Abstract
Today, we face difficulty in generating new hypotheses and understanding oral lichen planus due to the large amount of biomedical information available. In this research, we have used an integrated bioinformatics approach assimilating information from data mining, gene ontologies, protein–protein interaction and network analysis to predict candidate genes related to oral lichen planus. A detailed pathway analysis led us to propose two promising therapeutic targets: the stromal cell derived factor 1 (CXCL12) and the C-X-C type 4 chemokine receptor (CXCR4). We further validated our predictions and found that CXCR4 was upregulated in all oral lichen planus tissue samples. Our bioinformatics data cumulatively support the pathological role of chemokines and chemokine receptors in oral lichen planus. From a clinical perspective, we suggest a drug (plerixafor) and two therapeutic targets for future research.
Similar content being viewed by others
Introduction
Oral lichen planus (OLP) is a chronic immunologically mediated disease1. Histopathological features of OLP include hyperkeratosis, acanthosis, degeneration by liquefaction of the basal keratinocyte layer, and prominent lymphocyte infiltration at the connective tissue–epithelium interface2. OLP is not a homogeneous clinical entity, but a complex disease. A recent review proposes a new classification of this disease, taking into consideration a set of lesions with similar characteristics, including oral lichenoid contact lesions, oral lichenoid drug reactions, and lichen planus pemphigoides, among others3.The clinical findings vary from white reticular plaques to erythema, erosions, ulceration, and hyperkeratotic plaques in the oral mucosa4. Although reticular lesions are often asymptomatic, burning or pain often accompanies erosive/ulcerative manifestations of OLP. The severity of pain may vary from mild to severe and may even interfere with patients’ speech, feeding, and swallowing4. According to the WHO, this disease has a potential for cancerization5. This potential is presented in about 1% of patients6. Despite the efforts in biomedical research, the etiology of OLP is unknown, and its molecular basis is still unclear. This may explain why the treatment is palliative. For these reasons, exploration of the mechanisms that govern the disease’s pathophysiological process is needed.
A disease is rarely the result of an abnormality in a single gene, but it reflects disturbances in the complex intracellular and intercellular network that links tissue and organ systems7. Today, we face difficulty in generating new hypotheses and understanding OLP due to the large amount of biomedical information available. Encapsulating that information in an understandable way is one of the current challenges of network-based approaches to the study of human diseases8. The heterogeneity of pathologies, the difficulty of finding a gene or protein in a certain localization, and the cost involved in experimental studies have led to the development of several in silico approaches for the identification of genes and proteins associated with a disease9. These bioinformatic approaches address complexity by simplifying systems, summarizing them as interaction maps or interactomes, and containing components (nodes) and lines (interactions) between them7. To explore biological systems, analyses involve data mining10, protein–protein interactions11, gene ontology (GO)12, gene regulatory networks, and pathways for gene identification candidates13. Several publications have reported on the successful use of such techniques to prioritize disease-related genes and propose therapeutic targets14,15,16. However, these technologies have not been applied to the study of OLP.
In order to better understand the molecular basis of OLP and propose new therapeutic targets, we use data mining to obtain a large number of protein coding genes associated with this disease. Then, we describe the biological attributes of proteins and prioritize a small group of them using protein–protein interaction networks. With the most relevant proteins, we defined the OLP interactome, which allowed us to propose a new treatment for the disease and two therapeutic targets. Finally, we check our computational predictions by immunohistochemistry.
Methods
General design
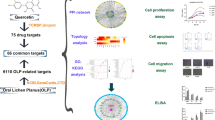
Figure 1 provide a brief description of our workflow. All procedures were in accordance with the Helsinki Declaration17. The Ethics Committee of UTALCA (protocol #2017-02-CR) approved this research (https://doi.org/10.5281/zenodo.3576206).
Experimental design. Initially, we used Génie web server to perform a ranking of protein-coding genes associated with oral lichen planus (OLP). Then, using PANTHER, we characterized them based on gene ontology (controlled vocabulary that describes the gene and the attributes of the gene product). The most important proteins of that set (which we call the OLP interactome) were prioritized by building interaction networks (CHAT). From them, we identified drug target proteins (STITCH). One of the proposed targets, a chemokine receptor, was evaluated (immunohistochemistry) in a series of cases of focal fibrous hyperplasia (a reactive hyperplastic lesion of the connective tissue in response to local irritation), head and neck cancer (malignant neoplasm), pemphigus and pemphigoid (blistering autoimmune diseases), and OLP.
