2017 (2017).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339, https://doi.org/10.1038/nature24489 (2017).
Luo, X. C. et al. Infection by the parasitic helminth Trichinella spiralis activates a Tas2r-mediated signaling pathway in intestinal tuft cells. Proc Natl Acad Sci U S A 116, 5564–5569, https://doi.org/10.1073/pnas.1812901116 (2019).
Bjerknes, M. et al. Origin of the brush cell lineage in the mouse intestinal epithelium. Dev Biol 362, 194–218, https://doi.org/10.1016/j.ydbio.2011.12.009 (2012).
Westphalen, C. B. et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest 124, 1283–1295, https://doi.org/10.1172/JCI73434 (2014).
Gagliardi, G. & Moroz, K. & Bellows, C. F. Immunolocalization of DCAMKL-1, a putative intestinal stem cell marker, in normal colonic tissue. Pathol Res Pract 208, 475–479, https://doi.org/10.1016/j.prp.2012.05.015 (2012).
Gilroy, D. W. et al. Inducible cyclooxygenase may have anti-inflammatory properties. Nat Med 5, 698–701, https://doi.org/10.1038/9550 (1999).
Kokrashvili, Z., Mosinger, B. & Margolskee, R. F. Taste signaling elements expressed in gut enteroendocrine cells regulate nutrient-responsive secretion of gut hormones. Am J Clin Nutr 90, 822S–825S, https://doi.org/10.3945/ajcn.2009.27462T (2009).
Kokrashvili, Z. et al. Release of endogenous opioids from duodenal enteroendocrine cells requires Trpm5. Gastroenterology 137(598-606), 606 e591–592, https://doi.org/10.1053/j.gastro.2009.02.070 (2009).
Mitrovic, S. et al. TRPM5-mediated calcium uptake regulates mucin secretion from human colon goblet cells. eLife 2, e00658, https://doi.org/10.7554/eLife.00658 (2013).
Marks, P. W., Arai, M., Bandura, J. L. & Kwiatkowski, D. J. Advillin (p92): a new member of the gelsolin/villin family of actin regulatory proteins. J Cell Sci 111, 2129–2136 (1998).
Toyoshima, K., Seta, Y., Takeda, S. & Harada, H. Identification of Merkel cells by an antibody to villin. J Histochem Cytochem 46, 1329–1334 (1998).
Hunter, D. V. et al. Advillin Is Expressed in All Adult Neural Crest-Derived Neurons. eNeuro 5, https://doi.org/10.1523/ENEURO.0077-18.2018 (2018).
Yoshie, S., Kumakura, M. & Toyoshima, K. Villin is a possible marker of receptor cells in frog taste organs. Histochem Cell Biol 119, 447–450 (2003).
Hofer, D. & Drenckhahn, D. Localisation of actin, villin, fimbrin, ezrin and ankyrin in rat taste receptor cells. Histochem Cell Biol 112, 79–86 (1999).
Sukumaran, S. K. et al. Taste cell-expressed alpha-glucosidase enzymes contribute to gustatory responses to disaccharides. Proc Natl Acad Sci U S A 113, 6035–6040, https://doi.org/10.1073/pnas.1520843113 (2016).
Maunoury, R. et al. Villin expression in the visceral endoderm and in the gut anlage during early mouse embryogenesis. Embo J 7, 3321–3329 (1988).
Harold, A. et al. Conversion of Sox2-dependent Merkel cell carcinoma to a differentiated neuron-like phenotype by T antigen inhibition. Proc Natl Acad Sci U S A 116, 20104–20114, https://doi.org/10.1073/pnas.1907154116 (2019).
Ye, W. et al. The K+ channel KIR2.1 functions in tandem with proton influx to mediate sour taste transduction. Proc Natl Acad Sci U S A 113, E229–238, https://doi.org/10.1073/pnas.1514282112 (2016).
Bornstein, C. et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559, 622–626, https://doi.org/10.1038/s41586-018-0346-1 (2018).
Sukumaran, S. K. et al. Whole transcriptome profiling of taste bud cells. Sci Rep 7, 7595, https://doi.org/10.1038/s41598-017-07746-z (2017).
