Abstract
GPRC6A is a widely expressed G-protein coupled receptor that regulates energy metabolism. Global deletion of Gprc6a in mice is reported to result in a metabolic syndrome-like phenotype and conditional deletion of Gprc6a in pancreatic β-cell and skeletal muscle respectively impair insulin secretion and glucose uptake. In the current study, we explore the hepatic functions of GPRC6A by conditionally deleting Gprc6a in hepatocytes by cross breeding Alb-Cre and Gprc6aflox/flox mice to obtain Gprc6aLiver-cko mice. Gprc6aLiver-cko mice on a normal diet showed excessive hepatic fat accumulation and glycogen depletion. These mice also exhibit impaired glucose and pyruvate tolerance, but normal insulin sensitivity. Decreased circulating FGF-21 levels and FGF-21 message expression in the liver were found in Gprc6aLiver-cko mice. Hepatic transcriptome analysis identified alterations in multiple pathways regulating glucose, fat and glycogen metabolism in Gprc6aLiver-cko mice. Taken together, our studies suggest that GPRC6A directly regulates hepatic metabolism as well as regulates the production and release of FGF-21 to control systemic energy homeostasis. GPRC6A’s unique regulation of β-cell, skeletal muscle and hepatic function may represent a new therapeutic target for treating disordered energy metabolism metabolic syndrome and type 2 diabetes.
Similar content being viewed by others
Introduction
GPRC6A is a family C G-protein coupled receptor (GPCR) that consists of an N-terminal venus fly trap motif (VFTM) homologous to the bacterial periplasmic L-amino acid sensor fused with a classical 7 transmembrane domain (7-TM). This structure allows GPRC6A to sense multiple ligands, including the bone-derived peptide, osteocalcin (Ocn), amino acids, L-Arginine, L-Ornithine, and L-Lysine1,2,3, testosterone (T)1,4, and natural products in green tea5. The bone-derived ligand Ocn regulates energy metabolism by regulating insulin secretion from pancreatic β-cells and testosterone production in Leydig cells6,7,8,9. Gprc6a−/− mice exhibit complex metabolic derangements that resemble metabolic syndrome (MetS), including glucose intolerance, insulin resistance, and fatty liver1,10,11,12,13,14. The similar phenotypes of Gprc6a−/− and Ocn−/− mice and the additive phenotypic effects in compound Gprc6a+/−/Ocn+/− mice support the presence of this endocrine network in mice15.
GPRC6A is expressed in key metabolic tissues, including β-cells, liver hepatocytes, skeletal muscle, fat, and Leydig cells. GPRC6A’s organ-specific functions have been partially examined in mice. Conditional deletion of Gprc6a in Leydig cells attenuates Ocn induced testosterone production by the testes6, in pancreatic β-cells selective deletion of Gprc6a impairs cell proliferation and insulin secretion11,13, and selective Gprc6a loss-of-function skeletal muscle impairs muscle glucose and fatty acid utilization and IL-6 production9,14,16. A potential function of GPRC6A in the liver is suggested by the presence of hepatosteatosis in Gprc6a−/− mice10 and pharmacological actions of Ocn administration to prevent high fat diet induced fatty liver disease in mice10,17,18,19. The direct functions of GPRC6A in the liver, an organ essential for regulating glucose and fat metabolism has not been determined.
In the current study, we advance our understanding of the metabolic functions of GPRC6A by conditionally deleting Gprc6a in hepatocytes in male mice. The phenotype in the hepatocyte loss-of-function mouse model establishes a key role of GPRC6A in regulating hepatic glucose and fat metabolism in mice.
Results
In vitro functions of GPRC6A in muscle and hepatic cells in vitro
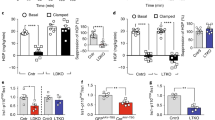
First, we compared GPRC6A signaling in mouse muscle and hepatic cells in vitro. Because conditional knockout of the mouse Gprc6a in skeletal muscle has been shown to regulate muscle glucose and fat metabolism14,16, we tested the expression, effects of GPRC6A ligands, and knock-down of Gprc6a in C2C12 cells. To establish the function of GPRC6A in hepatocytes we performed similar studies the Hepa1c1c7 cell line. Both C2C12 and Hepa1c1c7 express endogenous Gprc6a (Fig. 1a). siRNA treatment successfully reduced Gprc6a message and protein expression in both cell lines (Fig. 2b,c). T and Ocn, agonists for GPRC6A, also stimulated ERK activity in both cell lines (Fig. 1d,e). siRNA mediated knock down of Gprc6a resulted in an attenuation of T and Ocn-induced ERK activation in both muscle and hepatic cell lines, respectively (Fig. 1d,e). T and Ocn also stimulated glucose production in Hepa1c1c7 cells and this response was blocked by siRNA treatment of these cells (Fig. 1f).
Expression and function of GPRC6A in myocytes and hepatocytes in vitro. (a,b) Assessment of endogenous GPRC Gprc6a 6A message expression by RT-PCR in C2C12 and Hepa1c1c7 cells before (a) and after siRNA knockdown of Gprc6a (b). C2C12.ncsiRNA and Hepa1c1c7.ncsiRNA are C2C12 and Hepa1c1c7 cells transfected with negative control siRNA. (c) Western blot of GPRC6A protein in C2C12 and Hepa1c1c7 cells transfected with negative control siRNA or siRNA to GPRC6A. (d and e) ERK activation in response to testosterone (T) and osteocalcin (Ocn) stimulation. siRNA knockdown of Gprc6a inhibited these responses in both C2C12 (d) and Hepa1c1c7 cells (e). Quantification by ERK Elisa assay indicated in Methods section. Values represent the mean ± SEM. *P < 0.05 significant difference between control group and treated group (n = 3). (f) Glucose production in mouse Hepa1C1C7 hepatocytes with negative control siRNA or siRNA to knockdown of Gprc6a. Values represent the mean ± SEM. *significant difference between Hepa1C1C7 control and siRNA knockdown Gprc6a Hepa1C1C7 cells (P < 0.05; n = 3).
