Abstract
Here, we evaluate protocol requirements to mimic therapeutically relevant drug concentrations at the site of infection (i.e. lung lesion) in an in-vitro hollow fibre model of infection using pulmonary tuberculosis as a paradigm. Steady-state pharmacokinetic profiles in plasma, lung tissue and lung lesion homogenate were simulated for isoniazid, rifampicin and pyrazinamide and moxifloxacin. An R-shiny User Interface was developed to support conversion of in-vivo pharmacokinetic CMAX, TMAX and T1/2 estimates into pump settings. A monotherapy protocol mimicking isoniazid in lung lesion homogenate (isoniazid CMAX = 1,200 ng/ml, TMAX = 2.2 hr and T1/2 = 4.7 hr), and two combination therapy protocols including drugs with similar (isoniazid and rifampicin (CMAX = 400 ng/ml)) and different half-lives (isoniazid and pyrazinamide (CMAX = 28,900 ng/ml and T1/2 = 8.0 hr)) were implemented in a hollow-fiber system. Drug levels in the perfusate were analysed using ultra-high-performance liquid chromatographic-tandem mass spectrometric detection. Steady state pharmacokinetic profiles measured in the hollow fiber model were similar to the predicted in-vivo steady-state lung lesion homogenate pharmacokinetic profiles. The presented approach offers the possibility to use pharmacological data to study the effect of target tissue exposure for drug combinations. Integration with pharmacokinetics modelling principles through a web interface will provide access to a wider community interested in the evaluation of efficacy of anti-tubercular drugs.
Similar content being viewed by others
Introduction
The predictive value of nonclinical research for antimicrobial drug combinations, such as for tuberculosis, depends on how realistic experimental protocols reflect in-vivo conditions (e.g. nutrition, oxygen, drug exposure). This implies that experimental conditions must be tailored to the in-vivo disease conditions to ensure accurate translation of the findings. Whilst the use of the hollow fiber systems is not new, detailed research protocols are not readily available and it remains unclear whether such factors have been considered when results are reported in the literature. Of note are the challenges associated with regimens including drugs with different pharmacokinetic characteristics (e.g. different distribution, peak concentrations and different elimination half-lives) which need to be mimicked simultaneously within the experimental infection protocol using a single in one hollow fiber system. This oversight can have major implications for the dose rationale as well as the identification of suitable companion or partner compounds for combination therapy1,2.
This is relevant as combination therapy is common practice in the treatment of many infectious diseases, and essential in tuberculosis3. Yet, antibiotic combination regimens and doses have been based on empirical evidence of efficacy and safety. Attempts have been made to evaluate pharmacokinetics (drug concentration vs. time profiles) and pharmacodynamics (e.g. effect on mycobacterial load) in-vivo at the site of infection. However, experimental limitations make it difficult to fully characterise the concentration-effect relationship of each drug as well as their interaction when used as combination therapy. Serial pharmacokinetic and pharmacodynamic data may be difficult to obtain in a clinical setting and pulmonary tuberculosis is a good example of just such a challenging disease entitity. Data at lung lesions/granuloma level can usually only be obtained during a lung resection and not routinely due to the invasive nature of the procedure4,5,6.
Serial sampling for pharmacokinetics and pharmacodynamics could be achieved using the in-vitro hollow fiber model of infection7. Such protocols may enable the evaluation of a wide range of drug exposures8, mimicking drug concentrations likely to occur at the site of infection. This would be useful for novel antimicrobial compounds and combinations in pre-clinical stages and as a method to explore the efficacy of existing antimicrobial compounds being evaluated at different doses or in different combinations9.
The use of hollow fiber has been qualified by the EMA and the FDA as a technique that has value in the pre-clinical setting as complementary tool to existing anti-tuberculosis research methods7,10. As such, the method can be used to evaluate a variety of compounds in monotherapy or in differing combinations, allowing for a more comprehensive characterisation of bacterial killing effects based on factorial designs which remain rather complex, if not impossible, to conduct in-vivo11.
This investigation reports the protocol requirements to mimic therapeutically relevant drug concentrations at the site of infection (i.e. lung lesions). We have selected standard of care compounds (isoniazid, rifampicin, pyrazinamide and moxifloxacin) due to their clinical relevance and availability of pharmacokinetic and biopsy data5. We make the necessary settings and procedures available through a web-based application to the wider community.