Bezencon, C. et al. Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. J Comp Neurol 509, 514–525, https://doi.org/10.1002/cne.21768 (2008).
Goto, N. et al. Lineage tracing and targeting of IL17RB(+) tuft cell-like human colorectal cancer stem cells. Proc Natl Acad Sci U S A 116, 12996–13005, https://doi.org/10.1073/pnas.1900251116 (2019).
Miller, C. N. et al. Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature 559, 627–631, https://doi.org/10.1038/s41586-018-0345-2 (2018).
Nadjsombati, M. S. et al. Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 49, 33–41 e37, https://doi.org/10.1016/j.immuni.2018.06.016 (2018).
McKinley, E. T. et al. Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity. JCI Insight 2, https://doi.org/10.1172/jci.insight.93487 (2017).
Roy-Carson, S. et al. Defining the transcriptomic landscape of the developing enteric nervous system and its cellular environment. BMC Genomics 18, 290, https://doi.org/10.1186/s12864-017-3653-2 (2017).
Hockley, J. R. F. et al. Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut 68, 633–644, https://doi.org/10.1136/gutjnl-2017-315631 (2019).
Rao, J. et al. Advillin acts upstream of phospholipase C 1 in steroid-resistant nephrotic syndrome. J Clin Invest, https://doi.org/10.1172/JCI94138 (2017).
George, S. P., Chen, H., Conrad, J. C. & Khurana, S. Regulation of directional cell migration by membrane-induced actin bundling. J Cell Sci 126, 312–326, https://doi.org/10.1242/jcs.116244 (2013).
Hofer, D. & Drenckhahn, D. Identification of brush cells in the alimentary and respiratory system by antibodies to villin and fimbrin. Histochemistry 98, 237–242 (1992).
Pinson, K. I., Dunbar, L., Samuelson, L. & Gumucio, D. L. Targeted disruption of the mouse villin gene does not impair the morphogenesis of microvilli. Dev Dyn 211, 109–121 (1998).
Gerbe, F., Brulin, B., Makrini, L., Legraverend, C. & Jay, P. DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology 137, 2179–2180; author reply 2180–2171, https://doi.org/10.1053/j.gastro.2009.06.072 (2009).
Hofer, D. & Drenckhahn, D. Identification of the taste cell G-protein, alpha-gustducin, in brush cells of the rat pancreatic duct system. Histochem Cell Biol 110, 303–309 (1998).
Howitt, M. R. et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333, https://doi.org/10.1126/science.aaf1648 (2016).
Gerbe, F. et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230, https://doi.org/10.1038/nature16527 (2016).
Heier, J. A., Dickinson, D. J. & Kwiatkowski, A. V. Measuring Protein Binding to F-actin by Co-sedimentation. J Vis Exp, https://doi.org/10.3791/55613 (2017).
Sato, A. Tuft cells. Anat Sci Int 82, 187–199, https://doi.org/10.1111/j.1447-073X.2007.00188.x (2007).
Gerbe, F. et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol 192, 767–780, https://doi.org/10.1083/jcb.201010127 (2011).
Herring, C. A. et al. Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut. Cell Syst 6, 37–51 e39, https://doi.org/10.1016/j.cels.2017.10.012 (2018).
Aladegbami, B. et al. Epithelial cell specific Raptor is required for initiation of type 2 mucosal immunity in small intestine. Sci Rep 7, 5580, https://doi.org/10.1038/s41598-017-06070-w (2017).
Durand, A. et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc Natl Acad Sci U S A 109, 8965–8970, https://doi.org/10.1073/pnas.1201652109 (2012).
Mori-Akiyama, Y. et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology 133, 539–546, https://doi.org/10.1053/j.gastro.2007.05.020 (2007).
May, R. et al. Brief report: Dclk1 deletion in tuft cells results in impaired epithelial repair after radiation injury. Stem Cells 32, 822–827, https://doi.org/10.1002/stem.1566 (2014).
Qu, D. et al. Ablation of Doublecortin-Like Kinase 1 in the Colonic Epithelium Exacerbates Dextran Sulfate Sodium-Induced Colitis. PLoS One 10, e0134212, https://doi.org/10.1371/journal.pone.0134212 (2015).
