Abstract
Insects harbor a wide variety of microorganisms that form complex and changing communities and play an important role in the biology and evolution of their hosts. Aphids have been used as model organisms to study microorganism-insect interactions. Almost all aphids are infected with the obligate endosymbiont Buchnera aphidicola and can host different bacteria that allow them to acquire traits of agronomic importance, such as resistance to high temperatures and/or defense against natural enemies. However, the bacterial communities of most aphid species remain poorly characterized. In this study, we used high-throughput DNA sequencing to characterize the bacterial communities of Aphis gossypii and Myzus persicae from two cultivable pepper species, Capsicum frutescens (Tabasco variety) and C. annuum (Cayenne variety), in four localities of southwestern Colombia. In addition, we evaluated the dynamics of A. gossypii-associated microorganisms on a seasonal basis. Our results show that the bacterial communities of A. gossypii and M. persicae are dominated by the primary endosymbiont B. aphidicola, while the presence of the facultative symbiont Arsenophonus sp. was only detected in one A. gossypii population from cayenne pepper. In addition to these two known symbionts, eight bacterial OTUs were identified that presented a frequency of 1% or more in at least one of the analyzed populations. The results show that the bacterial communities of aphids associated with pepper crops appears to be structured according to the host aphid species and the geographical location, while no differences were observed in the diversity of bacteria between host plants. Finally, the diversity and abundance of the A. gossypii bacterial community was variable among the four sampling points evaluated over the year and showed a relation with the aphid’s population dynamics. This study represents the first approach to the knowledge of the bacterial community present in chili pepper aphids from Colombia. Nevertheless, more in-depth studies, including replicates, are required to confirm the patterns observed in the microbial communities of aphids from pepper crops.
Similar content being viewed by others
Introduction
Insects are associated with various microorganisms, many of which are capable of significantly affecting different aspects of their biology1. Through long periods of co-evolution, microorganisms and insects have developed complex interactions, ranging from pathogenesis to an obligate mutualism2. Although a great deal of attention has been devoted to pathogenic bacteria, it is likely that most insect species harbor a microbial community that even outnumbers their own cells, which mainly consists of non-pathogenic and free-living bacteria3.
Aphids (Hemiptera: Aphididae) are a diversified group of specialized insects that feed on the sap of plants and have been recognized as model organisms for studying microorganism-insect relationships4. Almost all aphids present a primary endosymbiont, Buchnera aphidicola, which is transmitted in a stable manner from mother to offspring and is essential to compensate for the nutritional deficiencies of these sap-sucking insects5. This endosymbiont has been shown to have a co-speciation pattern with aphids, the result of an ancestral infection and its vertical transmission along the host lineage5.
In addition to Buchnera, aphids can present a series of secondary or facultative symbionts that are inherited vertically but can also be transferred horizontally within and between host species4. Although these symbionts are not necessary for the survival of the host, they appear to replicate only in association with the host and can confer characteristics of agronomic, ecological and evolutionary importance to insects under certain environmental conditions4. For example, these bacteria can increase host resistance to pathogens and natural enemies6. The presence of certain species of bacteria seems to confer resistance to high temperatures, which could affect the range and variability of climates that a host organism tolerates and determine its range of geographical distribution7. These microorganisms can also be involved in determining the host plant that is attacked by aphids8 and can facilitate the transmission of viruses9,10,11, an effect of the symbiont-insect interaction that is of great relevance for agronomic management due to the role that aphids can play as virus vectors for different host plants12,13. The ability of facultative symbionts to introduce new hereditary traits in their hosts can affect both the abundance of symbionts and the success and evolution of aphids4.
Several molecular characterization studies have assessed the diversity of microorganisms in the pea aphid, Acyrthosiphon pisum, focusing on a limited range of symbionts in geographical regions such as Europe, Japan and the United States14,15,16,17,18,19. Studying the diversity of microorganisms based only on the search for known symbionts, limits our knowledge about the possible interactions between members of microbial communities and limits the possibility of finding new symbionts20, especially in non-model aphid species. High-throughput sequencing technologies are now replacing traditional PCR for the investigation of microbial communities21. These sequencing technologies have become an efficient tool to characterize the diversity and structure of bacterial communities in aphids because they do not rely on prior knowledge about the diversity of the communities investigated, allowing for a better understanding of the roles of microorganisms in these insects and the importance of these symbiotic associations18,22,23,25, specially in non-model aphids26,27,28.