Materials and Methods
In-vivo pharmacokinetic profiles
Steady-state total concentrations of isoniazid, rifampicin, pyrazinamide, and moxifloxacin were simulated using compartmental population pharmacokinetic models (Supplementary Material), which mimicked exposure profiles in plasma, lung tissue homogenate and lesion homogenate after standard oral doses (i.e. daily 600 mg rifampicin [450 mg for patients less than 50 kg in body weight], 300 mg isoniazid, 1500 mg pyrazinamide, and 400 mg moxifloxacin). A virtual population of 2,000 patients was simulated according to reported body weight distributions12. This sample size allowed us to evaluate the pharmacokinetic characteristics of the study drugs taking into account the covariate effects (i.e., body weight). For rifampicin, pyrazinamide and moxifloxacin 20 replicates of a virtual South Korean patient population of 100 patients were deemed sufficient to capture the overall variability in drug levels. For isoniazid, 20 replicates of a virtual population of 250 patients were used to ensure allocation of approximately 10% slow metabolisers across each body weight group13. Rifampicin auto-induction was accounted for by multiplying clearance estimates from a single dose model14 by a factor of 1.85. Geometric mean and 95% prediction intervals from the simulations were derived and visualised. Drug distribution from the systemic circulation to the site of infection was evaluated by comparing the ratio of lung/plasma exposure (Rtissue/plasma and Rlesion/plasma) for isoniazid, rifampicin and pyrazinamide or rate-constants ktissue-plasma, kplasma-tissue, klesion-plasma and kplasma-lesion for moxifloxacin (see appendix for further details).
R Shiny web application
An R-shiny web application, was developed (https://pkpdia.shinyapps.io/hfs_app/) to facilitate the conversion of in-vivo pharmacokinetic profiles into in-vitro pump settings in the hollow fiber system. The application supports the generation of pharmacokinetic profiles after monotherapy and combination therapy by converting key pharmacokinetic parameters CMAX, TMAX and T1/2 into pump settings using a published method running at the back end15.
The pump rate for the diluent, elimination, and titration reservoir (including scenarios in which two drugs with different half-lives are used) was calculated as: \({V}_{system}\times \frac{LN(2)}{{t}_{1/2}}\), where t1/2 represents the elimination half-life of the study drug and Vsystem represents the hollow fiber system volume (i.e. central reservoir, cartridge and tubes). Drug amounts required to mimic a certain CMAX were calculated by: CMAX × Vsystem where CMAX represents the maximum concentration. Oral absorption was based on syringe pump rates calculated by: \(\frac{{C}_{MAX}\times {V}_{system}}{{t}_{MAX}}\), where tMAX is the time associated with CMAX. Simultaneous simulation of a short and a long half-life drug in the in-vitro hollow fiber was achieved by topping up the amount in the long half-life drug in the diluent reservoir (i.e. serial setting) or by an additional reservoir (i.e. parallel setting). Subsequently, the pump-rate settings from the dilution reservoir were adjusted accordingly in order to ensure that the Rdilution reservoir + Rtitration reservoir was identical to Relimination reservoir (Fig. 1).
