Abstract
Disease susceptibility of dairy cows is greatest during the transition from pregnancy to lactation. Circulating exosomes may provide biomarkers to detect at-risk cows to enhance health and productivity. From 490 cows, animals at high- (n = 20) or low-risk (n = 20) of transition-related diseases were identified using plasma non-esterified fatty acid and β-hydroxybutyrate concentrations and liver triacylglyceride concentrations during the two weeks post-calving. We isolated circulating exosomes from plasma of dairy cows at low-risk (LR-EXO) and high-risk (HR-EXO), and analyzed their proteome profiles to determine markers for metabolic dysfunction. We evaluated the effects of these exosomes on eicosanoid pathway expression by bovine endometrial stromal (bCSC) and epithelial (bEEL) cells. HR-EXO had significantly lower yield of circulating exosomes compared with LR-EXO, and unique proteins were identified in HR-EXO and LR-EXO. Exposure to LR-EXO or HR-EXO differentially regulated eicosanoid gene expression and production in bCSC and bEEL cells. In bCSC, LR-EXO exposure increased PGE2 and PGD2 production, whereas HR-EXO exposure increased PTGS2 gene expression. In bEEL, HR-EXO exposure caused a decrease in PGE2, PGF2α, PGD2, PGFM and TXB2 production. The unique presence of serpin A3-7, coiled-coil domain containing 88A and inhibin/activin β A chain in HR-EXO, indicates potential biomarkers for cows at-risk for metabolic diseases. Our results are in line with the health status of the cow indicating a potential diagnostic role for exosomes in enhancing cows’ health and fertility.
Similar content being viewed by others
Introduction
The transition from pregnancy to lactation is one of the most challenging periods in a dairy cow’s reproductive cycle. The prevalence of potentially preventable diseases is greatest in the first few weeks after calving, resulting in significant economic losses within dairy industries1. During this period, significant metabolic, hormonal, and immunological changes occur, and the body requires an increase in the intake of energy, protein, and minerals1,2,3. When physiological processes fail to successfully coordinate and regulate these metabolic changes, maladaptation can occur, leading to immune dysfunction and increased risk of metabolic and infectious diseases4,5,6. Management programs aim to reduce peripartum diseases, thereby increasing the possibility of an early return to estrus, a successful pregnancy, and improved milk yields4. Despite the efforts of applying different nutritional strategies, the occurrence of metabolic diseases and impaired reproductive performance has not declined, and, in fact, may have increased6. Biomarkers that can identify pre-pathological diseases have the potential to enhance the effectiveness and profitability of management programs aimed at improving animal health, welfare, and productivity.
In the past decade, new technologies have been utilized to improve diagnostic approaches using exosomes. Exosomes are extracellular vesicles (EVs) 30–120 nm in diameter that are produced by all cell types7,8. Exosomes have been detected in biological fluids including plasma, urine, saliva, and milk9,10,11,12,13. Through the secretion of exosomes, cells can communicate with adjacent cells, or travel long-distances through the circulatory system to provide tissue-specific messages including proteins, lipids, mRNA, and miRNA14. The content of exosomes—and therefore their biological function—is influenced by specific signals from their cell of origin or cues from the cellular milieu10,15,16,17,18. Their interaction with the targeted cell can directly activate membrane receptors or alter the extracellular environment by releasing their contents into the targeted cell19. Circulating exosomes isolated from dairy cows may have potential as indicators of fertility status20, uterine diseases21, and metabolic diseases22, as they carry proteins that are unique to the state of the cow.
Dairy cows have only 12 weeks after calving before needing to reestablish a new pregnancy if they are to maintain a 365-day calving interval, and prostaglandins (PG) play a pivotal role during this period. The general role of endometrial PGF2α is associated with the luteolytic pathway and activation of luteolysis, while endometrial PGE2 is key in the luteotrophic pathway and maternal recognition of pregnancy23. However, if PG concentrations are altered, this period may also be altered resulting in reduced reproductive performance24. We have recently demonstrated that circulating exosomes derived from dairy cows with induced uterine infection decreased the production of PGF2α by endometrial cell lines, showing a direct effect of exosomes on the endometrium’s immune response25. However, the effect of exosomes derived from cows that are at-risk for subclinical and clinical diseases on endometrial cells is yet to be discovered.
We have previously investigated exosomal content from transition cows with high and low risk for metabolic diseases22 and its effect on kidney cell proliferation26; however, these earlier studies were underpowered, with only 5 animals in each group. Here, we greatly expand this research, providing proteome profiles of exosomes derived from the plasma of a larger number of transition cows at high-risk (n = 20) and low-risk (n = 20) for metabolic dysfunction. To investigate the effect of these exosomes on the endometrium, we also evaluated eicosanoid enzyme gene expression and PG production by endometrial cells in an in vitro co-incubation model. We hypothesize that during the transition period, circulating exosomes derived from cows at high-risk for metabolic dysfunction contain proteins that are different to those at low-risk. These differing exosomal cargos may be indicative of health, and, therefore, may provide novel biomarkers for disease risk. Additionally, endometrial cells would respond differently to exosomes from high- vs. low-risk animals, demonstrating direct biological effects on reproductive cells.
Results
Circulating exosomes derived from cows at low- and high-risk follow the accepted definition of exosomes
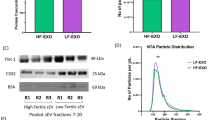
Exosomal fractions isolated from cows at either high- (HR-EXO) or low-risk (LR-EXO) for metabolic dysfunction were both enriched for the exosomal protein markers flotillin 1 (FLOT1) and tumor susceptibility gene 101 (TSG101) (representative immunoblots shown in Fig. 1A), and the vesicles possessed a classical rounded cup shaped morphology (representative TEM images shown in Fig. 1B).
Plasma exosomes from transition cows at low-risk (LR-EXO) and high-risk (HR-EXO) for metabolic dysfunction both confirmed exosomal characteristics and differ in exosome particle numbers. (A) Representative western blot for exosomal markers Flotillin 1 (FLOT1) and tumor susceptibility gene 101 (TSG101) of exosomal fractions 5–16 and their presence in exosome-enriched fractions, those exosome-enriched fractions (exosomal samples) were pooled within each animal. (B) Spherical shape was confirmed in electron micrographs of exosomes from exosomal samples. (C) The size of exosomes (nm) is within the defined size of exosome (30–120 nm), and particle numbers (particle per mL) of exosomal samples was higher in low-risk exosomes. Mann-Whitney test was used to identify the significant differences between groups. ***P ≤ 0.001.
The size of exosomes from LR-EXO and HR-EXO was within the exosome size range and was not significantly different (LR-EXO 85.5 ± 6.07 nm, HR-EXO 85.7 ± 6.68 nm), but exosomal number was reduced (P ≤ 0.001) in HR-EXO relative to LR-EXO (fold change 0.78; Fig. 1C).