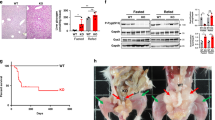
Generation and characterization of Gprc6aLiver-cko mice. (a) Efficiency of Gprc6a deletion by Alb-Cre in liver was tested by real-time PCR using specific Gprc6a primers as described in Materials and Methods. Expression was assessed by real-time PCR using total RNA derived from control wild type mice with unaltered phenotype and Gprc6aLiver-cko mouse tissues as indicated. Gprc6a expression is relative to the level of the cyclophilin A control gene. Values represent the mean ± SEM. *significant difference between control group and Gprc6aLiver-cko mice (P < 0.05; n = 5). (b) Western blot of GPRC6A protein in liver and pancreas from control and Gprc6aLiver-cko mice. (c) Comparison of the body weight in control and Gprc6aLiver-cko mice at age of 10 and 20 weeks. Values represent the mean ± SEM. *significant difference between control group and Gprc6aLiver-cko mice (P < 0.05; n = 6).
Conditional deletion of Gprc6a in hepatocytes in Gprc6a Liver-cko mice
Next, to test the in vivo hepatic function of GPRC6A, we mated Alb-Cre/+;Gprc6a+/- mice with homozygous Gprc6aflox/flox mice to generate Alb- Cre/+;Gprc6aflox/- (Gprc6aLiver-cko), harboring the liver tissue specific-Cre transgene. Deletion of Gprc6a occurred in the liver in Gprc6aLiver-cko mice, but not in other tissues tested, including testes, pancreas and muscle (Fig. 2a). Hepatic Gprc6a message levels were decreased by>70% in Gprc6aLiver-cko mice by quantitative RT-PCR (Fig. 2a). Prior studies that targeted Gprc6a in muscle found that a 50% reduction of Gprc6a was sufficient to achieve a measurable phenotype14. Western blot analysis shows Gprc6a protein expression was selectively lost in the liver of Gprc6aLiver-cko mice (Fig. 2b).
Gprc6aLiver-cko mice were born with the expected Mendelian frequency and had a normal gross appearance and survival. 10 and 20-week-old male Gprc6aLiver-cko mice exhibited a significant (p < 0.05) increase in body weight compared to control mice (Fig. 2b), but the body weight of female Gprc6aLiver-cko mice did not differ from the wild type control group (Fig. 2c).
The wild-type mice (Gprc6a+/+) did not differ from the various intermediate gene construct bearing mice, (i.e.,Gprc6aflox/+, Gprc6aflox/-, Alb-Cre/+;Gprc6a+/+and Alb-Cre/+;Gprc6aflox/+) with regards to fasting glucose and body weight and were combined to create the control group (Figure S1).
Serum biochemical alterations in Gprc6a Liver-cko mice
Gprc6aLiver-cko male mice had increased fasting blood glucose (Fig. 3a), and decreased serum insulin levels (Fig. 3b) compared to the control mice. We found that the serum level of the liver-derived hepatokine, fibroblast growth factor 21 (FGF-21), was significantly decreased in Gprc6aLiver-cko mice (80.7 ± 19.9 pg/ml) compared to control mice (225.9 ± 15.4 pg/ml) (Fig. 3c). To investigate effects of hepatic GPRC6A deficiency on lipid metabolism, we measured cholesterol, free fatty acid and triglyceride levels in serum from Gprc6aLiver-cko mice. The serum cholesterol and free fatty acid levels were increased in Gprc6aLiver-cko mice (Fig. 3d,e), but serum triglyceride levels were not changed (Fig. 3f).
Serum parameters in Gprc6aLiver-cko mice. Comparison of blood glucose (a), serum insulin (b) FGF-21 (c), cholesterol (d), free fatty acid (e) and triglyceride levels (f) in control group and Gprc6aLiver-cko male mice at age of 10 weeks. Data represent the mean ± SEM from more than 6 male mice in each group. *difference from control group and Gprc6aLiver-cko mice at P < 0.05.
Glucose and pyruvate intolerance, but not insulin resistance in Gprc6a Liver-cko mice
To determine the mechanisms for changes in serum glucose, we performed tolerance tests for glucose (GTT) (Fig. 4a) and insulin (ITT) (Fig. 4b) in Gprc6aLiver-cko and control mice. After injection of glucose (2 g/kg), Gprc6aLiver-cko male mice had a significantly higher (p < 0.05) serum glucose levels than controls, consistent with impaired glucose tolerance (Fig. 4a). In contrast, Gprc6aLiver-cko and control mice exhibited a similar sensitivity to insulin (0.75 U/kg) administration (Fig. 4b). In spite of higher fasting glucose, Gprc6aLiver-cko lowered serum glucose in response to insulin to the same levels as in control mice. This contrast to insulin resistance by ITT in Gprc6a−/− mice10, which indicates that loss of Gprc6a in multiple sites is required to impart insulin resistance.
Gprc6a deficiency results in glucose and pyruvate intolerance. (a–c) Shown is blood glucose (mg/dL) during glucose tolerance test (GTT) (a), insulin tolerance test (ITT) (b) and pyruvate tolerance test (PTT) (c) in 10-week-old control and Gprc6aLiver-cko male mice. Data represent the mean ± SEM from 6 male mice in each group. *difference from control group and Gprc6aLiver-cko mice at P < 0.05. (d) Glucose production in hepatocytes isolated from liver of the controls and Gprc6aLiver-cko mice. Glucose production was measured 12 hours after 100 nM testosterone (T) or 80 ng/ml osteocalcin (Ocn) treatment. Values represent the mean ± SEM. *significant difference between the liver cell of wild type and Gprc6aLiver-cko mice (P < 0.05; n = 6).
To further investigate glucose metabolism in the liver, we performed pyruvate tolerance test (PTT) to investigate GPRC6A’s role in regulating gluconeogenesis (Fig. 4c). PTT was performed a by IP injection of pyruvate sodium (2 g/kg body weight) to Gprc6aLiver-cko and control mice after fasting for 5 hours. Gprc6aLiver-cko mice had a significantly lower serum glucose levels during the PTT, consistent with impaired gluconeogenesis (Fig. 4c).
To confirm that GPRC6A regulates hepatic gluconeogenesis in vitro, we studied glucose production in the isolated hepatocytes from control and Gprc6aLiver-cko mice. We found that the GPRC6A ligands Ocn and T increased glucose production in primary hepatocytes from wild type mice (Fig. 4d). This response was significantly attenuated in isolated hepatocytes from Gprc6aLiver-cko mice. The results, which are like the above studies in Hepa1c1c7 cells expressing endogenous Gprc6a before and after siRNA knockdown (Fig. 1), show GPRC6A-dependent increases in glucose production by Ocn and T. Thus, the fasting hyperglycemia in Gprc6aLiver-cko is not due to enhanced gluconeogenesis or insulin resistance.