Protein identification and classification
To identify genes encoding proteins related to OLP from a large volume of information, we use the Génie web tool18. This tool analyzes relationships between genes and biomedical topics in all abstracts available on MEDLINE/PubMed. The topic of interest was defined by the search (carried out on July 19, 2018): “lichen planus, oral” [MeSH] and “biomarkers” [MeSH] and (risk ratio [Title/Abstract] or relative risk [Title/Abstract] or odds ratio [Title/Abstract] or risk [Title/Abstract]) and (“humans” [MeSH Terms]). With these criteria, 23 abstracts were identified as a training set. In order to find only significant coding genes, we established as a cut-off p-value of <0.01 for abstracts and a false positive discovery rate (FDR) of <0.01 for genes. We use Fisher’s exact test to define the relationship between genes and search topics.
The gene list was subjected to a gene ontology analysis using the PANTHER representation test (http://www.pantherdb.org)19 version 13.1 to determine classifications based on biological processes, cellular components, molecular functions, and metabolic pathways. We select the coding genes from the most overrepresented classifications.
Interactome construction
To build the OLP interactome, we use the Contextual Hub Analysis (CHAT) application of the Cytoscape program20 and the STITCH web tool (http://stitch.embl.de/)21. CHAT identifies the genes of greatest relevance within a list. For this, it builds a molecular network composed of (i) the list of genes (each gene is a node), (ii) the numerical values for each gene (for example, expression or belonging to a group), and (iii) a database with the genes that will establish interactions. The smallest p-values represent the centers of greatest activity within the biological network (hubs). We assign a numerical value of +3 (context attribute) to all coding genes included in the overrepresented PANTHER categories, and as a database for the interactions, we select IntAct22. To establish whether the resulting hubs corresponded to a biologically connected network, we used STITCH. These base reports known interactions as well as establishing predictions of interactions between proteins and chemicals. In STITCH, we choose to establish interactions with the maximum confidence level (0.9) using all available resources. We call the resulting network the “OLP interactome.”
Immunohistochemistry verification
We retrospectively collected focal fibrous hyperplasias (n = 10), head and neck cancers (n = 4), pemphigoid (n = 2), pemphigus (n = 2) and OLP lesions (n = 12) in paraffin-embedded tissues stored in the UTALCA Biobank/Oral Pathology Laboratory (https://medicinaoral.pro/biobanco). The information of patients is provided in Supplementary Table 1 (https://doi.org/10.5281/zenodo.3483255). For OLP diagnosis, we use both clinical and histopathologic criteria enumerated in the position paper by the American Academy of Oral and Maxillofacial Pathology2. Immunostaining of 3 μm histological sections was performed using EnVision FLEX target retrieval solution (High pH, Dako) according to our previously published protocols23,24. The primary antibody used was a CXCR4 antibody (1:1000 dilution, #PA3305, Invitrogen Inc., USA), used overnight at 4 °C. Two pathologists blinded to the clinical data provided a consensus opinion of staining patterns. We use oral cancer cases as positive controls. Also, evidence that CXCR4 antibody represent a specific staining can be consulted in our previous studies23,24.
Results
OLP proteins participate in inflammation mediated by chemokines and cytokines
With Génie, we obtained 872 statistically significant protein-coding genes extracted from 1,075,776 articles (Supplementary Dataset 1, https://doi.org/10.5281/zenodo.3483255). To discover patterns in that large volume of information, we describe the set using PANTHER. This tool allowed us to classify gene products (that is, proteins) into four categories: the cell zone in which they are found (cellular component), the molecular functions, biological processes, and metabolic pathways in which they participate. The visual interface offers pie charts, in which we click on the categories that incorporated a greater number of genes. This allowed us to “dive” to the root of each topic to find more specific information. Table 1 shows the most relevant processes, which are represented by 51 proteins. Considering the recognized role of immune mediation in OLP, the metabolic pathway corresponding to inflammation mediated by chemokines and cytokines stands out. The complete classification can be consulted in Supplementary Dataset 2 (https://doi.org/10.5281/zenodo.3483255).