Circulating exosomes derived from cows at low- and high-risk had different proteomic profiles
We identified a total of 184 bovine proteins in LR-EXO and 185 in HR-EXO, with 139 found in both groups, 45 uniquely present in the low-risk group, and 46 uniquely present in the high-risk group (Fig. 2A). Proteins present in both groups predominately had functions in signaling (75.9%), as well as being highly enriched for secreted (51.8%), exosome (49.1%) and microparticle (34.8%) components, and extracellular region (27.7%) (Table 1). A high proportion of proteins were also involved in immune response (12.5 to 25%; Table 1). The shared proteins detected are presented in Supplementary Table 1.
Proteomic analysis reveals that protein content differ between plasma exosomes from transition cows at low-risk (LR-EXO) and high-risk (EXO) for metabolic dysfunction. (A) Venn diagram representing unique bovine proteins identifed in LR-EXO (green) or HR-EXO (pink) and common proteins (shaded). Panther analysis revealed differences in bovine proteins from circulating exosomes isolated from LR-EXO cows (B,C) and HR-EXO (D,E) in gene ontology (GO) classifications.
The unique proteins were placed into different GO categories based on the PANTHER Bos taurus database for LR-EXO (Fig. 2B,C, Supplementary Table 2) and HR-EXO (Fig. 2D,E, Supplementary Table 3). Based on molecular function, PANTHER analysis revealed that most of the exosomal proteins had binding functions (LR-EXO = 50.00%; HR-EXO = 40.00%; Fig. 2B,D) and many had catalytic activity functions (LR-EXO = 25.00%; HR-EXO = 30.00%; Fig. 2B,D). In terms of the biological process categorization, most proteins were related to metabolic process (LR-EXO = 28.13%; HR-EXO = 25.00%) and cellular process (LR-EXO = 21.88%; HR-EXO = 14.29%) (Fig. 2C,E). Nearly double the number of proteins that relate to response to stimulus were detected in LR-EXO (6.25%) compared with HR-EXO (3.57%) (Fig. 2C,E).
Incubation of circulating exosomes from animals at low- or high-risk of metabolic diseases leads to changes in eicosanoid signaling in stromal and epithelial cells of the endometrium
To investigate the effect of circulating exosomes from low- and high-risk cows on reproductive tissues, bovine endometrial stromal (bCSC) and epithelial (bEEL) cells were exposed in vitro to exosomes isolated from each group. Eicosanoid enzyme function in bCSC and bEEL was assessed using real-time PCR (Figs 3A–F and 4A–F, respectively), and eicosanoid production by bCSC and bEEL in the cultured media was measured (Figs 3G–L and 4G–L, respectively).
Co-incubation with exosomes from cows at low-risk (LR-EXO) and high–risk (HR-EXO) for metabolic dysfunction lead to changes in eicosanoid expression and secretion in stromal cells. Bovine endometrial stromal cells (bCSC) eicosanoid enzyme gene expressions (A–F) and eicosanoid production (G–L). Values are presented as mean ± SEM. Mann-Whitney test was used to identify the significant differences between groups. *P ≤ 0.05, **P ≤ 0.01.
Co-incubation with exosomes from cows low-risk (LR-EXO) and high–risk (HR-EXO) for metabolic dysfunction lead to changes in eicosanoid expression and secretion in epithelial cells. Bovine endometrial epithelial cells (bEEL) eicosanoid enzyme gene expressions (A–F) and eicosanoid production (G–L). Values are presented as mean ± SEM. Mann-Whitney test was used to identify the significant differences between groups. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
When bCSC were co-incubated with LR-EXO, no differences were observed in gene expression in comparison to control, but the exposure to HR-EXO resulted in an increase in the gene expression of prostaglandin-endoperoxide synthase 2 (PTGS2) and a decrease in the expression of thromboxane A synthase 1 (TBXAS1) relative to the control and LR-EXO exposure, respectively (Fig. 3D,F). In terms of eicosanoid production, the co-incubation of LR-EXO with stromal cells resulted in increased production of PGE2 and PGD2 in comparison to both control and HR-EXO co-incubation (Fig. 3G,I), but thromboxane B2 (TXB2) and PGF2α ethanolamide (PGF2α-EA) production was decreased relative to the control (Fig. 3K,L).
The co-incubation of bEEL with LR-EXO did not alter the gene expression of eicosanoid enzymes, but HR-EXO decreased expression of prostaglandin E synthase 2 (PTGES2) in comparison to the co-incubation of LR-EXO (Fig. 4E). The production of PGE2 and PGF2α-EA was increased when bEEL were exposed to LR-EXO (Fig. 4G,L). HR-EXO resulted in a decrease in the production of PGE2, PGFM and TXB2 in comparison to LR-EXO, a decrease in the production of PGF2α and PGD2 in comparison to control and LR-EXO, and an increase in the production of PGF2α-EA in comparison to control (Fig. 4A–F).
Discussion
Circulating exosomes originate from several different tissues and travel through the blood to targeted tissues or cells, influencing cell behavior. Therefore, circulating exosomes provide a sensitive liquid biopsy of physiological status. In this paper, we provided a comparison of proteins identified in circulating exosomes isolated from dairy cows that were categorized either at high- or low-risk for metabolic diseases during the transition period. The number of circulating exosomes in low-risk cows was greater than in high-risk cows, and exosomal protein content differed between these groups. Additionally, exosomes differentially altered multiple gene expression of eicosanoid enzymes and eicosanoid production by endometrial cells in vitro. Our results suggest that exosomes have roles in postpartum dairy cow adaptation to the lactation period and that the adaptive changes in circulating exosomes may provide biomarkers of disease risk.
The research presented here demonstrates improved sensitivity and specificity in exosome isolation compared with previous research conducted in transition dairy cows. Here, we identified 45 and 46 proteins unique to circulating exosomes from cows at low-risk and high-risk, respectively. Previously, fewer than 15 proteins were identified unique to metabolic status of transition cows22. The coupling of ultracentrifugation method, to isolate EVs, followed by size exclusion chromatography method, to enrich exosomes, removes most of the overabundant free plasma proteins, thereby minimizing confounding proteins and enabling the identification of unique proteins by mass spectrometry. We confirmed that the exosomes isolated followed the accepted definition of an exosome27 by evaluating size (NTA), spherical shape (TEM), and the presence of the exosomal markers TSG101 and FLOT1 (immunoblot). Additionally, our proteomic results confirmed the presence of the plasma-derived exosomal biomarker, CD5 antigen-like (CD5L), in both groups. Common proteins evident in the two groups are involved in intracellular trafficking and vesicular transport, such as sorting nexin-8 (SNX8)28 and trafficking protein particle complex subunit (TRAPPC6B)29. Further, bioinformatics revealed that the common proteins found in LR-EXO and HR-EXO are largely signal, secreted and exosome proteins. The presence of these proteins from our mass spectrometric results, as well as the exosome characterization results, support the enrichment of exosomes from plasma of transition cows with high- and low-risk for diseases and may provide information for determining biomarkers unique to disease status.