The cotton aphid, Aphis gossypii (Glover 1877), is considered one of the most destructive aphid species in the world29. Although this insect has a cosmopolitan distribution, it is particularly abundant in the tropics and attacks at least 64 species of plants of economic importance, primarily of the families Cucurbitaceae, Malvaceae and Solanaceae30. Myzus persicae (Sulzer 1776), known as the green peach aphid, is a cosmopolitan aphid that infests several plants of economic importance, primarily dicots. It has been reported that this aphid can transmit more than 100 viral diseases to approximately 30 different plant families31. Severe infestations of A. gossypii and M. persicae in crops of commercial interest can cause chlorosis, necrosis, wilting, stunted growth, flower and fruit abortion, leaf distortion and defoliation, among other symptoms32. These two aphid species, especially A. gossypii, are considered representative pests of pepper crops29, causing several different production constraints32. The cultivation of this Solanaceae is extremely important to the horticultural production of Colombia, because its growth rate is higher than the average of other crops, with a 13% rate per year33. Three varieties of pepper, Capsicum frutescens (Tabasco var.), C. annuum (Cayenne var.) and C. chinense (Habanero var.), are the most cultivated for domestic consumption and for export purposes.
Despite the economic importance of A. gossypii and M. persicae worldwide, the diversity of their microorganism communities has remained poorly characterized, and no studies have identified factors that may affect these communities. In this study, we performed mass sequencing of the 16S rRNA gene using an Illumina platform to evaluate the effects of the aphid species, host plant, geographical location and the time of year in the structuring of bacterial communities in aphids associated with pepper crops in the Colombian southwest.
Results
Field sampling
Twelve aphid samples, each consisting of a pool of 15 individuals, collected from crops of C. frutescens (Tabasco var.) and C. annum (Cayenne var.) in five localities of southwestern Colombia, were subjected to 16s rRNA Illumina Miseq sequencing, covering variable regions V3 and V4 (Table 1, Fig. 1). Eight samples of A. gossypii and M. persicae were used to compare diversity among localities, aphid species and host plants, and four samples collected in the Yotoco locality were used to evaluate seasonal dynamics of the A. gossypii bacterial community.
Sequence analysis and taxonomic assignment
After filtering for sequence quality and size, between 15392 and 16721 reads were recovered for eight samples of A. gossypii and M. persicae (Table 2). The rarefaction curves tended toward saturation (Fig. S1), and the coverage value of the sequencing data, with a dissimilarity value of 0.03, was greater than 99% in all the samples (Table 2). These results suggest that the depth of sequencing used was adequate to detect most of the bacterial diversity of A. gossypii and M. persicae from pepper crops.
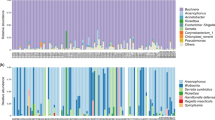
The obtained reads were distributed into 454 OTUs with an identity level of 97%. These OTUs included 22 phyla, with Proteobacteria being the most dominant phylum, with a relative abundance of 93.83% (Average values across all samples, Fig. S2). The dominant class of the samples was Gammaproteobacteria with a relative abundance of 91.3% (Fig. S3). At the family level, Enterobacteriaceae was the dominant family, with a relative abundance of 87.4% (Fig. S4). The relative abundances of the representative OTUs of the bacterial communities of A. gossypii and M. persicae (>1% relative frequency in at least one sample) are shown in Table 3. The values of relative abundances obtained correspond only to approximate proportions of the bacterial OTUs in each sample evaluated.
Two known endosymbionts, B. aphidicola and Arsenophonus sp., were observed, of which B. aphidicola was dominant in the bacterial communities of both aphid species and was observed in all samples, as well as an OTU belonging to an unreported genus of the family Enterobacteriaceae. The highest relative abundance of B. aphidicola was observed in the MpT2 and MpB2 samples of M. persicae, and the lowest relative abundance was observed in the AgD1 sample of A. gossypii (Table 3). In contrast, Arsenophonus sp. was only observed in the AgT2 sample, with a relative abundance of 2.01% (Table 3).