Gprc6a Liver-cko mice exhibit decreased glycogen content
Consistent with the abnormal pyruvate tolerance test, we found that the liver glycogen storage was significantly decreased in Gprc6aLiver-cko compared to control group mice as measured by PAS staining of glycogen (Fig. 5a). We also observed reduced liver glycogen content (Fig. 5b) in the liver of Gprc6aLiver-cko compared to control mice. We found that liver glucose-6-phosphate was significantly reduced in Gprc6aLiver-cko mice compared to wild type controls (Fig. 5c), consistent with the impaired uptake of glucose and conversion to glycogen.
Gprc6a deficiency in liver is associated with fatty liver and steatohepatitis. (a) PAS staining for glycogen in liver from control group and Gprc6aLiver-cko male mice. (b) Comparison of liver glycogen content in the liver from control group and Gprc6aLiver-cko male mice. Values represent the mean ± SEM. *significant difference between control group and Gprc6aLiver-cko mice (P < 0.05; n = 5). (c) Comparison of the content of glucose-6-phosphate (Glucose-6-P) in liver from control group and Gprc6aLiver-cko male mice at age of 10 weeks. Values represent the mean ± SEM. *significant difference between control group and Gprc6aLiver-cko mice (P < 0.05; n = 5). (d) Representative histology of Oil Red O staining. The result was shown increased hepatic steatosis in Gprc6aLiver-cko mice. (e,f) Comparison of the contents of cholesterol (D) and triglyceride (E) in liver from control group and Gprc6aLiver-cko male mice at age of 10 week-old. Values represent the mean ± SEM. *significant difference between control group and Gprc6aLiver-cko male mice (P < 0.05; n = 5). (g,h) Comparison of FGF-21 liver content (G) and gene expression (H) in control group and Gprc6aLiver-cko male mice at age of 10 weeks. Values represent the mean ± SEM. *significant difference between control group and Gprc6aLiver-cko mice (P < 0.05; n = 5).
Gprc6a Liver-cko mice develop hepatosteatosis
Consistent with activation of GPRC6A by Ocn to prevent high fat induced fatty liver, the histology in Gprc6aLiver-cko also showed increase fat accumulation by Oil Red O staining (Fig. 5d). Lipid positive droplets were present in hepatocytes of Gprc6aLiver-cko mice, but not in control group mice. The liver content of cholesterol and triglyceride were also increased in GPRC6A loss-of-function Gprc6aLiver-cko mice compared to the control mice (Fig. 5e,f).
Decreased FGF21 in the liver of Gprc6aLiver-cko mice
FGF-21 is primarily produced by the liver. Consistent with the reduced FGF-21 serum levels (Fig. 3c), the liver content of FGF-21 protein (Fig. 5g) was significantly decreased in Gprc6aLiver-cko mice (12.9 ± 2.1 ng/g) compared to control mice (22.4 ± 3.7 ng/g). We also found that Fgf-21 message expression level significantly decreased in the liver from Gprc6aLiver-cko mice compared to control group mice (Fig. 5h). These data indicate for the first time that GPRC6A regulates FGF-21 expression and production by the liver.
Liver transcriptome in Gprc6a Liver-cko mice
To investigate molecular basis for the observed hepatic phenotypes, we assessed the Gprc6a hepatic transcriptome by performing RNA-seq analysis on whole liver RNA obtained from Gprc6aLiver-cko and control mice. Volcano plot and heat map visualization of the hepatic transcriptome demonstrated distinct differences between wild-type and Gprc6aLiver-cko mouse (Fig. 6a,b). A total of 1208 (677 upregulated and 531 downregulated) genes were identified to be differentially expressed in livers from Gprc6aLiver-cko mice and controls (adjusted p < 0.05) (Tables S2 and 3). Biological process (GO) enrichment analysis of the differentially expressed genes (DEGs) revealed that Gprc6a specific knockout in liver resulted in differences in lipid and glucose metabolism (Fig. 6c). Genes induced in Gprc6aLiver-cko mice included genes involved in lipid homeostasis (10 genes), localization (36 genes), modification (24 genes), transport (29 genes), and storage (10 genes).
Hepatic gene expression in the liver of Gprc6aLiver-cko mice. (a,b) Volcano plot (a); Heat map (b) of regulated genes between WT and Gprc6aLiver-cko mice. Blue and red colors indicate high and low gene expression, respectively. Volcano plot and heat map visualization of the hepatic transcriptome demonstrated distinct differences between WT and Gprc6aLiver-cko mice. (c–e) Gene ontology (c), the Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) (d) and Mammalian phenotype (e) in the liver of Gprc6aLiver-cko mice. The top rank ordered processes, maps and networks are based on statistical significance.
In addition, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis also revealed genes significantly involved in insulin resistance (18 genes), insulin signaling pathway (21 genes), TNF signaling pathway (17 genes), AMPK signaling pathway (19 genes), PPAR signaling pathway (14 genes), and retinol metabolism (17 genes). More importantly, phenotype enrichment analysis of the DEGs revealed that a transcriptional profile pointing to abnormal glucose homeostasis (100 genes) and tolerance (54 genes) (Fig. 6d). Moreover, phenotype enrichment revealed 48 genes are contributed to abnormal triglyceride level such as Lipc, Abhd5, Lact, Dgat1, Gpihbp1, and Angptl4 (Fig. 6e).
Changes in expression of selected transcripts were confirmed by RT-PCR (Fig. 7). Specific genes altered in Gprc6aLiver-cko mice included genes involved in glycogen metabolism, such as Gys1, Slc37a4, Ppp1r3b, Ppp1r3c, and Ppp1r3g and genes involved in gluconeogenesis, such Pck2, Atf3, Gnmt, Mst1, Per2, and Ppargc1a.
Confirmation of the selected gene expression by real-time PCR. The gene expression was assessed by real-time PCR using total RNA derived from control group mice with unaltered phenotype and Gprc6aLiver-cko mice. Gprc6a expression is relative to the level of the cyclophilin A control gene. The real-time PCR using specific primers as described in Table S1. Values represent the mean ± SEM. *significant difference between control group and Gprc6aLiver-cko mice (P < 0.05; n = 5). G6p, glucose 6-phosphatase; Srebp1c, sterol Regulatory Element Binding Transcription Factor 1c; Phka1, phosphorylase kinase alpha 1; Gys1, glycogen synthase 1; Pygl, glycogen Phosphorylase, Liver Form; Ppara, Peroxisome proliferator-activated receptor alpha; Lpl, lipoprotein lipase.