In-vitro pharmacokinetic profiles
Pump settings for three antibiotics, which are commonly used as part of the standard tuberculosis treatment combination, were calculated by a web-application (https://pkpdia.shinyapps.io/hfs_app/) assuming a total system volume of 75 ml. Pharmacokinetic profile characteristics in lung lesions (i.e. CMAX, TMAX and T1/2) for the three antibiotic drugs, based on standard dosing regimens, were selected and accounted for plasma protein binding (i.e. 42% for isoniazid, 83% for rifampicin and 10% for pyrazinamide)16. Three experimental conditions were tested. One monotherapy experiment with isoniazid at CMAX = 1,200 ng/ml, TMAX = 2.2 hour and T1/2 = 4.7 hour was conducted. A combination therapy experiment with similar half-life drugs was conducted using isoniazid at CMAX = 1,200 ng/ml and rifampicin at CMAX = 400 ng/ml with TMAX = 2.2 hour and T1/2 = 4.7 hour. Lastly, a combination therapy experiment with two drugs showing different half-lives was conducted using isoniazid at CMAX = 1,200 ng/ml, TMAX = 2.2 hour and T1/2 = 4.7 hour and pyrazinamide at CMAX = 28,900 ng/ml, TMAX = 2.2 hour and T1/2 = 8.7 hour. All experiments were performed under uninfected conditions at 37 °C. Rifampicin and pyrazinamide TMAX were set to isoniazid TMAX in order to avoid an overly complex experimental setup even though clinical rifampicin and pyrazinamide TMAX are different. Rifampicin T1/2 was harmonised to isoniazid to serve an example of an experiment of drugs with similar half-lives and TMAX and T1/2 values were derived based on the pharmacokinetic model for isoniazid. Subsequent experimental dilution mistakes in the solution to be infused into the central reservoir (i.e. 10, 100 and 2-fold for isoniazid, rifampicin and pyrazinamide), only affecting CMAX, were accounted for as the pump settings were correctly set and therefore TMAX and T1/2 remained unaffected. The mean Prediction Error (MPE) was calculated (\(MPE=\frac{{\sum }_{i=1}^{n}\,D{V}_{i}-IPRE{D}_{i}}{n}\)) with the dependent variable (DV) and model predictions (IPRED) as numerical measure of bias of the pharmacokinetic profiles generated in the hollow fiber model of infection. The MPE was used together with visual inspection to assess how well the in-vitro pharmacokinetic profiles approximated the dashed line that represented those generated by the user. The simulated in-vivo lung lesion homogenate profiles for a typical patient with 95%-prediction intervals, taking into account plasma protein binding, were overlaid to ensure that the simulated in-vitro profiles fell within the prediction intervals. For isoniazid a typical patient with a clearance at the lower end of the distribution, at 13.1 l/hr, was used to mimic a slow metaboliser17.
The integrated hollow fiber experimental protocol comprised a FiberCell Systems Duet pump with a cellulosic cartridge (medium sized – C3008). Drugs were dissolved in 1 ml broth and infused into the central reservoir to mimic an oral absorption profile using a NE-4000 Programmable 2 Channel Syringe Pump. Flow rates, to generate in-vivo mimicking drug elimination profiles, were controlled using a Masterflex C/L Dual-Channel Pump and Masterflex SC0033-LT Tygon E-Lab ext tubing. Samples (2 ml) were taken just before drugs were added to the experiment and 2.2, 5, 8 and 24 hours after, both from the central reservoir and from the extra-capillary space of the cartridge (Fig. 1).
Ultra-high-performance liquid chromatographic-tandem mass spectrometric detection of analytes
Bioanalysis of drug concentrations was performed by chromatographic separation and mass spectrometric detection of three analytes using Acquity ultra-high-performance liquid chromatography (UPLC) system equipped with Waters TQ Detector (Waters, Milford, USA). The UPLC system consists of a binary solvent manager, a sample manager and a column thermostat. ESI-MS detection was carried out in positive ion detection mode. The assay was linear (r2 > 0.993) for isoniazid and rifampicin over the concentration range from 10 ng/mL to 10,000 ng/mL and from 10 ng/mL to 50,000 ng/mL for pyrazinamide. An appropriate dilution scheme was used for sample analysis to match the calibration concentrations. Detailed description on the sample preparation and a LC-MS/MS assay is presented in the Appendix section; Quantification of analytes in-vitro pharmacokinetic profiles.