Phylogenetic reconstruction
According to the Greengenes database, the taxonomic assignment agrees with the phylogenetic tree produced with 16S rRNA gene sequences obtained in this study and with GenBank sequences (Fig. 2). The clade formed by the primary endosymbiont, B. aphidicola, was monophyletic, with groups structured according to its host aphid (Fig. 2). Selenomonas sp., Sphingomonas sp. and Pseudomonas sp. grouped into monophyletic clades with bacteria of the same genus (Fig. 2). However, due to the high percentages of similarity (above 97%) with different species of bacteria, a taxonomic resolution at the species level was not achieved. For example, the bacterium Pseudomonas sp. reported in our study, with a relative abundance of 17.48% in the locality of Dagua, was grouped with the species Pseudomonas fulva (MF480331) and with an unknown species of Pseudomonas (MF103739) with 100% similarity (Fig. 2). The secondary endosymbiont Arsenophonus sp., reported in the AgT2 sample, formed a monophyletic clade with sequences of Arsenophonus sp. reported in other insects, such as Bemisia tabaci and Diaphorina citri (Fig. 2). The Bacterium belonging to the family Enterobacteriaceae reported in this study was in a clade close to that of B. aphidicola, as well as a bacterium belonging to the family Enterobacteriaceae reported by GenBank for Aphis glycines (KC097792). However, the two bacteria were not associated with any known symbiont (Fig. 2). The genetic distance (p-distance) between the Enterobacteriaceae reported in A. glycines (KC097792) and the Enterobacteriaceae observed in this study is 0.027. This genetic distance is similar to that observed between the B. aphidicola sequences of A. gossypii and M. persicae reported in this study (0.024–0.026).
Phylogenetic analysis of the representative bacterial OTUs (>1%) associated with A. gossypii and M. persicae from pepper crops (the names of the sequences from our work end in Gallo_Franco and are colored by genus) and related sequences obtained from GenBank (the accession number of each sequence is shown at the end of the name). The phylogenetic inference was made using maximum likelihood and the GTR model.
Comparative analysis of bacterial communities in aphids in localities and in host plants
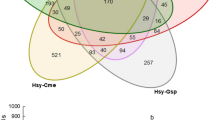
The samples that presented the lowest diversity indices were AgT2 (A. gossypii) and MpT2 (M. persicae), both from the Toro locality (Table 2), while the highest diversity indices were observed for the samples from A. gossypii, AgD1 and AgV1, in the localities of Dagua and Vijes, respectively (Table 2). In those localities (Toro and Vijes) where aphids were collected from the two varieties of pepper, the bacterial community associated with the Tabasco variety was the most diverse according to all measured diversity indices. In the localities of Toro and Bolívar, where both A. gossypii and M. persicae were collected from the Cayenne variety, A. gossypii presented the most diverse bacterial community (Table 2).
Although the diversity of bacteria seems to be greater in aphids collected from the Tabasco pepper variety (Table 2), the Principal Coordinates Analysis (PCoA), performed to determine the structure of the bacterial communities, did not show a clear clustering by host plant (Fig. 3). In the PCoA, Toro was separated from the other localities, and the Cayenne and Tabasco samples in this locality are distant from each other compared with the other localities (Fig. 3). Equally, the AMOVA (Analysis of Molecular Variance) showed that there are no significant differences in the microbial community among aphids from Cayenne and Tabasco (Fs = 1.306, p = 0.215).
The PCoA performed among aphid species showed two groups that differentiated between the M. persicae and A. gossypii samples, especially when the weighted UniFrac index was used (Fig. 4). This result shows that the bacterial communities are structured according to the host aphid. A microbial community composition different from the other localities was still observed in the locality of Toro (Fig. 4).
Seasonal dynamics of the A. gossypii bacterial community
The temporal dynamics analysis of the A. gossypii bacterial community was carried out using one sample for each of the four different times point of the year we studied, for which more than 14000 sequences of the 16S rRNA gene were obtained (Table 4). The rarefaction curves tended toward saturation (Fig. S5), and the coverage value of the sequencing data, with a dissimilarity value of 0.03, was above 99% (Table 4), indicating that the sampling of the microbial communities was adequate to identify most of the bacterial diversity of A. gossypii in the Yotoco experimental plot.
From the 16S rRNA gene sequences, a maximum of 731 OTUs were identified, with 97% identity. These OTUs were assigned to 24 phyla, among which the Proteobacteria phylum predominated with a relative abundance of 88.4% (Fig. S6) and 61 classes, with the class Gammaproteobacteria being predominant with a relative abundance of 84.9%. We identified 139 families, among which Enterobacteriaceae was the most abundant with a relative abundance of 84.6%. The relative abundances of the bacteria most representative of the bacterial community (>1% of relative frequency in at least one sample) are shown in Table 5. The dominant bacteria in all the samples was the primary endosymbiont B. aphidicola, with relative frequencies between 38.4 and 98.7%. The lowest relative abundance of B. aphidicola was observed in the sample taken in December 2016, and the highest relative abundance was observed in June 2017 (Table 5).