Gene expression analysis revealed that glucose transporter-2 (solute carrier family 2, member A2, Slc2a2 or Glut2), the predominant hepatic bi-directional glucose transporter, was increased in the liver of Gprc6aLiver-cko mice. Glucokinase (Gck), the first rate-limiting step in glucose metabolism, however, was decreased expression in the liver of Gprc6aLiver-cko consistent with the observed reduced glucose 6-phosphate levels and mild hyperglycemia associated with inactivation of GCK20; but genes that regulate glycolysis, including Gpi, Pdha1, Pdhb, Pklr, Slc2a3, Aldoa, Aldob, Pgam1, Pfkl, Pgm1, Pdhx and Creb1, expression were increased in the liver of Gprc6aLiver-cko.
The expression of gluconeogenesis pathway genes, including G6p, Pgk1, Pcx, Fbp1, Eno1, Eno3 and Gck, were decreased in the liver of Gprc6aLiver-cko mice, consistent with the impaired pyruvate tolerance test.
With regards to genes regulating glycogen metabolism, we found that the expression of glycogenesis pathway genes, including Hk1, Pgm2, Gys1, and glycogen synthase kinase, Gsk3a were decreased, consistent with the decreased glycogen stores in the liver of Gprc6aLiver-cko mice.
Surprisingly, the expression of glycogenolysis pathway genes, including Pygl, and branching enzymes, glycogen phosphorylase kinases (Gaa), phosphorylase kinase alpha 1 (Phka1) and phosphorylase kinase alpha 2 (Phka2), were also decreased in the liver of Gprc6aLiver-cko mice.
Gene transcripts related to fatty acid synthesis, including ATP-citrate lyase (Acl), acetylCoA carboxylase16, fatty acid synthase enzyme (Fas), fatty acid desaturase 2 (Fads2), an endoplasmic gene involved in fatty acid metabolism, and long-chain acyl-CoA synthetase (Acsl), elongation of long-chain fatty acids family member 6 (Elovl6), 3-hydroxyacyl-CoA dehydratase 1 (Hacd1), and mitochondrial trans-2-enoyl-CoA reductase (Mecr) were increased in the liver from Gprc6aLiver-cko mice. In contrast, the expression of fatty acid degradation or beta oxidation related genes, acyl-Coenzyme A dehydrogenase, medium chain (Acadm), enoyl Coenzyme A hydratase, short chain, 1, mitochondrial (Echs1), hydroxyacyl-Coenzyme A dehydrogenase (Hadh), and hydroxyacyl-Coenzyme A dehydrogenase, alpha subunit (Hadha) were decreased. In addition, the expression of ketogenesis pathway genes, including Hadha, Hadhb, 3-hydroxy-3-methylglutaryl-Coenzyme A lyase (Hmgcl), 3-hydroxybutyrate dehydrogenase, type 1 (Bdh1) and Bdh2, were decreased in the liver from Gprc6aLiver-cko mice. Srebp-1c and Chrebp transcripts were also increased in Gprc6aLiver-cko, as well as in the global Gprc6a−/− mice, consistent with increased de novo lipogenesis.
Discussion
This study establishes an important role of GPRC6A in the liver to regulate glucose and fat metabolism. Loss-of-Gprc6a function in hepatocytes resulted in several metabolic abnormalities in Gprc6aLiver-cko mice, including hyperglycemia, reduced serum insulin levels, increased circulating concentrations of free fatty acids and cholesterol, impaired glucose and pyruvate tolerance tests, and increased fat and decreased glycogen content in the liver (Fig. 8). These metabolic abnormalities likely resulted from both direct effects of GPRC6A on metabolic pathways regulating glucose and fat metabolism in hepatocytes (Fig. 8), as well as indirect effects mediated by increases in the hepatokine FGF-21.
Summary of metabolic pathways in hepatocytes in Gprc6aLiver-cko mice. Green designates increased gene expression, red indicates decreased gene expression from transcriptome studies in the liver of Gprc6aLiver-cko mice compared to control mice. Glucose is shunted toward triglyceride (TG) and away from glycogen storage.
Indeed, we show for the first time that GPRC6A regulates FGF-21 expression and production in the liver. FGF-21 was reduced 64.3% (p = 0.025) in the circulation and 43.3% in the liver (p = 0.031), a magnitude predicted to contribute to both the hepatic and systemic abnormalities in Gprc6aLiver-cko mice21. FGF-21 regulates glucose and lipid metabolism through systemic activation of fibroblast growth factor receptors complexed to the β-Klotho co-receptor in peripheral and neural tissues, as well as the liver22,23,24. FGF-21 has paracrine effects to regulate hepatic lipid oxidation, triglyceride clearance, ketogenesis, and gluconeogenesis25, systemic effects to increase fat browning, glucose and fatty acid utilization and insulin sensitivity in muscle, to increase insulin synthesis26 and central nervous system actions to regulated energy intake and sugar consumption17. The reduction of FGF-21 in Gprc6aLiver-cko mice confounds our ability to determine which of the observed alterations in glucose and fat metabolism are due to direct hepatic effects of GPRC6A or indirect systemic effects of these hormonal changes. FGF-21 deficiency, however, might have contributed to the observed weight gain and glucose intolerance in Gprc6aLiver-cko mice27. It will be instructive in future studies to see which of the liver loss-of-function phenotypes will be rescued if FGF-21 is supplemented.
In any event, our findings add FGF-21 to a growing list of hormones regulated by GPRC6A, which include insulin secretion in pancreatic β-cells3,11, testosterone (T) production in Leydig cells6,28, IL-6 secretion in skeletal muscle14,16, lipocalin 2 from adipocytes29 and glucagon-like peptide 1 (GLP-1) production from intestinal cells30,31,32. GPRC6A appears to be unique in the complement of the metabolically active hormones that it regulates.