Results
In-vivo pharmacokinetic profiles
Simulations using the population pharmacokinetic models for isoniazid, rifampicin and pyrazinamide displayed rapid distribution from the plasma into healthy lung tissue homogenate and lung lesion homogenate. It also suggested that lower exposure in the lung tended to remain within variability of the plasma data (Fig. 2). Total drug concentrations in the lung tissue and lung lesion homogenate are lower for isoniazid (9.3% [RSE: 23.5%] and 29.8% [RSE: 21.7%]), rifampicin (24.8% [RSE: 15%] and 52.2% [RSE: 13%]) and pyrazinamide (37.2% [RSE: 7%] and 36.8% [RSE: 8%]) as compared to plasma concentrations (Appendix Table 1). Unlike isoniazid, rifampicin and pyrazinamide, simulations of the profiles of moxifloxacin revealed a nearly doubled elimination half-life in lung tissue and lung lesion homogenate as compared to plasma. This resulted from a substantially faster distribution and diffusion of total moxifloxacin concentrations from plasma to lung tissue (k24 = 1.584 h−1 [37.3% RSE]) or lung lesion (k25 = 0.633 h−1 [79.8% RSE]) as compared to the re-distribution and clearance from lung tissue (k42 = 0.314 h−1 [50.1% RSE]) or lung lesion (k52 = 0.318 h−1 [73.3% RSE]) back to the plasma (Appendix Table 1). The geometric mean ± 1.96 SD of the simulated steady state profiles displayed in Fig. 2 show the between patient variability, which was determined primarily by the plasma data as time series data for lung tissue or lesion homogenate were not available (Appendix Fig. 1). Detailed information about the population pharmacokinetic model development and performance are summarised in the appendix (Appendix Table 1 and Fig. 1).
Facilitating modelling of pharmacokinetic profiles in-vitro
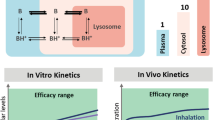
The web application was used to convert secondary pharmacokinetic parameter estimates CMAX, TMAX and T1/2 into pump settings to mimic lung lesion homogenate exposure taking into account protein binding. We have evaluated three antibiotics from the standard tuberculosis treatment combination, including isoniazid, isoniazid plus rifampicin (i.e. same half-lives) and isoniazid plus pyrazinamide (i.e. different half-lives) (Table 1). Prior to TMAX was reached (in all experiments) drugs were infused using a zero-order process into the central reservoir and after TMAX was reached flow rates between the diluent and central reservoir, and central and elimination reservoirs were set at an identical rate. The pharmacokinetic profiles of pyrazinamide (half-live 8.02 hour) and isoniazid (half-live 4.7 hour) were considerably different and this could be adjusted for through a serial top up setting (Fig. 1). The flow rate between the serial top-up reservoir and diluent reservoir was equal to the flow rate between the diluent reservoir and the central reservoir (Table 1). For the example with drugs showing similar half-lives, top-up reservoirs (neither serial nor parallel) were needed (Table 1). The in-vitro pharmacokinetic profiles, accounted for protein binding, tended to show over prediction for isoniazid and rifampicin at the last sampling time point. However, the general trend for the three experiments was acceptable and remained within the prediction intervals for lung lesion homogenate (Fig. 3). The isoniazid MPE was −178, −20.6, and −18.1 ng/ml for the monotherapy, rifampicin combination therapy and pyrazinamide combination therapy experiment, respectively. The rifampicin and pyrazinamide MPE was −37.9 and −1397 ng/ml, respectively. All three drugs displayed adequate penetration of the fibres yielding similar exposure in the central reservoir and extra-capillary space (Fig. 3).
Overview of in-vitro pharmacokinetic profiles obtained in the hollow fiber system for isoniazid monotherapy, isoniazid + rifampicin combination therapy and isoniazid + pyrazinamide combination therapy. Data relflect drug levels after correction for plasma protein binding. Circles and triangles in the concentration vs. time profile plots (top panels) represent single samples in the central reservoir and extra capillary space concentrations, respectively. Dashed lines represent the predicted in-vitro pharmacokinetic profiles obtained by the user interface, which is required for the selection of the pump settings. Shaded areas and solid lines in the concentration vs. time profile plots (top panels) represent the geometric mean and 95%-predicition intervals of a typical profile in humans. The bottom panels show the drug concentrations in the central reservoir and extra capillary space in the hollow fiber system.
Discussion
In this paper we report an in-vitro hollow fiber model of infection integrated with a pharmacokinetic model as a tool for the evaluation of target tissue exposure. Given the complexity of incorporating organ and plasma disposition data into a pharmacokinetic model and implications for infusion rates and titration steps, a user interface was also developed. This interface provides a simple way for investigators to use the hollow fiber in their own investigations.