Chandrakesan, P. et al. Intestinal tuft cells regulate the ATM mediated DNA Damage response via Dclk1 dependent mechanism for crypt restitution following radiation injury. Sci Rep 6, 37667, https://doi.org/10.1038/srep37667 (2016).
Nakanishi, Y. et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet 45, 98–103, https://doi.org/10.1038/ng.2481 (2013).
Heanue, T. A. & Pachnis, V. Expression profiling the developing mammalian enteric nervous system identifies marker and candidate Hirschsprung disease genes. Proc Natl Acad Sci U S A 103, 6919–6924, https://doi.org/10.1073/pnas.0602152103 (2006).
Rao, M. et al. Enteric glia express proteolipid protein 1 and are a transcriptionally unique population of glia in the mammalian nervous system. Glia 63, 2040–2057, https://doi.org/10.1002/glia.22876 (2015).
Ravenall, S. J., Gavazzi, I., Wood, J. N. & Akopian, A. N. A peripheral nervous system actin-binding protein regulates neurite outgrowth. Eur J Neurosci 15, 281–290 (2002).
Hasegawa, H., Abbott, S., Han, B. X., Qi, Y. & Wang, F. Analyzing somatosensory axon projections with the sensory neuron-specific Advillin gene. J Neurosci 27, 14404–14414 (2007).
Shibata, M. et al. Type F scavenger receptor SREC-I interacts with advillin, a member of the gelsolin/villin family, and induces neurite-like outgrowth. J Biol Chem 279, 40084–40090, https://doi.org/10.1074/jbc.M403844200 (2004).
Wang, Y. et al. A novel role for villin in intestinal epithelial cell survival and homeostasis. J Biol Chem 283, 9454–9464, https://doi.org/10.1074/jbc.M707962200 (2008).
Roy, S. et al. Villin-1 and Gelsolin Regulate Changes in Actin Dynamics That Affect Cell Survival Signaling Pathways and Intestinal Inflammation. Gastroenterology 154, 1405–1420 e1402, https://doi.org/10.1053/j.gastro.2017.12.016 (2018).
Wang, Y. et al. Both the anti- and pro-apoptotic functions of villin regulate cell turnover and intestinal homeostasis. Sci Rep 6, 35491, https://doi.org/10.1038/srep35491 (2016).
Zhai, L. et al. Regulation of actin dynamics by tyrosine phosphorylation: identification of tyrosine phosphorylation sites within the actin-severing domain of villin. Biochemistry 41, 11750–11760 (2002).
Kumar, N., Zhao, P., Tomar, A., Galea, C. A. & Khurana, S. Association of villin with phosphatidylinositol 4,5-bisphosphate regulates the actin cytoskeleton. J Biol Chem 279, 3096–3110 (2004).
George, S. P., Wang, Y., Mathew, S., Srinivasan, K. & Khurana, S. Dimerization and actin-bundling properties of villin and its role in the assembly of epithelial cell brush borders. J Biol Chem 282, 26528–26541, https://doi.org/10.1074/jbc.M703617200 (2007).
Acknowledgements
This study was supported by National Institutes of Diabetes and Digestive Kidney Diseases (Grant DK-117476 to S.K.); and the Public Health Service (Grant DK-56338). We would like to thank Ms. Saathvika Ravi, Mr. Jonathan Tao and Ms. Farnaz Naeemikia for their assistance with immunostaining of tissue sections.
Author information
Authors and Affiliations
Contributions
All in vitro studies to characterize the villin and advillin antibodies, and all animal studies to characterize tuft cells were performed by S.P.G and A.E. All enteroid studies were performed by A.E. and R.B. All experiments were conceived and designed by S.K. Data was analyzed by S.P.G., A.E. and S.K. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Esmaeilniakooshkghazi, A., George, S.P., Biswas, R. et al. Mouse intestinal tuft cells express advillin but not villin. Sci Rep 10, 8877 (2020). https://doi.org/10.1038/s41598-020-65469-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-65469-0
- Springer Nature Limited