GPRC6A also likely plays a direct role in regulating hepatic glucose and fat metabolism, similar and complementary to the role that GPRC6A plays in regulating catabolism of glucose and fatty acids in skeletal muscle14,16. Insights into possible direct hepatic effects of GPRC6A are derived from changes in the expression of key hepatic gene transcripts that control gluconeogenesis, glycolysis, glycogenesis, and glycogenolysis in Gprc6aLiver-cko mice (Fig. 8). Of the genes that regulate glucose conversion to glycogen, loss-of-GPRC6A decreases glycogenesis as well as glycogenolysis, with the net effects to reduce hepatic glycogen stores in in Gprc6aLiver-cko mice. In contrast, glycolysis is predicted to be increased leading to conversion of glucose to pyruvate, while gluconeogenesis is decreased in Gprc6aLiver-cko mice. Down-regulation of the expression of gluconeogenesis pathway genes predicts that de novo glucose synthesis form non-carbohydrate precursors is impaired in Gprc6aLiver-cko mice. A hepatic explanation for the elevation in fasting serum glucose in Gprc6aLiver-cko mice is not evident from the hepatic transcriptome analysis. Finally, the observed hepatosteatosis and increased hepatic triglyceride content in the liver of Gprc6aLiver-cko mice may result from multiple changes, including the enhanced conversion of glucose into triacylglycerol in Gprc6a-deficient hepatocytes from the increase in glycolysis and decrease in glycogen content in Gprc6aLiver-cko mice, as well as the increase in fatty acid uptake and synthesis and decrease in β-oxidation. Regardless, the direct hepatic effects of GPRC6A deficiency and reductions in paracrine effects of FGF-21 in the liver are concordant with regards to fatty acid metabolism and triglyceride accumulation.
Additional studies are needed to investigate the signaling and metabolic pathways regulated by GPRC6A in hepatocytes. Like insulin, GPRC6A activates PI3K/AKT/mTOR; but also activates cAMP, like glucagon and GLP-1, as well as ERK1,2,12. At the cellular level, the ability of GPRC6A to activate both PI3K/AKT and cAMP pathways creates potentially offsetting signaling mechanisms controlling nutrient uptake and utilization33,34.
The metabolic phenotype in Gprc6aLiver-cko mice is opposite to the reported effects of recombinant Ocn activation of GPRC6A to prevent high fat diet-induced hepatosteatosis, and to improve glucose tolerance, increase insulin sensitivity, reduce fat, and increase muscle mass, ostensibly through GPRC6A activation in multiple tissues1,2,3,4,6,7,12,13,18,35,36,37. Testosterone, another ligand for GPRC6A, has non-genomic effects to regulate fat metabolism in the liver19,38,39,40. These observations suggest that activation of GPRC6A may provide a method to treat metabolic syndrome (MetS), type 2 diabetes (T2D), and non-alcoholic fatty liver disease (NAFLD). To this end, novel small molecule agonists for GPRC6A have recently been developed that lower glucose in mice41 that may serve as therapeutic leads to develop GPRC6A agonists.
Though some laboratories questioned the function of the mouse GPRC6A, discrepancies in the phenotype of Gprc6a deficient mice are likely due to genetic, sex-dependent and environmental factors3,15,42,43,44,45,46,47. Our findings in the liver are consistent with the function of the ancestral ICL3_RKLP mouse variant that now has been confirmed by multiple groups both in vitro and in vivo3,15,42,43. Many studies now show that GPRC6ARKLP is localized to the cell surface and undergoes β-arrestin dependent internalization44,46,47. These observations, along with results from Gprc6a−/− mice, and conditional deletion of Gprc6a in β-cells, Leydig cells, and skeletal muscle, indicate that the ancestral Gprc6a in mice creates an integrative network for regulating energy metabolism involving direct actions to regulate organ-specific glucose and fat metabolism and indirect effects from the release of multiple hormones from different tissues.
In conclusion, the emerging function of GPRC6A in the liver and other tissues, along with its activation by amino acids, cations, Ocn, T and certain natural products defines a new systems biology in energy metabolism. These data in the liver, along with complementary finding in mice with the conditional deletion of Gprc6a in β-cells and skeletal muscle, indicate that GPRC6A plays a key role in glucose and fatty acid metabolism, through both direct tissue effects and the regulation of metabolically active hormones. The afferent and efferent limbs of these endocrine networks created by GPRC6A and its ligands differ from the classic hormonal and metabolic response model to feeding and fasting that are mediated by insulin and glucagon. Rather, variation in GPRC6A activity in response to nutrient, hormone and environmental factors directly controls the anabolic and catabolic functions of multiple organs as well as stimulates the release of an ensemble of hormones that further coordinate glucose and fat metabolism in these various organs.
Materials and Methods
Animals
We used global Gprc6a−/− and Gprc6aflox/flox mice that we have previously characterized10,11. We used Alb-Cre mice48 obtained from the Jackson Laboratory [B6.Cg-Tg (Alb-Cre) 21Mgn/J; Bar Harbor, ME, USA] to delete Gprc6a in hepatocytes by crossing with Gprc6aflox/flox mice. We used a strategy that pairs a heterozygous floxed allele with a “null” mutant allele (Gprc6a+/- mice) to reduce the risk of mosaicism caused by the less than 100% efficiency of Cre-recombinase to excise two floxed alleles (flox/flox) and achieves a >75% deletion of Gprc6a11,49,50. For genotyping, we isolated total DNA from mouse tails, and performed PCR using the primer sets listed in Table S1. Wild-type mice, +/+;Gprc6aflox/+, +/+;Gprc6aflox/-, Alb-Cre/+;Gprc6a+/+ and Alb-Cre/+;Gprc6aflox/+ were metabolically indistinguishable and were combined to create the control group.
All mouse strains were maintained in pure C57BL/6 J background more than 10 generations. Mice were maintained and used in accordance with recommendations as described (National Research Council. 1985; Guide for the Care and Use of Laboratory Animals DHHS Publication NIH 86-23, Institute on Laboratory Animal Resources, Rockville, MD, USA) and following guidelines established by the University of Tennessee Health Science Center Institutional Animal Care and Use Committee. The animal study protocol was approved by the institutional review boards at University of Tennessee Health Science Center Institutional Animal Care and Use Committee.
Metabolic Studies
The glucose tolerance test (GTT) was performed by injecting glucose (2 g/kg body weight) intraperitoneally51 after a 5 hours fast, and monitoring blood glucose using glucose strips and the Accu-Check glucometer at the indicated times52. For the insulin tolerance test (ITT), mice were fasted for 5 hrs, injected IP with insulin (0.75 U/kg body weight, Sigma; St. Louis, MO, USA), and blood glucose levels were measured at indicated times as described15. For the pyruvate tolerance test (PTT), samples were collected following IP injection with pyruvate sodium (2 g/kg bodyweight) to 5 hrs fasted mice. Insulin (mouse) ultrasensitive ELISA kit was obtained from ALPCO Immunoassays (Salem, NH, USA). Glycogen assay and cholesterol quantitation kits were purchased from Sigma (St. Louis, MO, USA). Triglyceride colorimetric assay kit was obtained from Cayman chemical (Ann Arbor, MI, USA). Free fatty acid assay kit was purchased from Fisher Scientific (Pittsburgh, PA, USA). Rat/Mouse FGF-21 Elisa kit purchased from EMD Millipore (Burlington, MA, USA).