Exosomal proteins unique to the risk groups may be indicative of increased or decreased susceptibility to disease. The total number of proteins detected in the risk groups was similar, 45 proteins were unique to LR-EXO and 46 were unique to HR-EXO; however, when we investigated the proteins that were uniquely present in circulating exosomes from cows with high-risk for metabolic diseases, there were proteins with known functions in disease states.
Peptides identified in circulating exosomes of high-risk cows may be indicative of dampened metabolic health. In the current study, we identified serum amyloid A-4 and serpin A3-7 proteins in the exosomes of transition dairy cows at high-risk of disease. Girolami et al. (2018) investigated serum biomarkers of contamination with dioxins and polychlorinated biphenyls (PCBs) in dairy cows and identified seven serum proteins associated with contamination. Of note, serum amyloid A-4, serpin A3-7, and fibrinogen β chain demonstrated a progressive decrease in concentration with decreasing contamination30. Additionally, serpin A3-7 was previously identified in exosomes derived from dairy cows with uterine infection21, and fibrinogen β chain was previously identified in exosomes of high-risk cows one week after calving22. Girolami et al. (2018) did not describe any other metabolic health parameters, such as non-esterified fatty acids (NEFA) or beta-hydroxybutyrate (BHBA), which would elucidate the metabolic health status of the contaminated animals. However, contamination with PCBs detrimentally affects (among other tissues and biological processes) the liver, immune system, and reproduction31. Collectively, the exosomal protein cargo suggests high-risk animals have overall detrimental health compared with low-risk animals.
Other proteins that were identified in exosomes of high-risk groups are associated with clinical diseases. Here, coiled-coil domain containing 88A (CCDC88A) was unique to exosomes of high-risk cows and was previously reported in the kidney cell proteome when co-incubated with exosomes from transition cows at high-risk for metabolic diseases26. Coiled-coil interactions are essential in mammalian cells for vesicular trafficking32 and vesicle-tethering33, which may be one reason why coiled-coil proteins are present in exosomes. However, these proteins have been associated with the fusion machinery of enveloped viral infection27. Our results indicate to the possibility that the presence of CCDC88A in the exosomes of high-risk transition cows and their delivery at infectious sites may increase the cow’s susceptibility to infectious diseases.
Inhibin or activin β A chain (INHBA) was also unique in exosomes from cows categorized at high-risk of metabolic disease. INHBA is a protein that functions as an inhibitor to secretion of follicle stimulating hormone by the pituitary gland28 and has local regulatory roles in various tissues29. Increased expression of INHBA was reported in granulosa cells in bovine cystic ovarian disease, which is a critical cause of infertility in dairy cows34. The known roles of INHBA in influencing fertility of dairy cows and its unique presence in exosomes from transition cows at high-risk for metabolic dysfunction, may warrant further study of this protein in blood and exosomes as an indicator of infertility.
On the other hand, proteins unique to low-risk cows may be indicative of cows that are at low risk of transition diseases. Toll-like receptor 4 (TLR4) was identified uniquely to the low-risk exosomes. TLRs are critical for the innate immune system to recognize pathogen associated molecules and TLR4, in particular, recognizes lipopolysaccharide (LPS) from Gram-negative bacteria35,36, such as Escherichia coli, one of the most common microbes associated with bovine endometrial diseases37. The unique presence of TLR4 in exosomes derived from transition cows at low-risk for diseases might indicate that the targeted cells will receive additional immunity compared with cows designated as high-risk of disease and, as such, be able to better defend themselves against uterine infection. The differences between the two groups may reflect, at least to some extent, the role that exosomes might play in protecting cows from the long-term effects of transition period metabolic disease.
Following the proteomic analysis of the circulating exosomes, we examined their effects on eicosanoid expression and production by endometrial stromal and epithelial cell lines. The biosynthesis of eicosanoids is driven by PTGS1 and 2 by converting arachidonic acid (AA) to prostaglandin endoperoxide H2 (PGH2), which converts rapidly to its metabolites, PGs and TXs, which, together, are called prostanoids. The dominant source of prostanoids is PTGS1, which is constitutively expressed in most cells to maintain homeostasis, but PTGS2 is stimulated in the presence of inflammation to activate eicosanoid pathway38,39,40. However, PTGS2 metabolizes other molecules; anandamide (AEA) and 2-arachidonoylglycerol (2-AG) to prostaglandin-like products, prostamides (PMs) or glycerol esters41,42. In our study, endometrial cells expressed PTGS1 in equal levels in all treatments, which is consistent with its housekeeping function. In endometrial stromal cells, the exposure of HR-EXO increased the expression of PTGS2, but did not stimulate production of downstream PGs, which remained at basal level. HR-EXO were derived from cows that are at high-risk for metabolic dysfunction, and the increase in endometrial PTGS2 expression indicates presence of inflammatory stimuli, which is in line with the health status of cows. LR-EXO also showed a tendency to increase PTGS2, but it was not significant.
Endometrial stromal cell production of PGE2 and PGD2 increased in the presence of LR-EXO signals. Eicosanoids, particularly PGE2, have crucial roles in physiological tissue repair and healing, and is always readily responsive to endometrial inflammation and infection43,44,45,46,47. This suggests that exosomes in cows at low-risk for metabolic dysfunction contain information that makes eicosanoids readily available to heal tissue damage and to respond to inflammation or infection. In endometrial epithelial cells, high production of PGE2 was observed in the presence of LR-EXO; however, HR-EXO caused a more pronounced effect by decreasing PGE2, PGF2α, PGD2, PGFM and TXB2 production. The general pattern of increased eicosanoid production in the presence of LR-EXO and the decrease in the presence of HR-EXO suggests that cows at low-risk may respond faster to inflammation, while the same response in cows at high-risk will be relatively delayed.
A previous study that examined postpartum cows with uterine Escherichia coli infection indicated an increase in circulating PGE2 but not PGFM, and a switch from PGF2α to PGE2 in endometrial explants in response to LPS43, which suggests that PGE2 is the main PG driver. In our study, endometrial stromal and epithelial cells had similar responses to LR-EXO, but not HR-EXO, where PGE2 was increased and PGF2α or PGFM remained unchanged in comparison to basal production. This effect suggests a role for exosomes in a cows’ adaptation to the transition period and quicker recovery responses in cows that are at low-risk for metabolic dysfunction in comparison to those at high-risk.