OLP interactome reveals two promising therapeutic targets for plerixafor
For a better interpretation of the data in a biological context, we evaluate the proteins using the Cytoscape program and its CHAT application. CHAT identifies central nodes (proteins with many connections) that interact with more “contextual” nodes (i.e., 51 proteins obtained from the previous step). CHAT calls these nodes “contextual hubs.” These hubs have topological and functional relevance, since their elimination causes great damage in a network, so they are the best representatives of a biological system25. The application built an interaction network of 1,045 nodes using the IntAct database as a source (Supplementary Fig. 1, https://doi.org/10.5281/zenodo.3483255). From this network, we selected the most important hubs (17 significant p-values reported by CHAT, Supplementary Dataset 3, https://doi.org/10.5281/zenodo.3483255). Next, we identified the interactions between chemicals and proteins. Using STITCH, we identified a network composed of 21 proteins with a high clustering coefficient (0.97), which indicates that it is highly feasible that these proteins are a biologically interconnected community. We call that network the OLP interactome. In that network, two proteins interact with plerixafor: the stromal cell–derived factor 1 (CXCL12) and the C-X-C type 4 chemokine receptor (CXCR4) (Fig. 2).
Interactome of oral lichen planus. Network with 21 proteins processed by STITCH and its pharmacological relationship with plerixafor. The clustering coefficient is 0.968. Thicker lines represent stronger associations. Protein–protein interactions are shown in gray, and chemical–protein interactions in green. FDR = false discovery rate. The original network can be consulted at http://stitch.embl.de/cgi/network.pl?taskId=L9CBnAWsnpeX.
CXCR4 is overexpressed in connective tissue of OLP lesions
To confirm our bioinformatic predictions, we used immunohistochemistry. We detected higher expression of CXCR4 in OLPs than in other lesions (fibrous hyperplasia, cancer, pemphigoid and pemphigus, Fig. 3). Epithelial staining reactivity were positive in all samples. In the subepithelial connective tissue, the differences are remarkable. A high marking intensity for CXCR4 is present in all OLP cases. CXCR4 staining coincides with inflammatory areas. These results may indicate the presence of actively infiltrating immune cells, which are positive for this receptor.
CXCR4 is overexpressed in the connective tissue of oral lichen planus. Representative microphotographs of CXCR4 receptor immunohistochemical staining in oral tissues (40× objective). Positive staining is shown in brown. Compared to the controls, oral lichen planus lesions show high intensity and reactivity to CXCR4 in areas of active inflammatory infiltrate. All images (n = 99) can be reviewed at, https://doi.org/10.5281/zenodo.3352836.
Discussion
The identification of genes involved in diseases is an important tool to reveal molecular mechanisms for disease development and for the establishment of new therapies26. Here, we have predicted and prioritized a group of proteins associated with OLP along with proposing two possible therapeutic targets for the disease, CXCL12 and its receptor, CXCR4.
Connecting genes and proteins with the diseases, in which they are involved, is the heart of molecular medicine27. Several applications have been developed to allow genes to be linked with complex diseases26,27,28,29,30,31. One of them is Génie, which has been previously used in the determination of risk genes for non–small cell lung cancer32. Using this tool, we obtained a set close to 1,000 genes coding for proteins related to OLP, among which chemokine and cytokine-mediated inflammation stand out. These findings are in accordance with the current evidence that define this disease focused on its chronic and immunological basis33.
With our prioritization analysis, we identify the oral lichen planus interactome, consisting of two clusters of 21 proteins: CXCL10, CXCL12, CCL5 (RANTES), CCL19, CCL20, CCL21, CXCR1, CXCR2, CXCR3, CXCR4, CCR1, CCR2, CCR3, CCR5, CCR6, CCR7, IL-8, MTHFR, MTR, MTRR, and MTHFD1. The main cluster includes chemokines and chemokine receptors participating in dendritic cell chemotaxis. In addition, these proinflammatory molecules are produced by cells primarily to recruit leukocytes at sites of infection or injury34. For example, CXCL10 and CXCL12 are chemoattractors of T lymphocytes and monocytes35,36; CCL5 of monocytes, memory T-helper lymphocytes, and eosinophils37; CCL19 of T and B lymphocytes38; and IL-8 of neutrophils, basophils, and T lymphocytes39. The fact that 17 lichen planus interactome proteins are chemokines or their receptors explains why the lymphocytic infiltrate in this disease is intense in the connective tissue, resembling a band40. The intensity of this infiltrate leads to overlying keratinization and degeneration due to liquefaction of the basal layer41, the latter directed by CD8 + auto-cytotoxic T-lymphocytes1. Our interactome suggests that cell-mediated histological features are triggered by a large flow of chemokines.