Hepatocyte glucose production
Primary mouse hepatocytes or Hepa1C1C7 cells on 12-well plates (1 × 105 cells/well) were maintained in DMEM medium supplemented with 10% fetal bovine serum (Atlanta Biologicals; Norcross, GA, USA) and 10 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Rockville, MA, USA) and 100 nM dexamethasone (Sigma; St. Louis, MO, USA) for 16 hours prior to the measurement of glucose production. Hepatocytes were washed once with PBS, and glucose production was determined after a 12-hour incubation period in glucose-free DMEM containing lactate/pyruvate (10:1 mM) alone or with 100 ng/ml osteocalcin or 100 nM testosterone. At the end of the incubation period, 50 μl of condition medium was collected and glucose level was measured by Glucose assay kit (Sigma; St. Louis, MO, USA).
Measurement of Total and Phospho‐ERK by ERK Elisa Analysis
Briefly, HEK‐293 cells transfected with/without mouse GPRC6A cDNA plasmid were starved by overnight incubation in serum‐free DMEM/F12 containing 0.1% bovine serum albumin (BSA) and stimulated with various ligands at different doses. ERK activation were assessed 20 min after treatment by using ERK1/2 (phospho‐T203/Y204) ELISA Kit (Invitrogen) corrected for the amount of total ERK using ERK1/2 (Total) ELISA Kit (Invitrogen) to measure ERK levels.
Histology
Mouse liver tissues were embedded into Tissue-Tek OCT compound from Sakura Finetek USA, Inc. (Torrance, CA, USA). Cryostat sections (6 μm) were air-dried, fixed in 10% buffered formalin for 5 min, and washed in phosphate-buffered saline (PBS) for 10 min. For Oil Red O staining, the cryostat sections were rinsed in H2O twice, and wash with 2 ml 60% isopropanol for 5 min at room temperature. After slides were completely dry, 1 ml Oil Red O working solution (0.36% Oil Red O in 60% isopropanol) was added to slide for 15 min at room temperature. Then the slides were rinsed with 60% isopropanol and H2O, and dehydrated and mounted. For Periodic Acid-Schiff (PAS) staining, after rinsing in H2O, the slide was immersed in 1% Periodic acid solution for 5 min and rinsed in H2O. The slide was immersed in Schiff’s reagent (1% pararosaniline HCl, and 4% sodium metabisulfite in 0.25 mol/l hydrochloric acid) for 15 min at room temperature. The slide was counterstained in Hematoxylin solution and mounted.
RNA extraction
Total RNA was extracted from mouse liver tissue (~30 mg for each sample) using QIAGEN RNeasy Mini Kit (Frederick, MD, USA). QIAGEN RNase-free DNase Set (Frederick, MD, USA) were used for RNA cleanup. RNA quantity was determined by Qubit fluorometer and RNA integrity were determined by Agilent 2100 bioanalyzer (Santa Clara, CA, USA). The qualified samples (RNA integrity number, RIN > 9) were subjected to RNA-seq analysis.
Real-time RT-PCR
We used 2.0 μg of total RNAs for reverse transcription using cDNA synthesis kit (Bio-Rad). PCR reactions were described in previously publications10,11. The primers for mouse Gprc6a (NM_153071) consisted of mGPRC6A.For135: catgattggtggcttgtttg and mGprc6a/Rev353: gctgctgtgacttcggtaca, and for the Cyclophilin A (NM_008907) consisted of CycA.For: ctgcactgccaagactgaat and CycA. Rev: ccacaatgttcatgccttct.
siRNA Suppression of Gprc6a Gene Expression
For Gprc6a knockdown experiments, the short interfering RNAs (siRNA) (CTCAAGG-ATGCTGAACTTA) has been designed from the mGPRC6A sequence (NM_153071). The siRNA hairpins were synthesized and cloned into a pSilencer 4.1-CMV neo vector (Ambion). A circular pSilencer 4.1-CMV neo vector that expresses a hairpin siRNA with limited homology to any known sequence was used as a negative control. The construct of siRNA duplexes has been stably transfected into C2C12 and Hepa1c1c7 cells using Lipofectamine (Invitrogen) and were selected by G418 (Invitrogen). Successful knock down of GPRC6A will be confirmed by assessing RT-PCR analysis of Gprc6a RNA and Western blot of GPRC6A protein expression.
Library preparation and sequencing
The library preparation and sequencing were carried out by Novogene Co., Ltd. (Chula Vista, CA, USA). Briefly, mRNA was first enriched using oligo53 beads and fragmented randomly by adding fragmentation buffer. Then the cDNA was synthesized by using mRNA template and random hexamers primer, after which a custom second-strand synthesis buffer (Illumina; Mountain View, CA, USA), dNTPs, RNase H, and DNA polymerase I were added to initiate the second-strand synthesis. Second, after terminal repair, a ligation and sequencing adaptor ligation, the double-stranded cDNA library was completed through size selection and PCR enrichment. The library quality was accessed by Qubit 2.0, Agilent 2100, and Q-PCR. The DNA from the qualified libraries are fed into Illumina sequencers at an average depth of 42 million reads per sample.
RNA-seq data analysis
Raw reads were quality filtered with NGS QC Toolkit version 2.354 to remove adaptor contaminated reads or reads containing >20% low-quality (Q < 20) bases. Filtered reads were aligned to the mouse reference sequence (GRCm38/mm10) using STAR aligner version 2.5.0a55. Raw read count was quantified across all annotated mm10 transcript using FeatureCounts version 1.6.3 implemented in Subread package56, then submitted to DeSeq. 2 version57 to identify the differentially expressed genes between KO and WT groups (three replicates for each group). Differentially expressed genes were defined as having an adjusted p value <0.05. The final significant differential gene list was used for gene enrichment analysis58 including Gene Ontology (Biological Process), KEGG pathway59, and Mammalian Phenotype Ontology. The heatmap and barplot were prepared with R and Excel program respectively.
Statistics
We evaluated differences between groups by Studet’s t test, and for multiple groups by two-way ANOVA, followed by a post-hoc Tukey’s test. Significance was set at p < 0.05. All values are expressed as means ± SEM. All computations were performed using the Statgraphic statistical graphics system (STSC Inc., Rockville, MD, USA).