The samples from the month of December presented the highest diversity index and the highest number of bacterial OTUs. The other three samplings presented diversity indices similar to each other but lower than that observed in the sample from December. When we compare this seasonal variation in the A. gossypii bacterial community with the average rainfall and temperature data in the Yotoco locality, no relationship was observed between bacterial diversity and environmental conditions (Fig. 5).
Variation in the diversity of the Aphis gossypii bacterial community in four temporal sampling points compared to fluctuations in temperature and precipitation in an experimental plot in the locality of Yotoco (Data taken from the Cenicaña-Yotoco weather station). The gray bars represent the alpha diversity present of the four samplings made throughout the year 2017.
Discussion
In this study, the microbial diversity of A. gossypii and M. persicae from pepper crops of southwestern Colombia was characterized. These two aphid species are of global economic importance, and little is known regarding their interactions with microorganisms, which have proven to be key elements in the evolution and ecology of insects4,34. The results showed that the bacterial communities of A. gossypii and M. persicae are dominated by the endosymbiont B. aphidicola. The dominant presence of the primary endosymbiont in both aphid species is in agreement with the obligate mutualistic relationship that has been reported between these insects and their primary endosymbiont. This relationship is crucial, since aphids are dependent on this microorganism for the production of the essential amino acids, vitamins and sterols that are necessary for their normal development and reproduction35. Similarly, aphids provide nutrients for B. aphidicola, including non-essential amino acids and carbohydrates that are abundant in the aphid diet, which is based on the phloem of plants, or that are produced by the aphids themselves35,36,37.
In addition to the primary endosymbiont B. aphidicola, we found the secondary endosymbiont Arsenophonus sp. in A. gossypii specimens collected from the Cayenne pepper variety from Toro locality. Former studies have found that natural populations of A. gossypii can be associated with six genera of secondary or facultative endosymbionts (Hamiltonella, Arsenophonus, Regiella, Rickettsia, Serratia and Wolbachia)24,38,39,40,41,42,43,44, while natural populations of M. persicae with three genera of symbionts (Regiella, Serratia and Hamiltonella)27. However, despite the biological importance that these endosymbionts may have, they are only partially distributed in the populations of some aphid species and are even completely absent in others24,26,27. The absence of different secondary symbionts in our samples of aphids, may be because maintaining a secondary symbiont, despite being a benefit, can infer a fitness cost to the host45, which suggests that the persistence of a symbiont is determined by the balance between cost and benefit46. Fukatsu et al.47, proposed a simple model to explain the presence or absence of a symbiont in natural populations, in which the infection frequency of an endosymbiont in a host population is determined mainly by three parameters: fidelity of vertical transmission, fitness effect on the host and frequency of horizontal transmission. These patterns of variation of secondary symbionts in natural populations are consistent with previous studies realized in A. gossypii populations. Najar-Rodríguez et al.41, evaluated the bacterial diversity of A. gossypii in localities of Australia and Japan, and reported the presence of the endosymbiont Arsenophonus in some of the evaluated populations. In Chinese populations of A. gossypii, Zhao et al.24 report the presence of the symbionts Arsenophonus and Hamiltonella in all the analyzed populations, nevertheless the other three symbionts were not detected in their study.
Unlike what was believed a few years ago, when Arsenophonus was considered an absent endosymbiont in aphids4,48, recent studies have shown that it is a bacterium widely distributed in various insect species, including those of the family Aphididae49. Jousselin et al.44, in a study of 86 aphid species, reported an Arsenophonus infection incidence of 7%. These researchers reported an especially high incidence of Arsenophonus in the Aphis genus, more than 31% of the species were infected, but absence of Arsenophonus in the Myzus genus. Our results confirm these findings because Arsenophonus sp. was only detected in A. gossypii in populations where both A. gossypii and M. persicae coexisted on the same crop. We report the presence of Arsenophonus sp. in populations of A. gossypii infesting C. annuum plants (Cayenne var.), which had not been reported by Jousselin et al.44 in their study.