The switch that starts the mechanisms of oral lichen planus is still unknown. Revealing the molecules that constitute the major centers of activity or disturbance points in the OLP network can provide a chance to find new therapies. Surprisingly, using STITCH, we predict that plerixafor has two therapeutic targets, CXCR4 and CXCL12.
The CXCL12-CXCR4 axis regulates leukocyte chemotaxis in inflammatory conditions and autoimmune diseases. It has significantly been studied in numerous cancers and autoimmune diseases42. This axis modulates effects on cells in an autoimmunity context, which may be important for the development or severity of various diseases, including psoriasis, multiple sclerosis, rheumatoid arthritis, systemic lupus erythematosus, idiopathic inflammatory bowel diseases, and type 1 diabetes43. Recent experimental evidence shows that the CXCL12-CXCR4 axis participates in inappropriate retention of activated innate inflammatory cells at inflammatory sites. This is highly relevant for chronic diseases, such as chronic obstructive pulmonary disease and asthma44. The broad participation of the CXCL12-CXCR4 axis in several diseases justifies the recognition of antagonistic drugs.
Plerixafor, also known as AMD3100, was originally developed as a drug against human immunodeficiency virus and then characterized as a CXCR4 antagonist45. Plerixafor has proven to be useful for inhibiting the CXCL12-CXCR4axis in patients with leukemia46. Although there is no evidence of the usefulness of this drug in OLP, a biological basis supports this therapeutic use.
Previous evidence shows that the microdissected oral epithelium of OLP patients presents an increase in gene expression of 258% for CXCL12 and 629% for CXCR447. In addition, our immunohistochemistry corroborated the predictions. We tested the expression of CXCR4 for two reasons: first, because our research group has previous studies with this protein23,24 and second, the immunohistochemistry method is poorly quantitative and can suffer from low sensitivity for detection of secreted proteins (for example, CXCL12)48.
We observed that CXCR4 is highly expressed in the connective tissue of OLP patients. It is known that CXCR4 and CXCL12 are expressed in dendritic cells (Langerhans cells)49,50. Since dendritic cells are highly activated in OLP, this may be an important event for disease development. Dendritic cells are the most potent antigen presenting cells for lymphocytes51,52. These cells present the antigen to memory T cells, the predominant phenotype in OLP53. It is known that CXCR4 is highly-expressed in resting T cells, including naïve and memory T cells, and is downregulated during T cell activation54. We believe that the presence of high amounts of CXCR4 may represent populations of dendritic cells, memory T cells, or a combination of both.
An initial event in the disease mechanism may involves the expression or presentation of the keratinocyte antigen (still unknown) to trigger the immune response1. Therefore, it is logical to think that an intervention that interrupts the triggering of lymphocytic infiltrate could ensure epithelial integrity. Although topical steroids are considered the first-line treatment for symptomatic oral lichen planus, there is no evidence to support the effectiveness of these drugs55,56, which we believe is an invitation to explore new options.
The initial search for our design does not distinguish between the clinical variants of OLP; however, our analyses are inspired by the erosive/ulcerative forms that are accompanied by painful symptomatology. Our results are limited to the performance of our applications in silico, and new studies are needed to provide experimental data for our analysis. In the future, we should test whether the high expression of CXCR4 can be verified using other techniques, such as PCR experiments (after tissue microdissection) or salivary ELISA.
In this investigation, bioinformatics data cumulatively support the pathological role of chemokines and chemokine receptors in OLP. From a clinical perspective, we suggest a drug and two therapeutic targets for future research. Additionally, we demonstrate that it is possible to comprehensively analyze a large volume of biomedical information in order to better understand the molecular complexity of the OLP, even confirming a bioinformatics prediction using immunohistochemistry. We believe in the need for future studies to verify and monitor – experimentally and clinically – the role of the proteins listed in this research to move towards more precise therapies.
Data availability
All supplementary files are included in this article and are publicly available at: https://doi.org/10.5281/zenodo.3483255.
References
Lavanya, N., Jayanthi, P., Rao, U. K. & Ranganathan, K. Oral lichen planus: An update on pathogenesis and treatment. J. Oral. Maxillofac. Pathol. 15, 127–132, https://doi.org/10.4103/0973-029x.84474 (2011).