References
Pi, M. et al. Structural and Functional Evidence for Testosterone Activation of GPRC6A in Peripheral Tissues. Molecular endocrinology 29, 1759–1773, https://doi.org/10.1210/me.2015-1161 (2015).
De Toni, L. et al. Osteocalcin and Sex Hormone Binding Globulin Compete on a Specific Binding Site of GPRC6A. Endocrinology, en20161312 https://doi.org/10.1210/en.2016-1312 (2016).
Wei, J., Hanna, T., Suda, N., Karsenty, G. & Ducy, P. Osteocalcin promotes beta-cell proliferation during development and adulthood through Gprc6a. Diabetes 63, 1021–1031, https://doi.org/10.2337/db13-0887 (2014).
Pi, M., Parrill, A. L. & Quarles, L. D. GPRC6A mediates the non-genomic effects of steroids. J Biol Chem 285, 39953–39964, https://doi.org/10.1074/jbc.M110.158063 (2010).
Pi, M. et al. GPCR6A Is a Molecular Target for the Natural Products Gallate and EGCG in Green Tea. Molecular nutrition & food research 62, e1700770, https://doi.org/10.1002/mnfr.201700770 (2018).
Oury, F. et al. Endocrine regulation of male fertility by the skeleton. Cell 144, 796–809, https://doi.org/10.1016/j.cell.2011.02.004 (2011).
Ferron, M. et al. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell 142, 296–308, https://doi.org/10.1016/j.cell.2010.06.003 (2010).
Pi, M., Nishimoto, S. K. & Quarles, L. D. GPRC6A: Jack of all metabolism (or master of none). Molecular metabolism 6, 185–193, https://doi.org/10.1016/j.molmet.2016.12.006 (2017).
Karsenty, G. & Olson, E. N. Bone and Muscle Endocrine Functions: Unexpected Paradigms of Inter-organ Communication. Cell 164, 1248–1256, https://doi.org/10.1016/j.cell.2016.02.043 (2016).
Pi, M. et al. GPRC6A null mice exhibit osteopenia, feminization and metabolic syndrome. PLoS One 3, e3858, https://doi.org/10.1371/journal.pone.0003858 (2008).
Pi, M. et al. Evidence for Osteocalcin Binding and Activation of GPRC6A in beta-Cells. Endocrinology 157, 1866–1880, https://doi.org/10.1210/en.2015-2010 (2016).
Pi, M., Wu, Y., Lenchik, N. I., Gerling, I. & Quarles, L. D. GPRC6A mediates the effects of L-arginine on insulin secretion in mouse pancreatic islets. Endocrinology 153, 4608–4615, https://doi.org/10.1210/en.2012-1301 (2012).
Ferron, M., Hinoi, E., Karsenty, G. & Ducy, P. Osteocalcin differentially regulates beta cell and adipocyte gene expression and affects the development of metabolic diseases in wild-type mice. Proceedings of the National Academy of Sciences of the United States of America 105, 5266–5270, https://doi.org/10.1073/pnas.0711119105 (2008).
Mera, P., Laue, K., Wei, J., Berger, J. M. & Karsenty, G. Osteocalcin is necessary and sufficient to maintain muscle mass in older mice. Molecular metabolism 5, 1042–1047, https://doi.org/10.1016/j.molmet.2016.07.002 (2016).
Lee, N. K. et al. Endocrine regulation of energy metabolism by the skeleton. Cell 130, 456–469, https://doi.org/10.1016/j.cell.2007.05.047 (2007).
Mera, P. et al. Osteocalcin Signaling in Myofibers Is Necessary and Sufficient for Optimum Adaptation to Exercise. Cell metabolism 23, 1078–1092, https://doi.org/10.1016/j.cmet.2016.05.004 (2016).
Gillum, M. P. Parsing the Potential Neuroendocrine Actions of FGF21 in Primates. Endocrinology 159, 1966–1970, https://doi.org/10.1210/en.2018-00208 (2018).
Gupte, A. A. et al. Osteocalcin protects against nonalcoholic steatohepatitis in a mouse model of metabolic syndrome. Endocrinology 155, 4697–4705, https://doi.org/10.1210/en.2014-1430 (2014).
Du, J. et al. Osteocalcin improves nonalcoholic fatty liver disease in mice through activation of Nrf2 and inhibition of JNK. Endocrine 53, 701–709, https://doi.org/10.1007/s12020-016-0926-5 (2016).
Adeva-Andany, M. M., Perez-Felpete, N., Fernandez-Fernandez, C., Donapetry-Garcia, C. & Pazos-Garcia, C. Liver glucose metabolism in humans. Biosci Rep 36, https://doi.org/10.1042/BSR20160385 (2016).
Lin, X., Liu, Y. B. & Hu, H. Metabolic role of fibroblast growth factor 21 in liver, adipose and nervous system tissues. Biomed Rep 6, 495–502, https://doi.org/10.3892/br.2017.890 (2017).
Markan, K. R. et al. Circulating FGF21 is liver derived and enhances glucose uptake during refeeding and overfeeding. Diabetes 63, 4057–4063, https://doi.org/10.2337/db14-0595 (2014).
Kliewer, S. A. & Mangelsdorf, D. J. A Dozen Years of Discovery: Insights into the Physiology and Pharmacology of FGF21. Cell metabolism 29, 246–253, https://doi.org/10.1016/j.cmet.2019.01.004 (2019).
Lee, S. et al. Structures of beta-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nature 553, 501–505, https://doi.org/10.1038/nature25010 (2018).
Badman, M. K. et al. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell metabolism 5, 426–437, https://doi.org/10.1016/j.cmet.2007.05.002 (2007).
Inagaki, T. Research Perspectives on the Regulation and Physiological Functions of FGF21 and its Association with NAFLD. Frontiers in endocrinology 6, 147, https://doi.org/10.3389/fendo.2015.00147 (2015).
Badman, M. K., Koester, A., Flier, J. S., Kharitonenkov, A. & Maratos-Flier, E. Fibroblast growth factor 21-deficient mice demonstrate impaired adaptation to ketosis. Endocrinology 150, 4931–4940, https://doi.org/10.1210/en.2009-0532 (2009).
Oury, F. et al. Osteocalcin regulates murine and human fertility through a pancreas-bone-testis axis. J Clin Invest 123, 2421–2433, https://doi.org/10.1172/JCI65952 (2013).
Mosialou, I. et al. MC4R-dependent suppression of appetite by bone-derived lipocalin 2. Nature 543, 385–390, https://doi.org/10.1038/nature21697 (2017).