Arsenophonus sp. can have different effects on its hosts, including obligate mutualism in blood-sucking insects50, improving the performance of whiteflies51, or through facultative mutualism by protecting psyllids against parasitoid attacks52. Although the effect of Arsenophonus on aphids is still not fully understood, some studies have shown, particularly for the genus Aphis, that Arsenophonus could be involved in important aspects of aphid ecology. Wagner et al.53 observed that Arsenophonus appears to be involved in the food specialization that the polyphagous aphid Aphis craccivora has developed on one of its host plants. Wulff & White54 observed that the presence of Arsenophonus in individual A. glycines aphids improved the performance of this insect on aphid-resistant soybean plants.
In addition to the symbionts B. aphidicola and Arsenophonus sp., eight bacterial OTUs were observed at a frequency of 1% or more in at least one sample. After B. aphidicola, Pseudomonas sp. presented the highest frequency in the locality of Dagua (17.48%) and was observed at a frequency of higher than 1% in two additional samples. The OTU of the genus Pseudomonas observed in this study was phylogenetically grouped with the species Pseudomonas fulva reported in GenBank. P. fulva has been reported as a symbiotic bacterium in Hypothenemus hampei (Coleoptera: Curculionidae), one of the primary pests of coffee. By using caffeine from plants to produce nitrogen, this bacterium allows the coffee borer beetle to survive in coffee plants55. Microorganisms of the genus Pseudomonas, have also been reported as common members of the bacterial community of insects41,56 and it has been suggested that these bacteria have superficial associations with insects, for example, on the surface of the body or, if in the gut, then close to gut orifices. Likewise, Pseudomonas and Sphingomonas, another genus reported in our study, have been previously described in associations with phloem-feeding insects, in low abundances17,23,57,58. Conversely, Selenomonas, genus that was found in all of our samples, it is a group of bacteria uncommon in insects. However, this bacterial genus has been reported as part of the intestinal microbiota of the tick, Amblyomma maculatum59.
Several studies of non-model aphid species have shown that there are bacteria of the family Enterobacteriaceae that have not been described but that may be important in the ecology of aphids22,43. For example, Guidolin & Cônsoli60 suggest that an unknown genus of this family, which they named Cluster B, can play a key role in helping Aphis citricidus to exploit less suitable host plants, complementing the contributions of B. aphidicola. In our study, the presence of a bacterial OTU belonging to the Enterobacteriaceae family was also reported in all the samples of A. gossypii and M. persicae. According to the phylogenetic tree made with sequences of the genebank and genus of known symbionts, this bacterium is closely related to the obligate symbiont Buchnera, as well as another genus of the family Enterobacteriaceae reported for the aphid A. glycines.
The bacterial OTUs found in this study, including the bacterium of the family Enterobacteriaceae, may be playing an important role in the aphid biology that has not yet been reported due to the lack of studies of non-model aphid species. To evaluate this, it is necessary to perform laboratory-based experiments that compare the response of aphids, in terms of performance, in presence or absence of these bacteria, evaluate the location of the symbionts inside the aphids, presence and absence in populations, among others.
By the other hand, there is the possibility that some of these bacteria could be contaminants. Although heritability cannot be discarded, it is likely that this microbes participate in opportunistic associations with these insects, perhaps as gut associates, pathogens, or they could be contaminants from soil, plants or human management. Jousselin et al.61, report that contaminants accounted for a small proportion of the bacteria identified in their samples, but, the removal of sequences accounting for <1% of the reads in aphid samples would eliminate most of the sequences from contaminants present at low frequency in the aphid samples.
The bacterial communities of A. gossypii and M. persicae seem to be structured according to the species of host aphid. The association between the aphid and its bacterial community is affected by a large number of abiotic and biotic factors and may involve the immune system, nutrition, reproduction, communication and many other host systems62. According to Henry et al.27, there is a relationship between the life history traits of the aphids and the cooperative relationships they establish with the symbionts, which may determine the presence of symbionts in certain aphid species or in certain populations within a species. Recent studies have even highlighted that the microbiome is involved in the ability of a given host to transmit a pathogen63.
According to our results, geography can be another factor that influence the composition and frequency of bacteria associated with insects64. In our study, the locality of Toro showed a microbial community that was differentiated from the other localities and was the only one that presented the endosymbiont Arsenophonus. Natural populations of aphids may experience different selection pressures according to geographical location. For example, agricultural management practices65, the dynamics of natural enemies and environmental conditions can affect the diversity and frequency of associated bacteria64. Tsuchida et al.14 observed a geographic cline in the distribution and frequency of secondary endosymbionts, in the pea aphid, A. pisum, which in turn may be related to the host plant species, temperature and precipitation. In the psyllid species Glycaspis brimblecombei, significant geographic variation of the endosymbiont Arsenophonus was also reported52. It is necessary to carry out a greater number of samplings or replicates in each of the localities evaluated in this study to confirm the existing geographical variation, as well as, the prevalence of the Arsenophonus symbiont in the locality of Toro.