Although we examined the effects of exosomes on endometrial and epithelial cells, this method is not without limitations. To completely address the roles of exosomes in endometrial eicosanoid interactions during transition period, lipidome analysis of exosome content should be addressed in the future. There is evidence that exosomes can carry eicosanoids or compounds related to their pathways such as phospholipases, arachidonic acid, PGs, leukotrienes, regardless of the species and their involvement in reproduction48,49,50,51. Not only PGs were carried by exosomes but also PTGS1 and 2, demonstrating that exosomes can be a production site for eicosanoids48. Therefore, the lipid content of the exosome could influence the cellular responses of endometrial cell lines after the incubation of the exosomes from the two groups.
Circulatory exosomes can be used to identify potential biomarkers at risk of metabolic disease. Here, we described exosomal cargo that is indicative of decreased liver function, immune function, and overall health in high-risk animals, and a possible link with fertility status. The identification of exosomal proteins, such as Serpin A3-7, CCDC88A and INHBA may be harbingers of additional factors that exacerbate infectious disease and metabolic dysfunction, which have long-been associated with reproductive failure. Overall, the unique proteins identified in cows divergent in metabolic health, indicate the potential for exosomal cargo to be used as biomarkers of health status in transition dairy cows. The current study is a basic discovery approach for the identification of potential biomarkers for cows at risk; however, validation cohorts of the potential candidate proteins through a targeted quantification approach would lead to the development of those proteins as markers. A thorough investigation into the functions of exosomal cargo on eicosanoid production by endometrial cells, may reveal key biological signals that would aid the assessment of health and fertility status of dairy cows. The changes in exosomal content identified here may provide novel targets for the future development of diagnostic biomarkers of metabolic health in cattle; however, additional research will be required to determine if biomarkers can perform better than current methods. We aim to deliver robust and replicable findings to the point at which scalability becomes possible. Further investment may take our initial discoveries to a commercial test.
Materials and Methods
Animals
Holstein-Friesian primiparous cows used in this study were part of a larger experiment (approved by the Ruakura Animal Ethics Committee AEC#13574, #14200) and were managed in a pasture-based, spring-calving dairy system. All experiments were performed in accordance with relevant guidelines and regulations.
From the larger group of calved cows (n = 490), 40 cows were identified as being at either high-risk (n = 20) or low-risk (n = 20) of metabolic dysfunction. This was based on plasma BHBA (mmol/L), plasma NEFA (mmol/L), and liver triacylglycerol (TAG, % of liver wet weight) concentrations within the first two weeks of calving. Risk category of individual cows was based on their metabolic health status 7 and 14 days (plasma BHBA and NEFA) and 10 days (liver TAG) after calving (Table 2). The remaining 450 animals with intermediate risk of metabolic disease were not investigated further.
Blood samples for metabolite analysis (day 7 and day 14) and exosome extraction (day 10) were collected by coccygeal venipuncture into evacuated blood tubes containing either heparin or EDTA anticoagulant, respectively. Blood was placed immediately on ice and centrifuged at 1, 500 × g for 12 min at 4 °C; then, the plasma was aspirated and stored at −20 °C until analysis. Approximately 1 g (wet weight) of liver tissue was collected, on average, 10 days (SD 0.98 d) post-calving for TAG analyses as previously outlined by Roche et al.52. The collected liver samples were snap-frozen in liquid nitrogen and stored at −80 °C until analyses. Gribbles Veterinary Pathology Laboratory undertook the analyses; plasma BHBA in Hamilton (NZ) and plasma NEFA and liver TAG in Dunedin (NZ), respectively. Plasma BHBA was assayed with a Roche reagent kit using colorimetric technique on a Hitachi Modular P800 analyses at 37 °C (Roche Diagnostics), plasma NEFA concentration were measured using a Wako Chemicals kit (Osaka Japan), and liver TAG content analyzed using a Wako LabAssay Triglyceride kit (290–63701, Wako Chemicals USA Inc, USA).
Extracellular vesicle isolation from plasma by ultracentrifugation
Extracellular vesicles (EVs) were isolated from a total of 40 plasma samples by successive differential centrifugation steps, conducted as previously described53. Briefly, the plasma was centrifuged (4 °C) at 2,000 × g for 30 min and then 12,000 × g for 30 min. The supernatant was filtered through a 0.22-μm filter, and centrifuged at 100,000 × g for 2 h at 4 °C using a fixed angle ultracentrifuge rotor Type 70.1 Ti (Beckman Coulter, California, USA). The 100,000 × g pellet (EVs) was reconstituted in 500 µL PBS (Gibco, Thermo Fisher Scientific Australia Pty Ltd, Scoresby Vic) and stored at −80 °C.
Exosome separation and purification by size exclusion columns (SEC)
Extracellular vesicles obtained from ultracentrifugation were loaded onto qEV size exclusion columns (Izon, Oxford, UK) for exosome isolation, as per manufacturer instructions. This technique allows the separation of particles from EV pellet based on their size into 16 fractions. The 16 fractions were concentrated using a vacuum centrifuge for 1.5 h at room temperature. Plasma exosomes of each animal were stored at −80 °C for exosome characterization and further analysis.
Protein quantification
Quantification of protein concentration of exosomal fractions was evaluated using Bicinchoninic Acid (BCA) assay (Sigma-Aldrich, St Louis, MO, USA) and bovine serum albumin (Sigma-Aldrich, St Louis, MO, USA) dilution was used as standards.
Immunoblotting
Immunoblotting followed the same method done by Almughlliq, et al.25 to identify the exosomal fractions, which were then pooled accordingly. Exosomes (10 µg protein) were further characterized using gel electrophoresis (NuPAGE Novex 4–12% Bis-Tris, Thermo Fisher Scientific Australia Pty Ltd, Scoresby Vic) for confirmation of exosomal markers Flotillin 1 (FLOT1) and tumor susceptibility gene 101 (TSG101). The gel was then transferred to a PVDF membrane (Bio-Rad Laboratories Pty., Ltd, Australia) using Trans-Blot Turbo system (Bio-Rad Laboratories Pty., Ltd, Australia). After blocking (5% skim milk powder), membranes were probed overnight with primary antibodies anti-FLOT1 (1:500; Abcam, Cambridge, United Kingdom) and, anti-TSG101 (1:500; Abcam, Cambridge, United Kingdom) at 4 °C, followed by secondary anti-rabbit IgG (1:1000 Sigma–Aldrich, St Louis, MO, USA) and secondary anti-goat (1:1000; Sigma–Aldrich, St Louis, MO, USA), respectively. Membranes were then covered with SuperSignal West Dura-Extended Duration Substrate (Thermo Fisher Scientific Australia Pty Ltd, Scoresby Vic). Targeted proteins were visualized on X-ray films using Konica SRX101A processor (Konica Minolta medical and graphic INC, Japan).