The In-vitro pharmacokinetic profiles for lung lesion homogenate generated in the hollow fiber system fell within the intervals predicted by the population pharmacokinetic models taking into account plasma protein binding in-vivo. The in-vitro pharmacokinetic profiles generally followed the simulated in-vivo steady-state lung lesion homogenate profiles. However, a trend of higher in-vitro generated concentrations was observed for the later measurements of isoniazid and rifampicin, as compared to the web-application based simulations (Fig. 3). The discrepancies could probably be attributed to the variability in the peristaltic pump and sampling handling. Reproducibility of pharmacokinetic profiles (i.e. between experiment variability) was acceptable, as isoniazid levels in all three experiment were comparable. One way to account for between lab/setup variability, in addition to ensuring accurate calibration of the pumps, is to analyse the in-vitro pharmacokinetic profiles in conjunction to bacterial load level data from the same experimental protocol. Simultaneous modelling of pharmacokinetic and pharmacodynamic data may be especially beneficial when experimental variability is a concern.
In order to avoid over complicating the experimental setup with series of syringe drivers at different infusion rates we have simulated drug levels that correspond to the terminal elimination phase, even though isoniazid displays bi-phasic disposition pharmacokinetics in-vivo. This resulted in a monophasic pharmacokinetic profile in-vitro. Future experiments may consider more realistic scenarios, including repeated rifampicin dosing, which displays auto-induction. Whilst more complex, these scenarios imply further modification of the pump setting during the course of the experiment. Furthermore, our method allowed to standardise TMAX for companion drugs to the treatment backbone, i.e. in this example rifampicin and pyrazinamide were coupled to the same value of TMAX of isoniazid to simplify the experimental setup. Although in-vivo oral absorption is a first order process, for simplification we have adopted an experimental setup for in-vitro oral absorption with a zero-order infusion.
Mimicking representative environmental parameters remains key for the interpretation of data arising from the hollow-fiber model for mycobacterial infections. In the current study, we did not include the contribution of the inoculum as factor to be considered, but the approach proposed here can be used with different drugs and any other aspect of an experimental infection protocol. Moreover, tuberculosis for example has three metabolic states, the log-phase, acid-phase and non-replicative persister phenotype-phase which all tend to reside in different environments. An acid-phase organism often stays within the macrophage whereas the non-replicative persister phenotype-phase tends to reside in the caseum, which renders the environmental pH important. This can be modelled in the hollow-fibre model of infection by modifying the broth, oxygen levels in the incubator, flow rates and adding macrophages. However, this also has implications for drug levels to be simulated. When one uses homogenate levels it is not possible to distinguish and quantify intra- and extra-cellular levels. It is also difficult to assess whether drug levels were similar in the caseum or at the edges of the lesion.
Serial in-vivo exposure vs. time data remains key to inform dose selection for experimental protocols using the hollow fiber system. Drug level data is, however, often quantified as total drug concentrations (i.e. protein unbound plus protein-bound) even though it remains questionable whether total drug levels are the most suitable metrics for further characterisation of pharmacokinetic-pharmacodynamic relationships. Depending on the mechanism of action, possibly free drug concentrations at the site of infection might be more relevant for the antibacterial activity, e.g., rifampicin that prevents Mycobacterium tuberculosis to grow by inhibiting DNA-dependent RNA synthesis18. However, unlike in plasma, little6 or no in-vivo information is available on drug protein binding or non-specific tissue binding at the site of infection in patients (i.e. lung lesions in the case of pulmonary tuberculosis). Total homogenate concentrations could also be misleading in that it does not represent a demarcated physiological compartment, hence in homogenates all cell boundaries have been destroyed, which results in mixed intra- and extra-cellular concentrations19. Often, as only total drug concentrations are available, assumptions are required to conduct a hollow-fiber experiment to ensure other factors such as protein- and/or tissue-drug binding are taken into consideration. First, one could assume that due to poor vascularisation of the lung lesion the equilibrium of free drug between plasma and lung lesions is impaired. A best guess for this scenario is complex, if not impossible, as free drug levels at lung lesion homogenate would be needed. To evaluate this scenario in the current study, total lesion homogenate concentrations were used, after correction for plasma protein binding. Another scenario would assume that free drug can cross the blood vessel and cell membranes yielding free drug concentrations in plasma and lung lesion to be in equilibrium. However, this assumption might not be representative of necrotic foci of the lesion. Alternatively, physiochemical characteristics could be taken into account (e,g, sum of the hydrophobicity and number of aromatic rings), to describe drug distribution and binding to caseum20. In fact, very similar fractions of unbound drug in plasma and caseum for isoniazid, rifampicin and pyrazinamide, were found in in-vitro experiments mimicking caseum and lung lesion and homogenate from rabbits. These findings seem in agreement with our results. Clearly, the scenario in this study where total lesion homogenate concentrations is used might lack translational value, but one needs to acknowledge that each of the alternative proposed methods also have their drawbacks. Only ex-vivo microdialysis data may provide information on free drug levels at the site of infection, although this type of data is rarely available6.