According to the AMOVA and PCoA results, there are no significant differences between the A. gossypii microbial communities from the two evaluated pepper species (Cayenne and Tabasco). However, the PCoA shows that the populations of A. gossypii from the Tabasco and Cayenne varieties in Toro are separated by the composition of their bacterial communities and the symbiont Arsenophonus sp. was only detected in the Cayenne variety. In addition, in the Vijes and Toro localities, where both pepper varieties were sampled, and thereby could be compared, simultaneously, it was found the three indices of diversity were higher in the Tabasco variety. These pieces of evidence of possible differences between bacterial communities of Cayenne and Tabasco, can be the starting point for further studies that test the effects of the host plant in aphids.
The bacterial community of A. gossypii showed fluctuations in diversity and frequency between the different seasons of sampling, suggesting that the microbial community of aphids is dynamic over time. When comparing our results with reports of population dynamics of A. gossypii by Melo & Manzano66, in the same experimental plot of Yotoco, and during the same time period of our study, we observed that only the December sampling, which had the highest bacterial diversity index, correspond with a growth in aphid density as reported by these authors. These results propose that the dynamics of the bacterial community depend on the population dynamics of A. gossypii. Smith et al.19 observed that the frequency of bacteria present in aphids can decrease drastically, even over periods as short as three weeks, after seasonal peaks or depending on the frequency of natural enemies. These sudden changes appear to be associated with potentially large costs for aphids, as they have a community of bacteria under certain environmental conditions21. Another hypothesis for obtaining greater diversity in one of the samples (December sampling), is the presence of contaminants, especially because there are no replicates of that same season to compare. Diverse studies have shown that bacterial DNA contamination in extraction kits and laboratory reagents can significantly influence the results of microbiota studies, especially samples containing a low microbial load67. Among bacterial OTUs reported for the sample taken in December, we found the families Comamonadaceae and Chitinophagaceae reported as possible environmental contaminants61 and the family Pelagibacteraceae, reported as commonly present in reagents for the DNA extraction68.
This study represents the first approach to the knowledge of the bacterial community present in chili pepper aphids from Colombia. Our results showed that in addition to the primary endosymbiont B. aphidicola, found in the studied aphids, the secondary symbiont Arsenophonus sp, was also found in one population of A. gossypii. Interesting was finding several genera of bacteria, such as Pseudomonas and a genus of the Enterobacteriaceae family, which have not been reported as symbionts. Although differences were found in the bacterial community of A. gossypii and M. persicae at the locality, plant and season level, it is possible that these differences are related to contamination events or can be the result of stochastic processes, such as genetic drift, founder effect or isolated cases of symbionts acquisition. Due to this, more in-depth studies, including replicates, are required to confirm whether the differences observed persist both seasonally and in the geographical context.
Materials and Methods
Sampling of aphids
Aphid sampling was carried out for crops of C. frutescens Linneo (Tabasco var.) and C. annum Linneo (Cayenne var.), located in five localities of southwestern Colombia (Table 1, Fig. 1). Colonies of aphids were collected systematically, taking into account the edges and center of each plot. Each sampling point was separated by at least 20 m to minimize the probability of collecting the offspring of the same mother. To guarantee that the aphids were free of parasitoids, they were collected alive and brought to the laboratory where they were reared under controlled conditions in an environmental chamber (SANYO-MLR-351H) with a photoperiod of 12 hours of light and 12 hours of darkness at a temperature of 25 °C for fifteen days. Captive-born nymphs were excluded from the study. Next, from the colonies of aphids collected in the field, groups of 15 adult apterous surviving aphids were formed from each of the plots sampled (each individual from a different colony) to form a single sample, which was preserved in 96% ethanol at −20 °C until the extraction of DNA. In those localities where A. gossypii and M. persicae were observed, 15 aphids of each species were collected. The morphological identification of the aphids was carried out using the key from Blackman & Eastop13.