Mizokami, A. et al. Osteocalcin induces release of glucagon-like peptide-1 and thereby stimulates insulin secretion in mice. PLoS One 8, e57375, https://doi.org/10.1371/journal.pone.0057375 (2013).
Mizokami, A. et al. Oral administration of osteocalcin improves glucose utilization by stimulating glucagon-like peptide-1 secretion. Bone 69, 68–79, https://doi.org/10.1016/j.bone.2014.09.006 (2014).
Oya, M. et al. The G protein-coupled receptor family C group 6 subtype A (GPRC6A) receptor is involved in amino acid-induced glucagon-like peptide-1 secretion from GLUTag cells. J Biol Chem 288, 4513–4521, https://doi.org/10.1074/jbc.M112.402677 (2013).
Nauck, M. & Wilhelm, B. [Why is a combination of basal insulin with a GLP-1 receptor agonist useful in many patients with type 2 diabetes?]. MMW. Fortschritte der Medizin 159, 7–15, https://doi.org/10.1007/s15006-017-9803-2 (2017).
Newgard, C. B. et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell metabolism 9, 311–326, https://doi.org/10.1016/j.cmet.2009.02.002 (2009).
Ferron, M., McKee, M. D., Levine, R. L., Ducy, P. & Karsenty, G. Intermittent injections of osteocalcin improve glucose metabolism and prevent type 2 diabetes in mice. Bone, https://doi.org/10.1016/j.bone.2011.04.017 (2011).
Levy-Litan, V. et al. Autosomal-recessive hypophosphatemic rickets is associated with an inactivation mutation in the ENPP1 gene. Am J Hum Genet 86, 273–278, https://doi.org/10.1016/j.ajhg.2010.01.010 (2010).
Pi, M. & Quarles, L. D. GPRC6A regulates prostate cancer progression. Prostate, https://doi.org/10.1002/pros.21442 (2011).
Jia, Y. et al. Testosterone protects high-fat/low-carbohydrate diet-induced nonalcoholic fatty liver disease in castrated male rats mainly via modulating endoplasmic reticulum stress. American journal of physiology. Endocrinology and metabolism 314, E366–E376, https://doi.org/10.1152/ajpendo.00124.2017 (2018).
Haider, A., Gooren, L. J., Padungtod, P. & Saad, F. Improvement of the metabolic syndrome and of non-alcoholic liver steatosis upon treatment of hypogonadal elderly men with parenteral testosterone undecanoate. Experimental and clinical endocrinology & diabetes: official journal, German Society of Endocrinology [and] German Diabetes Association 118, 167–171, https://doi.org/10.1055/s-0029-1202774 (2010).
Nikolaenko, L. et al. Testosterone replacement ameliorates nonalcoholic fatty liver disease in castrated male rats. Endocrinology 155, 417–428, https://doi.org/10.1210/en.2013-1648 (2014).
Pi, M. et al. Computationally identified novel agonists for GPRC6A. PLoS One 13, e0195980, https://doi.org/10.1371/journal.pone.0195980 (2018).
Jorgensen, C. V. et al. Metabolic and skeletal homeostasis are maintained in full locus GPRC6A knockout mice. Scientific reports 9, 5995, https://doi.org/10.1038/s41598-019-41921-8 (2019).
Jorgensen, S. et al. Genetic Variations in the Human G Protein-coupled Receptor Class C, Group 6, Member A (GPRC6A) Control Cell Surface Expression and Function. J Biol Chem 292, 1524–1534, https://doi.org/10.1074/jbc.M116.756577 (2017).
Jacobsen, S. E. et al. Delineation of the GPRC6A receptor signaling pathways using a mammalian cell line stably expressing the receptor. The. Journal of pharmacology and experimental therapeutics 347, 298–309, https://doi.org/10.1124/jpet.113.206276 (2013).
Lambert, L. J. et al. Increased trabecular bone and improved biomechanics in an osteocalcin-null rat model created by CRISPR/Cas9 technology. Disease models & mechanisms 9, 1169–1179, https://doi.org/10.1242/dmm.025247 (2016).
Pi, M. et al. Identification of a novel extracellular cation-sensing G-protein-coupled receptor. J Biol Chem 280, 40201–40209 (2005).
Rueda, P. et al. Murine GPRC6A Mediates Cellular Responses to L-Amino Acids, but Not Osteocalcin Variants. PLoS One 11, e0146846, https://doi.org/10.1371/journal.pone.0146846 (2016).
Postic, C. et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274, 305–315 (1999).
Liu, S. et al. Pathogenic role of Fgf23 in Hyp mice. American journal of physiology. Endocrinology and metabolism 291, E38–49, https://doi.org/10.1152/ajpendo.00008.2006 (2006).
Kwan, K. M. Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis 32, 49–62 (2002).
Echeverri, C. J. et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods 3, 777–779, https://doi.org/10.1038/nmeth1006-777 (2006).
Phillips, C. M. et al. Leptin receptor polymorphisms interact with polyunsaturated fatty acids to augment risk of insulin resistance and metabolic syndrome in adults. J Nutr 140, 238–244, https://doi.org/10.3945/jn.109.115329 (2010).
Taylor, C. R., Schmidt-Nielsen, K. & Raab, J. L. Scaling of energetic cost of running to body size in mammals. The American journal of physiology 219, 1104–1107 (1970).
Patel, R. K. & Jain, M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PloS one 7, e30619 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome biology 15, 550 (2014).
Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z. & Zhang, B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic acids research (2019).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research 28, 27–30 (2000).
Acknowledgements
This work was supported by grants from NIH grant R01-AR37308 and ADA grant 1-13-BS-149-BR (to LDQ), and NIH grant R01 DK120567 (to LDQ and LL).
Author information
Authors and Affiliations
Contributions
M.P., F.X., L.L. and L.D.Q. contributed to study design. M.P., F.X., S.K.N., L.L. and L.D.Q. wrote the manuscript with input from all authors and contributed discussion and data analysis. M.P. and R.Y. performed all experiments related to mouse phenotypic characterization and RNA sample preparation. F.X., L.L., and R.W.W. contributed to bioinformatics analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pi, M., Xu, F., Ye, R. et al. Role of GPRC6A in Regulating Hepatic Energy Metabolism in Mice. Sci Rep 10, 7216 (2020). https://doi.org/10.1038/s41598-020-64384-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-64384-8
- Springer Nature Limited