It is important to highlight that assumptions will have to be made on the drug concentration vs. time profiles when the hollow fiber system is used to evaluate novel antibiotic drug combinations for which human pharmacokinetic data is unavailable. Lung lesion drug levels from animal experiments can and should be used in these circumstances. Alternatively, when only plasma level data are available in healthy subjects, predictions of lesion and tissue exposure may be considered using physiochemical properties and physiological-based pharmacokinetic models20.
In summary, we have shown how pharmacokinetic profiles of drugs combinations in humans can be reproduced in the hollow fiber system, mimicking drug levels at the target tissue. We have created an R-shiny interface for other users. Our approach offers an opportunity for drug combinations to be evaluated more thoroughly in the early phases of drug development eliminating some of the uncertainties that limit the evaluation of relevant doses during clinical trials.
Data Availability
Data generated and analysed during the current study are available from the corresponding author on reasonable request.
References
Della Pasqua, O. Translational pharmacology: from animal to man and back. Drug Discov Today Technol 10, e315–317 (2013).
Muliaditan, M., Davies, G. R., Simonsson, U. S. H., Gillespie, S. H. & Della Pasqua, O. The implications of model-informed drug discovery and development for tuberculosis. Drug Discov Today 22, 481–486, https://doi.org/10.1016/j.drudis.2016.09.004 (2017).
Moellering, R. C. Jr. Rationale for use of antimicrobial combinations. Am J Med 75, 4–8 (1983).
Kempker, R. R. et al. Cavitary penetration of levofloxacin among patients with multidrug-resistant tuberculosis. Antimicrobial agents and chemotherapy 59, 3149–3155, https://doi.org/10.1128/AAC.00379-15 (2015).
Prideaux, B. et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nature medicine 21, 1223–1227, https://doi.org/10.1038/nm.3937 (2015).
Heinrichs, M. T. et al. Moxifloxacin target site concentrations in patients with pulmonary TB utilizing microdialysis: a clinical pharmacokinetic study. The Journal of antimicrobial chemotherapy 73, 477–483, https://doi.org/10.1093/jac/dkx421 (2018).
In-vitro hollow fiber system model of tuberculosis (HSF-TB). (European Medicines Agency, London, United Kingdom, 2015).
Drusano, G. L. et al. Analysis of combination drug therapy to develop regimens with shortened duration of treatment for tuberculosis. PLoS One 9, e101311, https://doi.org/10.1371/journal.pone.0101311 (2014).
Boeree, M. J. et al. High-dose rifampicin, moxifloxacin, and SQ109 for treating tuberculosis: a multi-arm, multi-stage randomised controlled trial. Lancet Infect Dis 17, 39–49, https://doi.org/10.1016/S1473-3099(16)30274-2 (2017).
Romero, K., Clay, R. & Hanna, D. Strategic Regulatory Evaluation and Endorsement of the Hollow Fiber Tuberculosis System as a Novel Drug Development Tool. Clin Infect Dis 61(Suppl 1), S5–9, https://doi.org/10.1093/cid/civ424 (2015).
Pasipanodya, J. G., Nuermberger, E., Romero, K., Hanna, D. & Gumbo, T. Systematic Analysis of Hollow Fiber Model of Tuberculosis Experiments. Clin Infect Dis 61(Suppl 1), S10–17, https://doi.org/10.1093/cid/civ425 (2015).