Transmission electron microscopy
Transmission electron microscopy was used to visualize exosome particles from exosomal fractions. Exosomes (5 μL) were added onto formvar coated copper grids for 2 min, then briefly washed in ultrapure water and negatively stained with 1% uranyl acetate. The samples were than visualized using the JEOL 1010 transmission electron microscope operated at 80 kV, and images captured using an Olympus Soft Imaging Veleta digital camera53.
Nanoparticle tracking analysis
Based on the presence of exosomal markers, exosomal fractions 10–16 were pooled. The size distribution and total exosome particle number (particles/mL) of each plasma exosome sample were determined using the nanoparticle tracking instrument (NanoSight NTA 3.1 Nanoparticle Tracking and Analysis Release Version Build 0064) as previously described10.
Proteomic analysis of plasma-derived exosomes by tandem mass spectrometry
Proteomic was processed and analysed following the method done by Almughlliq, et al.21. Each plasma exosome sample (10 μg of protein) was sonicated to expose proteins from the inside of the exosome. Samples were then treated with 50 mM ammonium bicarbonate and 20 mM dithiothreitol and reduced for 1 h at 65 °C. For alkylation, samples were then incubated with 100 mM iodoacetamide in the dark for 1 h at 37 °C, followed by an overnight incubation with trypsin at a 1:50 ratio (Sigma–Aldrich, Castle Hill, NSW, Australia) at 37 °C. The following day, 0.1% formic acid in H2O was added to each sample to stop the trypsin reaction and to acidify the samples. Samples were then desalted according to a modified version of the stage tip protocol54. Briefly, a 3-mm piece of an Empore C18 (Octadecyl) SPE Extraction Disk was excised and placed in a gel loader tip and POROS slurry (5 μL) was added to form a microcolumn. Trifluoroacetic acid (1 volume, 0.1% in water) was added to the sample and loaded onto the microcolumn. The microcolumn was washed with trifluoroacetic acid (20 μL, 0.1% in water). Peptides were eluted from the microcolumn by three washes of acetonitrile (20 μL × 3, 0.1% formic acid). The digested protein samples were dried in a vacuum centrifuge (Eppendorf Concentrator Plus, NSW, Australia) before spectral acquisition. Samples were reconstituted in formic acid (50 μL, 0.1%), vortexed for 10 sec and centrifuged for 2 min at 10, 000 × g to remove particulates10,54,55,56.
Protein samples were then analyzed using the TripleTOF® 5600 mass spectrometer (ABSciex, Redwood City, CA) coupled to an Eksigent nano ultra 1D + HPLC system. The ChromXP C18-CL TRAP (10 mm × 0.3 μm, 120 Å) and analytical ChromXP C18 columns (0.075 µm × 150 mm; 3 µm, 120 Å) (Eksigent, Redwood City, CA) were used to separate the digested proteins. A 5 μL aliquot of digested material was injected onto the column and separated with a linear gradient of 2% to 40% Buffer B for 60 min (Buffer A: 0.1% Formic acid/water; Buffer B: acetonitrile/0.1% formic acid), 40 to 65% Buffer B (5 min), 65 to 95% Buffer B (5 min), with a flow rate of 300 nL/min. The column was flushed with 95% Buffer B for 9 min and re-equilibrated with 2% Buffer B for 10 min. The in-depth proteomic analysis was performed using the Information Dependent Acquisition experiments on the TripleTOF® 5600 System interfaced with a nanospray ion source. The source parameters were as follows: Curtain gas value was 25 (arbitrary units), ion source gas 10 and declustering potential 70 (arbitrary units). A 250 msec accumulation time was set for the TOFMS survey scan and from this scan, the 12 most intense precursor ions were selected automatically for the MS/MS analysis (accumulation time of 150 msecs per MS/MS scan).
MS results were analyzed using the ProteinPilot™ Software (v5.0 beta, AB Sciex, Redwood City, CA), with the Paragon algorithm against UniProt bovine species database (http://www.uniprot.org/). False discovery rate (FDR) was estimated using a reversed sequence database. For a discovery approach, all samples were run separately then protein calls from found uniquely in each group (low or high risk) or shared between groups were combined into three data sets for in silico analysis. Three or more peptides were required to positively identify a protein as present. To further analyze the proteins identified by MS, PANTHER (PANTHER; http://www.pantherdb.org/) was used to place those proteins into different gene ontology (GO) categories on the basis of the PANTHER Bos taurus database57. The data were processed and categorized based on “biological process” and “molecular function” with the ontology and pathway analysis using the “protein analysis through evolutionary relationships” tool.
Endometrial cell culture
Bovine endometrial stromal (bCSC) and epithelial (bEEL) cell lines were kindly gifted by (Université Laval, Québec)58. They were grown in RPMI media (Gibco, Thermo Fisher Scientific Australia Pty Ltd, Scoresby Vic) containing exosome depleted 10% fetal bovine serum (Bovorgen, Interpath Services Pty Ltd, Australia), 1000 U/mL antibiotic-antimycotic solution (Gibco, Thermo Fisher Scientific Australia Pty Ltd, Scoresby Vic) and incubated at 37 °C and 5% CO259. Experiments were conducted in media without fetal bovine serum.
Functional studies of exosomes on endometrial cells
For cell culture, bCSC (seeding density of 8,000 cells per well) and bEEL (seeding density of 35,000 cells per well) were incubated for 24 h grown in RPMI media (Gibco, Thermo Fisher Scientific Australia Pty Ltd, Scoresby Vic) containing 10% fetal bovine serum (Bovorgen, Interpath Services Pty Ltd, Australia) and 1000 U/mL antibiotic-antimycotic solution (Gibco, Thermo Fisher Scientific Australia Pty Ltd, Scoresby Vic). For the co-incubation experiment (treatment with exosomes), FBS free RPMI media, containing 1000 U/mL antibiotic-antimycotic solution (Gibco, Thermo Fisher Scientific Australia Pty Ltd, Scoresby Vic) was used25. Cells were incubated with RPMI media with no addition of exosomes (No EXO control, for baseline measurements), or treated with HR-EXO (n = 20) or LR-EXO (n = 20) with 1 × 108 particles per well for 24 h to analyze gene expression and in separate wells at 36 h to measure eicosanoid expression in culture media at 37 °C and 5% CO2. We performed 2 well replicates per individual cow (n = 40). Exosome concentration for co-incubation was chosen in reference to literature60 and incubation time from Srinivasan et al.61. Cell culture experiments were performed in duplicate per cell line. Cells and cultured media was collected and stored at −80 °C until required for further analyses.