Cheng, Y. S., Gould, A., Kurago, Z., Fantasia, J. & Muller, S. Diagnosis of oral lichen planus: a position paper of the American Academy of Oral and Maxillofacial Pathology. Oral. Surg. Oral Med. Oral Pathol. Oral Radiol. 122, 332–354, https://doi.org/10.1016/j.oooo.2016.05.004 (2016).
Carrozzo, M., Porter, S., Mercadante, V. & Fedele, S. Oral lichen planus: A disease or a spectrum of tissue reactions? Types, causes, diagnostic algorhythms, prognosis, management strategies. Periodontol 2000 80, 105–125, https://doi.org/10.1111/prd.12260 (2019).
Olson, M. A., Rogers, R. S. III & Bruce, A. J. Oral lichen planus. Clin. Dermatol. 34, 495–504, https://doi.org/10.1016/j.clindermatol.2016.02.023 (2016).
Peng, Q., Zhang, J., Ye, X. & Zhou, G. Tumor-like microenvironment in oral lichen planus: evidence of malignant transformation? Expert. Rev. Clin. Immunol. 13, 635–643, https://doi.org/10.1080/1744666x.2017.1295852 (2017).
Giuliani, M. & Troiano, G. Rate of malignant transformation of oral lichen planus: A systematic review. Oral. Dis. 25, 693–709, https://doi.org/10.1111/odi.12885 (2019).
Barabasi, A. L., Gulbahce, N. & Loscalzo, J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12, 56–68, https://doi.org/10.1038/nrg2918 (2011).
Cheung, W. A., Ouellette, B. F. & Wasserman, W. W. Inferring novel gene-disease associations using Medical Subject Heading Over-representation Profiles. Genome Med. 4, 75, https://doi.org/10.1186/gm376 (2012).
Kumar, R., Samal, S. K., Routray, S., Dash, R. & Dixit, A. Identification of oral cancer related candidate genes by integrating protein-protein interactions, gene ontology, pathway analysis and immunohistochemistry. Sci. Rep. 7, 2472, https://doi.org/10.1038/s41598-017-02522-5 (2017).
Piro, R. M. & Di Cunto, F. Computational approaches to disease-gene prediction: rationale, classification and successes. Febs J. 279, 678–696, https://doi.org/10.1111/j.1742-4658.2012.08471.x (2012).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504, https://doi.org/10.1101/gr.1239303 (2003).
Ortutay, C. & Vihinen, M. Identification of candidate disease genes by integrating Gene Ontologies and protein-interaction networks: case study of primary immunodeficiencies. Nucleic Acids Res. 37, 622–628, https://doi.org/10.1093/nar/gkn982 (2009).
Rivera, C. et al. Agrin has a pathological role in the progression of oral cancer. Br. J. Cancer 188, 1628–1638, https://doi.org/10.1038/s41416-018-0135-5 (2018).
Siddani, B. R., Pochineni, L. P. & Palanisamy, M. Candidate gene identification for systemic lupus erythematosus using network centrality measures and gene ontology. PLoS One 8, e81766, https://doi.org/10.1371/journal.pone.0081766 (2013).
Jamal, S., Goyal, S., Shanker, A. & Grover, A. Integrating network, sequence and functional features using machine learning approaches towards identification of novel Alzheimer genes. BMC Genomics 17, 807, https://doi.org/10.1186/s12864-016-3108-1 (2016).
Sun, Y. et al. Combining genomic and network characteristics for extended capability in predicting synergistic drugs for cancer. Nat. Commun. 6, 8481, https://doi.org/10.1038/ncomms9481 (2015).
World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. Jama 310, 2191–2194, https://doi.org/10.1001/jama.2013.281053 (2013).
Fontaine, J.-F., Priller, F., Barbosa-Silva, A. & Andrade-Navarro, M. A. Genie: literature-based gene prioritization at multi genomic scale. Nucleic acids Res. 39, W455–W461 (2011).
Mi, H. et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–d189, https://doi.org/10.1093/nar/gkw1138 (2017).
Muetze, T. et al. Contextual Hub Analysis Tool (CHAT): A Cytoscape app for identifying contextually relevant hubs in biological networks. F1000Res. 5, 1745, https://doi.org/10.12688/f1000research.9118.2 (2016).
Szklarczyk, D. et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44, D380–384, https://doi.org/10.1093/nar/gkv1277 (2016).