Sahota, T. & Della Pasqua, O. Feasibility of a fixed-dose regimen of pyrazinamide and its impact on systemic drug exposure and liver safety in patients with tuberculosis. Antimicrobial agents and chemotherapy 56, 5442–5449, https://doi.org/10.1128/AAC.05988-11 (2012).
Lee, S. Y. et al. Complete sequencing of a genetic polymorphism in NAT2 in the Korean population. Clin Chem 48, 775–777 (2002).
Smythe, W. et al. A semimechanistic pharmacokinetic-enzyme turnover model for rifampin autoinduction in adult tuberculosis patients. Antimicrobial agents and chemotherapy 56, 2091–2098, https://doi.org/10.1128/AAC.05792-11 (2012).
Blaser, J. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. The Journal of antimicrobial chemotherapy 15(Suppl A), 125–130 (1985).
Lakshminarayana, S. B. et al. Comprehensive physicochemical, pharmacokinetic and activity profiling of anti-TB agents. The Journal of antimicrobial chemotherapy 70, 857–867, https://doi.org/10.1093/jac/dku457 (2015).
Seng, K. Y. et al. Population pharmacokinetic analysis of isoniazid, acetylisoniazid, and isonicotinic acid in healthy volunteers. Antimicrobial agents and chemotherapy 59, 6791–6799, https://doi.org/10.1128/AAC.01244-15 (2015).
Schmidt, S., Barbour, A., Sahre, M., Rand, K. H. & Derendorf, H. PK/PD: new insights for antibacterial and antiviral applications. Curr Opin Pharmacol 8, 549–556, https://doi.org/10.1016/j.coph.2008.06.010 (2008).
Mouton, J. W. et al. Tissue concentrations: do we ever learn? The Journal of antimicrobial chemotherapy 61, 235–237, https://doi.org/10.1093/jac/dkm476 (2008).
Sarathy, J. P. et al. Prediction of Drug Penetration in Tuberculosis Lesions. ACS Infect Dis 2, 552–563, https://doi.org/10.1021/acsinfecdis.6b00051 (2016).
Acknowledgements
We would like to thank Véronique Dartois for providing us with individual bodyweight data in addition to publicly available individual patient concentration-time data which was published as an appendix5. This research was supported by a British Society of Antimicrobial Chemotherapy Grant (GA2015-172R). FK has conducted the research as part of a Medical Research Council fellowship (MR/P014534/1) and consumables to develop the ultra-high-performance liquid chromatographic-tandem mass spectrometric detection method and analyse the samples were paid to Analytical Services International Ltd. from a Medical Research Council fellowship (MR/P014534/1). Neither the British Society of Antimicrobial Chemotherapy nor the Medical Research Council had a role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
F.K. developed the population pharmacokinetic models, the R-shiny User Interface, designed the in-vitro hollow fiber experiments and wrote the manuscript. R.H. conducted all in-vitro experiments and K.K. conducted the drug quantification assays. R.H., K.K., S.H.G. and O.D.P. reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
Oscar Della Pasqua is also Senior Director Clinical Pharmacology at GlaxoSmithKline but declares no financial nor non-financial interests in this research. Karin Kipper is employee at Analytical Services International Ltd. but declares no financial nor non-financial interests in this research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41598_2019_49556_MOESM1_ESM.docx
Supplementary material for: Mimicking in-vivo exposures to drug combinations in-vitro: anti-tuberculosis drugs in lung lesions and the hollow fiber model of infection.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kloprogge, F., Hammond, R., Kipper, K. et al. Mimicking in-vivo exposures to drug combinations in-vitro: anti-tuberculosis drugs in lung lesions and the hollow fiber model of infection. Sci Rep 9, 13228 (2019). https://doi.org/10.1038/s41598-019-49556-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-49556-5
- Springer Nature Limited
This article is cited by
-
Emergence of phenotypic and genotypic antimicrobial resistance in Mycobacterium tuberculosis
Scientific Reports (2022)
-
Microfluidic dose–response platform to track the dynamics of drug response in single mycobacterial cells
Scientific Reports (2022)
-
Clinical Pharmacokinetics and Pharmacodynamics of Imipenem–Cilastatin/Relebactam Combination Therapy
Clinical Pharmacokinetics (2020)