Eicosanoid enzymes gene expression analysis
Gene expression was analyzed after 24 h of LR-EXO and HR-EXO treatments on bEEL and bCSC. Following Almughlliq, et al.62, the bovine custom RT2 Profiler PCR Array (CAPA9696-24; Qiagen, VIC, Australia) was used to quantify the gene expression of eicosanoid enzymes (NAA20 (PPB11826A), FAAH (PPB11713A), PTGS1 (PPB00825A), PTGS2 (PPB00826A), PTGES2 (PPB01910A) and TBXAS1 (PPB04022A)) by reverse-transcriptase quantitative PCR. The PCR arrays were cycled using the following parameters: 1 × (10 min at 95 °C) followed by 40 × (15 sec at 95 °C and 1 min at 60 °C) cycles using the QuantStudio ™ 3 Real-Time PCR System (Applied Biosystems ™, Foster City, CA). The endogenous control genes included on the array were TATA box binding protein (TBP) and hypoxanthine phosphoribosyltransferase 1 (HPRT1). Real-time PCR specificity was confirmed by analyzing the melting curves. PCR reproducibility, reverse transcription efficiency and the presence of genomic DNA contamination were verified before analyzing further. Housekeeper genes were not changed with treatment. Gene expression results were normalized to the endogenous control genes HPRT1 and TBP. Real-time PCR data were analyzed using comparative CT method26,63.
Eicosanoid extraction from cell culture media
Eicosanoid metabolites were extracted from cell culture media for each sample using the following method described in Mitchell et al.64. Briefly, extraction solution was made up using 99% methanol (Sigma-Aldrich, Australia) and 1% of 0.1% formic acid in water containing 50 μL (250 fmol) of each internal standard (IS; 1 μM); for prostaglandin E2-d4 (PGE2-d4), prostaglandin F2α-d4 (PGF2α-d4), PGD2-d4, 13,14-dihydro-15-keto-PGF(2alpha) -d4 (PGFM-d4), prostaglandin E2-ethanolamide-d4 (PGE2-EA-d4), and prostaglandin F2α-ethanolamide-d4 (PGF2α-EA-d4) (Cayman chemicals, USA). In a 96-well extraction plate (Phenomenex, USA), 100 μL of each sample/standard and 250 µL of extraction solution was added and vortexed (900 rpm, 5 min) followed by addition of 750 μL chilled water, place Teflon mat on, and vortexed (900 rpm, 5 min). A Solid Phase Extraction plate (SPE; Phenomenex Australia Pty Ltd) was pre-equilibrated consecutively using 500 μL methanol (Sigma-Aldrich, Australia) and 2 × 500 μL water (Sigma-Aldrich, Australia) via the use of a vacuum manifold. The prepared standards and samples were loaded on to the SPE plate and left to bind for 1 min. The SPE plate was then washed twice with chilled water via the use of a vacuum manifold vacuum (<5 mm Hg). The bound lipid extract preparation was eluted by loading 500 μL acetonitrile and dried using a vacuum concentrator (overnight at room temperature). The dried lipids were then reconstituted in 20% methanol and prepared in triplicate (sample replicates).
Eicosanoid measurements using liquid chromatography tandem mass spectrometry (LC-MS/MS)
For separation of the sample analytes, we used UPLC (Shimadzu, Japan) coupled with a Kinetek C8 column (Phenomenex, USA) attached to a guard column (Phenomenex, USA). Oven temperature was set to 60 °C and 15 min gradient was set up using aqueous and organic mobile phase solvent preparations. A multiple reaction monitoring (MRM) method was carried out for the analyses and quantitation of 6 analytes (PGE2, PGF2α, PGD2, PGFM, TXB2 and PGF2α-EA,) and the deuterated versions (PGE2-d4, PGD2-d4, PGF2α-d4, PGFM-d4, and PGF2α-EA-d4) using negative and positive modes (Table 3). 20 uL of each sample was injected into the mass spectrometer (QTRAP 5500 LCMS system, SCIEX, USA)64.
MultiQuant Software (SCIEX, USA) was used to analyze individual peaks for each analyte. Peaks were selected using +/−0.5 min retention time window. The MultiQuant data was used to generate area and peak area ratio (PAR) information for both standards and samples. Standard curves were plotted with individual concentrations against the PARs, and the line equation was used to extrapolate the concentrations (pg/mL) for the samples.
Statistical analysis
Mann-Whitney test was used to identify the statistically significant differences between control and each treatment. Prism Software (prism7, GraphPad Inc., La Jolla, CA) was used for the statistical test. Data are presented as sample means ± SEM. Statistical significance was defined as P < 0.05.
Data Availability
All data needed to evaluate the conclusion in the paper are present in the paper.
References
Sordillo, L. M. & Raphael, W. Significance of metabolic stress, lipid mobilization, and inflammation on transition cow disorders. Vet Clin North Am Food Anim Pract 29, 267–278, https://doi.org/10.1016/j.cvfa.2013.03.002 (2013).
Bell, A. W. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J Anim Sci 73, 2804–2819 (1995).
Drackley, J. K. ADSA Foundation Scholar Award. Biology of dairy cows during the transition period: the final frontier? J Dairy Sci 82, 2259–2273 (1999).
Roche, J. R. et al. Fertility and the transition dairy cow. Reprod Fertil Dev 30, 85–100, https://doi.org/10.1071/RD17412 (2017).
Goff, J. P. & Horst, R. L. Physiological Changes at Parturition and Their Relationship to Metabolic Disorders1,2. J Dairy Sci 80, 1260–1268, https://doi.org/10.3168/jds.S0022-0302(97)76055-7 (1997).
Roche, J. R., Bell, A. W., Overton, T. R. & Loor, J. J. Nutritional management of the transition cow in the 21st century – a paradigm shift in thinking. Anim Prod Sci 53, 1000–1023, https://doi.org/10.1071/AN12293 (2013).
Sohel, M. M. et al. Exosomal and non-Exosomal transport of extra-cellular microRNAs in follicular fluid: Implications for bovine oocyte developmental competence. PLoS One 8, e78505, https://doi.org/10.1371/journal.pone.0078505 (2013).
Raposo, G. & Stoorvogel, W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200, 373–383, https://doi.org/10.1083/jcb.201211138 (2013).
Koh, Y. Q. et al. Characterization of exosomes from body fluids of dairy cows. J Anim Sci, https://doi.org/10.2527/jas.2017.1727 (2017).
Mitchell, M. D. et al. Plasma exosome profiles from dairy cows with divergent fertility phenotypes. J Dairy Sci 99, 7590–7601, https://doi.org/10.3168/jds.2016-11060 (2016).
Yamada, T., Inoshima, Y., Matsuda, T. & Ishiguro, N. Comparison of methods for isolating exosomes from bovine milk. J Vet Med Sci 74, 1523–1525 (2012).
Sharma, S., Gillespie, B. M., Palanisamy, V. & Gimzewski, J. K. Quantitative nanostructural and single-molecule force spectroscopy biomolecular analysis of human-saliva-derived exosomes. Langmuir 27, 14394–14400, https://doi.org/10.1021/la2038763 (2011).