Kerrien, S. et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40, D841–846, https://doi.org/10.1093/nar/gkr1088 (2012).
Domingueti, C. B. et al. Prognostic value of immunoexpression of CCR4, CCR5, CCR7 and CXCR4 in squamous cell carcinoma of tongue and floor of the mouth. Med. Oral. Patol. Oral Cir. Bucal 24, e354–e363, https://doi.org/10.4317/medoral.22904 (2019).
Gonzalez-Arriagada, W. A., Lozano-Burgos, C., Zuniga-Moreta, R., Gonzalez-Diaz, P. & Coletta, R. D. Clinicopathological significance of chemokine receptor (CCR1, CCR3, CCR4, CCR5, CCR7 and CXCR4) expression in head and neck squamous cell carcinomas. J. Oral. Pathol. Med. 47, 755–763, https://doi.org/10.1111/jop.12736 (2018).
Muetze, T. & Lynn, D. J. Using the Contextual Hub Analysis Tool (CHAT) in Cytoscape to Identify Contextually Relevant Network Hubs. Curr. Protoc. Bioinforma. 59, 8.24.21–28.24.13, https://doi.org/10.1002/cpbi.35 (2017).
Kim, J., Kim, J. J. & Lee, H. An analysis of disease-gene relationship from Medline abstracts by DigSee. Sci. Rep. 7, 40154, https://doi.org/10.1038/srep40154 (2017).
Pletscher-Frankild, S., Palleja, A., Tsafou, K., Binder, J. X. & Jensen, L. J. DISEASES: text mining and data integration of disease-gene associations. Methods 74, 83–89, https://doi.org/10.1016/j.ymeth.2014.11.020 (2015).
Xu, D. et al. DTMiner: identification of potential disease targets through biomedical literature mining. Bioinformatics 32, 3619–3626, https://doi.org/10.1093/bioinformatics/btw503 (2016).
Barbosa-Silva, A. et al. PESCADOR, a web-based tool to assist text-mining of biointeractions extracted from PubMed queries. BMC Bioinforma. 12, 435, https://doi.org/10.1186/1471-2105-12-435 (2011).
Fontaine, J. F., Priller, F., Barbosa-Silva, A. & Andrade-Navarro, M. A. Genie: literature-based gene prioritization at multi genomic scale. Nucleic Acids Res. 39, W455–461, https://doi.org/10.1093/nar/gkr246 (2011).
Liu, Y., Liang, Y. & Wishart, D. PolySearch2: a significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Res. 43, W535–542, https://doi.org/10.1093/nar/gkv383 (2015).
Teng, Y. et al. Genome-wide haplotype association study identifies risk genes for non-small cell lung cancer. J. Theor. Biol. 456, 84–90 (2018).
Chiang, C.-P. et al. Oral lichen planus–Differential diagnoses, serum autoantibodies, hematinic deficiencies, and management. J. Form. Med. Assoc. (2018).
Turner, M. D., Nedjai, B., Hurst, T. & Pennington, D. J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta 1843, 2563–2582, https://doi.org/10.1016/j.bbamcr.2014.05.014 (2014).
Marshall, A., Celentano, A., Cirillo, N., McCullough, M. & Porter, S. Tissue-specific regulation of CXCL9/10/11 chemokines in keratinocytes: Implications for oral inflammatory disease. PLoS one 12, e0172821 (2017).
Sugiyama, T., Kohara, H., Noda, M. & Nagasawa, T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25, 977–988, https://doi.org/10.1016/j.immuni.2006.10.016 (2006).
Soria, G. & Ben-Baruch, A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 267, 271–285 (2008).
Marsland, B. J. et al. CCL19 and CCL21 induce a potent proinflammatory differentiation program in licensed dendritic cells. Immunity 22, 493–505 (2005).
Koch, A. E. et al. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science 258, 1798–1801 (1992).
Krupaa, R. J., Sankari, S. L., Masthan, K. M. & Rajesh, E. Oral lichen planus: An overview. J. Pharm. Bioallied Sci. 7, S158–161, https://doi.org/10.4103/0975-7406.155873 (2015).
Ghaleno, M. N., Shahrekipour, M. & Mahdavifard, H. Evaluation of blood groups in patients with oral lichen planus. Dent. Clin. Exp. J., https://doi.org/10.5812/dcej.9386 (2018).