In the locality of Yotoco (Table 1, Fig. 1), the dynamics of the bacterial communities associated with the aphid species A. gossypii were evaluated on a seasonal basis. For this analysis, four samplings were carried out in an insecticide-free experimental plot between September 2016 and June 2017 with intervals of three months (Table 1). The date and the number of samples taken were chosen considering the average temperature and precipitation recorded by IDEAM (the Institute of Hydrology, Meteorology and Environmental Studies) for 2014 and 2015 (http://www.ideam.gov.co/web/tiempo-y-clima/climatologico-mensual, visited in November 2016). The two peaks of maximum precipitation and minimum temperature occurred during the months of March-April and October-November, and the two peaks of minimum precipitation and maximum temperature occurred during the months of December-January and June-July.
DNA extraction, amplification and sequencing of the 16S rRNA gene
Prior to DNA extraction, aphid samples were washed for 5 min in 70% ethanol and rinsed three times with sterile water to remove surface contamination. DNA was extracted from surface sterilized aphids (15 per sample) using a commercial DNeasy® Blood & Tissue kit (QIAGEN) kit, following the manufacturer’s protocol. The amplification of the 16S rRNA gene was carried out using the universal primers Bakt_341F (CCTACGGGNGGCWGCAG) and Bakt_805R (GACTACHVGGGTATCTAATCC)69, which amplify the V3 and V4 hypervariable regions. The sequencing was obtained on a MiSeq Illumina platform generating 300-bp paired reads. Additionally, the NCBI Primer-BLAST tool was used to verify that the primers sequences were present on the genome of the most common symbionts in aphids (S. symbiotica, H. defensa and R. insecticola). The entire amplification and sequencing process was performed by Macrogen Corp. (http://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html).
Data analysis
Sequence analysis and taxonomic assignments
Twenty-thousand sequences per sample were processed using the program Mothur (v 1.39.5)70 (http://www.mothur.org/) according to the protocol described in the MiSeq Standard Operating Procedure71,72. The maximum length of the allowed sequences was 465 bp. We used the Uchime algorithm73, implemented in Mothur to detect chimeric sequences (sequences resulting from the recombination of two different taxa sequences due to jumping events in the PCR). This procedure was carried out for each sample and the identified chimeric sequences were excluded from the data set. The operational taxonomic units (OTUs) were defined at a level of 97% similarity, and a taxonomic assignment was subsequently made using the Greengenes reference taxonomy database as a reference and a minimum confidence value of 0.8 (https://www.mothur.org/wiki/Greengenes-formatted_databases).
Phylogenetic reconstruction
Representative sequences of OTUs with frequencies equal to or greater than 1% in the different samples were selected, as this threshold allowed us to reduce the probability of processing sequences from contaminating bacteria61. Subsequently, these sequences were used to search for symbiotic and free-living bacteria at the databases using the BLASTn tool at NCBI web interface and the database Nucleotides Collection (nr/nt). A phylogenetic analysis of the whole set of sequences was carried out using the maximum likelihood method, which also included GenBank sequences. The alignment was made using the ClustalW algorithm74 in MEGA 775. Phylogenetic reconstruction was achieved using the RAxML-HPC BlackBox program (8.2.8)76 through the CIPRES platform77 with automatic bootstrapping and the other criteria applied by default.
Analysis of bacterial communities among aphids, localities and host plants
From the taxonomic allocation file and their respective absolute frequencies, the relative frequencies were calculated and the OTUs were selected with a representation of at least 1% in some of the samples. Rarefaction curves were generated and diversity descriptors were calculated, including Simpson’s index, the Shannon index and the Chao index using Mothur, to quantify the diversity and dominance of bacteria by aphid species, locality, host plant and season. To explore differences in the structure of the bacterial communities between the aphid species and between the host plants (in the latter comparison, only the A. gossypii samples were included, since they have greater representation in both host plants), a PCoA was realized using Mothur with the weighted (includes relative abundance of each OTU) and unweighted (only absence or presence of each OTU) UniFrac distances. UniFrac is a measure of distance that is used to calculate differences between bacterial communities based on phylogenetic information78. Finally, an AMOVA69,78,79,80,81 was carried out in Mothur to evaluate if there were significant differences between the bacterial communities of the A. gossypii aphid, taking into account the host plant.
References
Bahrndorff, S., Alemu, T., Alemneh, T. & Nielsen L. J. The Microbiome of Animals: Implications for Conservation Biology. International Journal of Genomics. Article ID 5304028 (2016).