Alvarez, M. L., Khosroheidari, M., Kanchi Ravi, R. & DiStefano, J. K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int 82, 1024–1032, https://doi.org/10.1038/ki.2012.256 (2012).
Rashed, H. et al. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int J Mol Sci 18, https://doi.org/10.3390/ijms18030538 (2017).
Silverman, J. M. et al. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J Cell Sci 123, 842–852, https://doi.org/10.1242/jcs.056465 (2010).
Sabapatha, A., Gercel-Taylor, C. & Taylor, D. D. Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am J Reprod Immunol 56, 345–355, https://doi.org/10.1111/j.1600-0897.2006.00435.x (2006).
King, H. W., Michael, M. Z. & Gleadle, J. M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 12, 421, https://doi.org/10.1186/1471-2407-12-421 (2012).
Lee, M. J., Park, D. H. & Kang, J. H. Exosomes as the source of biomarkers of metabolic diseases. Ann Pediatr Endocrinol Metab 21, 119–125, https://doi.org/10.6065/apem.2016.21.3.119 (2016).
Mathivanan, S. & Simpson, R. J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000, https://doi.org/10.1002/pmic.200900351 (2009).
Koh, Y. Q. et al. Proteome profiling of exosomes derived from plasma of heifers with divergent genetic merit for fertility. J Dairy Sci, https://doi.org/10.3168/jds.2017-14190 (2018).
Almughlliq, F. B. et al. Proteomic content of circulating exosomes in dairy cows with or without uterine infection. Theriogenology 114, 173–179, https://doi.org/10.1016/j.theriogenology.2018.03.024 (2018).
Crookenden, M. A. et al. Short communication: Proteins from circulating exosomes represent metabolic state in transition dairy cows. J Dairy Sci 99, 7661–7668, https://doi.org/10.3168/jds.2015-10786 (2016).
Arosh, J. A., Parent, J., Chapdelaine, P., Sirois, J. & Fortier, M. A. Expression of cyclooxygenases 1 and 2 and prostaglandin E synthase in bovine endometrial tissue during the estrous cycle1. Biol Reprod 67, 161–169, https://doi.org/10.1095/biolreprod67.1.161 (2002).
Ranasinghe, R. M. et al. Characteristics of prolonged luteal phase identified by milk progesterone concentrations and its effects on reproductive performance in Holstein cows. J Dairy Sci 94, 116–127, https://doi.org/10.3168/jds.2010-3213 (2011).
Almughlliq, F. B. et al. Effect of exosomes from plasma of dairy cows with or without an infected uterus on prostaglandin production by endometrial cell lines. J Dairy Sci 100, 9143–9152, https://doi.org/10.3168/jds.2017-13261 (2017).
Crookenden, M. A. et al. Effect of circulating exosomes from transition cows on Madin-Darby bovine kidney cell function. J Dairy Sci 100, 5687–5700, https://doi.org/10.3168/jds.2016-12152 (2017).
Strauss, H. M. & Keller, S. In Protein-Protein Interactions as New Drug Targets (eds Enno Klussmann & John Scott) 461–482 (Springer Berlin Heidelberg, 2008).
De Jong, F. H. I. Physiol Rev 68, 555–607, https://doi.org/10.1152/physrev.1988.68.2.555 (1988).
Campbell, B. & Baird, D. Inhibin A is a follicle stimulating hormone-responsive marker of granulosa cell differentiation, which has both autocrine and paracrine actions in sheep. J Endocrinol 169, 333–345 (2001).
Girolami, F. et al. Identification of candidate biomarkers of the exposure to PCBs in contaminated cattle: A gene expression- and proteomic-based approach. Sci Total Environ 640–641, 22–30, https://doi.org/10.1016/j.scitotenv.2018.05.284 (2018).
Crinnion, W. J. Polychlorinated biphenyls: persistent pollutants with immunological, neurological, and endocrinological consequences. Altern Med Rev 16, 5–13 (2011).
Wong, M. & Munro, S. Membrane trafficking. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science 346, 1256898, https://doi.org/10.1126/science.1256898 (2014).
Witkos, T. M. & Lowe, M. The Golgin Family of Coiled-Coil Tethering Proteins. Front Cell Dev Biol 3, 86, https://doi.org/10.3389/fcell.2015.00086 (2015).
Stangaferro, M. L. et al. Role of activin, inhibin, and follistatin in the pathogenesis of bovine cystic ovarian disease. Anim Reprod Sci 148, 97–108, https://doi.org/10.1016/j.anireprosci.2014.06.005 (2014).
Wira, C. R., Grant-Tschudy, K. S. & Crane-Godreau, M. A. Epithelial cells in the female reproductive tract: a central role as sentinels of immune protection. Am J Reprod Immunol 53, 65–76, https://doi.org/10.1111/j.1600-0897.2004.00248.x (2005).
Herath, S. et al. Expression and function of Toll-like receptor 4 in the endometrial cells of the uterus. Endocrinology 147, 562–570, https://doi.org/10.1210/en.2005-1113 (2006).
Cronin, J. G., Turner, M. L., Goetze, L., Bryant, C. E. & Sheldon, I. M. Toll-like receptor 4 and MYD88-dependent signaling mechanisms of the innate immune system are essential for the response to lipopolysaccharide by epithelial and stromal cells of the bovine endometrium. Biol Reprod 86, 51, https://doi.org/10.1095/biolreprod.111.092718 (2012).
Smith, W. L., Garavito, R. M. & DeWitt, D. L. Prostaglandin endoperoxide H synthases (cyclooxygenases)-1 and -2. J Biol Chem 271, 33157–33160, https://doi.org/10.1074/jbc.271.52.33157 (1996).
Williams, C. S., Mann, M. & DuBois, R. N. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene 18, 7908–7916, https://doi.org/10.1038/sj.onc.1203286 (1999).
Smith, W. L., DeWitt, D. L. & Garavito, R. M. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem 69, 145–182, https://doi.org/10.1146/annurev.biochem.69.1.145 (2000).
Yu, M., Ives, D. & Ramesha, C. S. Synthesis of prostaglandin E2 ethanolamide from anandamide by cyclooxygenase-2. J Biol Chem 272, 21181–21186 (1997).
Kozak, K. R., Rowlinson, S. W. & Marnett, L. J. Oxygenation of the endocannabinoid, 2-arachidonylglycerol, to glyceryl prostaglandins by cyclooxygenase-2. J Biol Chem 275, 33744–33749, https://doi.org/10.1074/jbc.M007088200 (2000).
Herath, S. et al. Bacterial lipopolysaccharide induces an endocrine switch from prostaglandin F2alpha to prostaglandin E2 in bovine endometrium. Endocrinology 150, 1912–1920, https://doi.org/10.1210/en.2008-1379 (2009).