Mousavi, A. CXCL12/CXCR4 signal transduction in diseases and its molecular approaches in targeted-therapy. Immunol. Lett. 217, 91–115, https://doi.org/10.1016/j.imlet.2019.11.007 (2019).
Garcia-Cuesta, E. M. et al. The Role of the CXCL12/CXCR4/ACKR3 Axis in Autoimmune Diseases. Front. Endocrinol. 10, 585, https://doi.org/10.3389/fendo.2019.00585 (2019).
Isles, H. M. et al. The CXCL12/CXCR4 Signaling Axis Retains Neutrophils at Inflammatory Sites in Zebrafish. Front. Immunol. 10, 1784, https://doi.org/10.3389/fimmu.2019.01784 (2019).
Liu, T., Li, X., You, S., Bhuyan, S. S. & Dong, L. Effectiveness of AMD3100 in treatment of leukemia and solid tumors: from original discovery to use in current clinical practice. Exp. Hematol. Oncol. 5, 19, https://doi.org/10.1186/s40164-016-0050-5 (2015).
Burger, J. A. & Peled, A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia 23, 43–52, https://doi.org/10.1038/leu.2008.299 (2009).
Ichimura, M. et al. Expression profile of chemokines and chemokine receptors in epithelial cell layers of oral lichen planus. J. oral. Pathol. Med. 35, 167–174 (2006).
Amsen, D., de Visser, K. E. & Town, T. Approaches to determine expression of inflammatory cytokines. Methods Mol. Biol. 511, 107–142, https://doi.org/10.1007/978-1-59745-447-6_5 (2009).
Pablos, J. L. et al. Stromal-cell derived factor is expressed by dendritic cells and endothelium in human skin. Am. J. Pathol. 155, 1577–1586, https://doi.org/10.1016/s0002-9440(10)65474-0 (1999).
Sallusto, F. et al. Rapid and coordinated switch in chemokine receptor expression during dendritic cell maturation. Eur J Immunol 28, 2760-2769, https://doi.org/10.1002/(sici)1521-4141 (1998).
Caux, C. et al. Dendritic cell biology and regulation of dendritic cell trafficking by chemokines. Springer Semin. Immunopathol. 22, 345–369 (2000).
Banchereau, J. & Steinman, R. M. Dendritic cells and the control of immunity. Nature 392, 245–252, https://doi.org/10.1038/32588 (1998).
Devi, M., Vijayalakshmi, D., Dhivya, K., Janane, M. & Memory, T. Cells (CD45RO) Role and Evaluation in Pathogenesis of Lichen Planus and Lichenoid Mucositis. J. Clin. Diagn. Res. 11, Zc84–zc86, https://doi.org/10.7860/jcdr/2017/26866.9930 (2017).
Arieta Kuksin, C., Gonzalez-Perez, G. & Minter, L. M. CXCR4 expression on pathogenic T cells facilitates their bone marrow infiltration in a mouse model of aplastic anemia. Blood 125, 2087–2094, https://doi.org/10.1182/blood-2014-08-594796 (2015).
Thongprasom, K., Carrozzo, M., Furness, S. & Lodi, G. Interventions for treating oral lichen planus. Cochrane Database Syst. Rev., Cd001168, https://doi.org/10.1002/14651858.CD001168.pub2 (2011).
Lodi, G., Carrozzo, M., Furness, S. & Thongprasom, K. Interventions for treating oral lichen planus: a systematic review. Br. J. Dermatol. 166, 938–947, https://doi.org/10.1111/j.1365-2133.2012.10821.x (2012).
Acknowledgements
Funding was provided by Fondo Nacional de Desarrollo Científico y Tecnológico (Fondecyt; grant no. 11180170) and Red Estatal de Odontología (grant no. REO19–012).
Author information
Authors and Affiliations
Contributions
C. Rivera, contributed to conception, design, data acquisition, performed all statistical analyses, and interpretation, drafted and critically revised the manuscript; M.F. Crisóstomo, C. Peña, contributed to data acquisition, and interpretation, critically revised the manuscript; P. González-Díaz, W.A. González-Arriagada, contributed to data acquisition, critically revised the manuscript. All authors gave their final approval and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rivera, C., Crisóstomo, M.F., Peña, C. et al. Oral lichen planus interactome reveals CXCR4 and CXCL12 as candidate therapeutic targets. Sci Rep 10, 5454 (2020). https://doi.org/10.1038/s41598-020-62258-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62258-7
- Springer Nature Limited