Shen, Y. et al. PGE2 downregulates LPS-induced inflammatory responses via the TLR4-NF-κB signaling pathway in bovine endometrial epithelial cells. Prostag Leukotr Ess 129, 25–31, https://doi.org/10.1016/j.plefa.2018.01.004 (2018).
Davies, D. et al. Toll-like receptor and antimicrobial peptide expression in the bovine endometrium. Reprod Biol and Endocrin 6, 53, https://doi.org/10.1186/1477-7827-6-53 (2008).
Esser-von Bieren, J. Eicosanoids in tissue repair. Immunol Cell Biol, https://doi.org/10.1111/imcb.12226 (2019).
Kalish, B. T., Kieran, M. W., Puder, M. & Panigrahy, D. The growing role of eicosanoids in tissue regeneration, repair, and wound healing. Prostag Oth Lipid M 104–105, 130–138, https://doi.org/10.1016/j.prostaglandins.2013.05.002 (2013).
Subra, C. et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res 51, 2105–2120, https://doi.org/10.1194/jlr.M003657 (2010).
Deng, Z. B. et al. Exosome-like nanoparticles from intestinal mucosal cells carry prostaglandin E2 and suppress activation of liver NKT cells. J Immunol 190, 3579–3589, https://doi.org/10.4049/jimmunol.1203170 (2013).
Lin, M. C. et al. PGE2 /EP4 Signaling Controls the Transfer of the Mammary Stem Cell State by Lipid Rafts in Extracellular Vesicles. Stem Cells 35, 425–444, https://doi.org/10.1002/stem.2476 (2017).
Majumdar, R., Tavakoli Tameh, A. & Parent, C. A. Exosomes Mediate LTB4 Release during Neutrophil Chemotaxis. PLoS Biol 14, e1002336, https://doi.org/10.1371/journal.pbio.1002336 (2016).
Roche, J. R. et al. Effects of precalving body condition score and prepartum feeding level on production, reproduction, and health parameters in pasture-based transition dairy cows. J Dairy Sci 98, 7164–7182, https://doi.org/10.3168/jds.2014-9269 (2015).
Koh, Y. Q., Almughlliq, F. B., Vaswani, K., Peiris, H. N. & Mitchell, M. D. Exosome enrichment by ultracentrifugation and size exclusion chromatography. Front Biosci 23, 865–874 (2018).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75, 663–670 (2003).
Koh, Y. Q. et al. Characterization of exosomal release in bovine endometrial intercaruncular stromal cells. Reprod Biol Endocrinol 14, 78, https://doi.org/10.1186/s12958-016-0207-4 (2016).
Vaswani, K. et al. Applying SWATH Mass Spectrometry to Investigate Human Cervicovaginal Fluid During the Menstrual Cycle1. Biol Reprod 93(39), 31-10-39, 31-10, https://doi.org/10.1095/biolreprod.115.128231 (2015).
Lopera-Vasquez, R. et al. Extracellular Vesicles from BOEC in In Vitro Embryo Development and Quality. PLoS One 11, e0148083, https://doi.org/10.1371/journal.pone.0148083 (2016).
Fortier, M. A., Guilbault, L. A. & Grasso, F. Specific properties of epithelial and stromal cells from the endometrium of cows. J Reprod Fertil 83, 239–248, https://doi.org/10.1530/jrf.0.0830239 (1988).
Asselin, E., Goff, A. K., Bergeron, H. & Fortier, M. A. Influence of sex steroids on the production of prostaglandins F2 alpha and E2 and response to oxytocin in cultured epithelial and stromal cells of the bovine endometrium. Biol Reprod 54, 371–379 (1996).
Franzen, C. A. et al. Characterization of uptake and internalization of exosomes by bladder cancer cells. Biomed Res Int 2014, 619829, https://doi.org/10.1155/2014/619829 (2014).
Srinivasan, S. et al. TLR-exosomes exhibit distinct kinetics and effector function. Sci Rep 7, 41623, https://doi.org/10.1038/srep41623 (2017).
Almughlliq, F. B. et al. Eicosanoid pathway expression in bovine endometrial epithelial and stromal cells in response to lipopolysaccharide, interleukin 1 beta, and tumor necrosis factor alpha. Reprod Biol 18, 390–396, https://doi.org/10.1016/j.repbio.2018.10.001 (2018).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108 (2008).
Mitchell, M. D., Rice, G. E., Vaswani, K., Kvaskoff, D. & Peiris, H. N. Differential Regulation of Eicosanoid and Endocannabinoid Production by Inflammatory Mediators in Human Choriodecidua. PLoS One 11, e0148306, https://doi.org/10.1371/journal.pone.0148306 (2016).
Acknowledgements
We would like to thank Professor Michel A. Fortier (Université Laval, Québec) for providing us with Bovine endometrial stromal (bCSC) and epithelial (bEEL) cell lines. F.B.A. is supported by a student scholarship from Shimadzu Corporation (2164345), Kyoto, Japan. Y.Q.K. is supported by a student scholarship from a partnership fund (DRCX1302) between the New Zealand Ministry of Business, Innovation and Employment and New Zealand dairy farmers through DairyNZ Inc. The laboratory experiments were funded, in part, by the Australian Research Council (Grant Nos ARC DP170104273 and ARC LP160101854) and the tissues investigated originated from a research platform funded by the aforementioned partnership fund (DRCX1302). We also thank the technical and farm staff at DairyNZ Ltd for collecting the plasma samples, and the scientific and technical assistance of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy and Microanalysis at the University of Queensland.
Author information
Authors and Affiliations
Contributions
F.B.A. performed laboratory experiments, collected and interpreted data, performed statistical analysis and wrote the manuscript. F.B.A., Y.Q.K., S.R., B.J.A., H.N.P. and K.V. performed mass spectrometry analyses and reviewed the data generated. F.B.A., Y.Q.K., S.R., B.J.A., H.N.P., K.V. and M.D.M. designed the experiments. S.M., J.R.R., C.R.B. and M.A.C. designed, conducted and analyzed the animal model. Y.Q.K., H.N.P., K.V., O.H., S.M., J.R.R., C.R.B., M.A.C. and M.D.M. revised and approved the final version of manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41598_2019_50244_MOESM1_ESM.pdf
Supplementary Information: The full unedited blots used in Fig. 1A of exosomal fractions 5–16. The cropped blots presented in Fig. 1A are shown in the boxes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Almughlliq, F.B., Koh, Y.Q., Peiris, H.N. et al. Circulating exosomes may identify biomarkers for cows at risk for metabolic dysfunction. Sci Rep 9, 13879 (2019). https://doi.org/10.1038/s41598-019-50244-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-50244-7
- Springer Nature Limited