Jurkevitch, E. Riding the Trojan horse: Combating pest insects with their own symbionts. Microbial. Biotechnology. 4, 620–627 (2011).
Dillon, R. J. & Dillon, V. M. The gut bacteria of insects: Nonpathogenic interactions. Annual Review of Entomology. 49, 71–92 (2004).
Oliver, K. M., Degnan, P. H., Burke, G. R. & Moran, N. A. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annual Review of Entomology. 55, 247–266 (2010).
Douglas, A. E. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annual Review of Entomology. 43, 17–37 (1998).
Oliver, K. M. & Moran, N. A. Defensive symbionts in aphids and other insects. In Defensive Mutualism in Microbial Symbiosis, ed. White, J. F. & Torres, M. S. 129–47 (2009).
Montllor, C. B., Maxmen, A. & Purcell, A. H. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecological Entomology. 27, 189–95 (2002).
Tsuchida, T., Koga, R. & Fukatsu, T. Host plant specialization governed by facultative symbiont. Science. 303, 1989 (2004).
Gottlieb, Y. et al. The transmission efficiency of Tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. Journal of Virology. 84, 9310–9317 (2010).
Rana, V. S., Singh, S. T., Priya, N. G., Kumar, J. & Rajagopal, R. Arsenophonus GroEL interacts with CLCuV and is localized in midgut and salivary gland of whitefly B. tabaci. Plos One. 7, e42168 (2012).
Su, Q. et al. Insect symbiont facilitates vector acquisition, retention, and transmission of plant virus. Science Reports. 3, 1367 (2013).
Eastop, V. F. The biology of the principle virus vectors, In: Plant virus epidemiology. Ed by Plumb, R. T. & Thresh, J. M. Oxford: Blackwell Scientific Publications. 115–132 (1983).
Blackman, R. L. & Eastop, V. F. Aphids on the World’s Crops: An Identification and Information Guide. John Wiley and Sons, Chichester (2000).
Tsuchida, T., Koga, R., Shibao, H., Matsumoto, T. & Fukatsu, T. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Molecular Ecology. 11(10), 2123–2135 (2002).
Tsuchida, T., Kogal, R., Mengl, X. Y., Matsumoto, T. & Fukatsu, T. Characterization of a Facultative Endosymbiotic Bacterium of the Pea Aphid Acyrthosiphon pisum. Microbial ecology. 49, 126–133 (2005).
Ferrari, J., West, J. A., Via, S., Charles, H. & Godfray, J. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution. 66(2), 375–390 (2011).
Russell, J. A. et al. Uncovering symbiont-driven genetic diversity across North American pea aphids. Molecular Ecology. 22(7), 2045–2059 (2013).
Gauthier, J. P., Outreman, Y., Mieuzet, L. & Simon, J. C. Bacterial communities associated with host-adapted populations of pea aphids revealed by deep sequencing of 16S ribosomal DNA. Plos One. 10, e0120664 (2015).
Smith, A. H. et al. Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Molecular ecology. 25(5), 1135–1149 (2015).
Williamson, S. J. & Yooseph, S. From bacterial to microbial ecosystems (metagenomics). Methods of molecular biology. 804, 35–55 (2012).
Degnan, P. H. & Ochman, H. Illumina-based analysis of microbial community diversity. The ISME Journal. 6, 183–194 (2012).
Chen, D. Q. & Purcell, A. H. Occurrence and transmission of facultative endosymbionts in aphids. Current Microbiology. 34, 220–225 (1997).
Haynes, S. et al. Diversity of Bacteria Associated with Natural Aphid Populations. Applied and environmental microbiology. 69(12), 7216–7223 (2003).
Henry, L. M., Maiden, M. C., Ferrari, J. & Godfray, H. C. Insect life history and the evolution of bacterial mutualism. Ecology Letters. 18(6), 516–25 (2015).
Zytynska, S. E. & Weisser, W. W. The natural occurrence of secondary bacterial symbionts in aphids. Ecological Entomology. 41, 13–26 (2016).
Bansal, R., Mian, M. A. R. & Michel, A. P. Microbiome diversity of Aphis glycines with extensive superinfection in native and invasive populations. Environmental Microbiology Reports. 6, 57–69 (2014).
Jing, X. et al. The bacterial communities in plant phloem-sap-feeding insects. Molecular Ecology. 23, 1433–1444 (2014).
Zhao, Y. et al. Bacterial communities of the cotton aphid Aphis gossypii associated with Bt cotton in northern China. Scientific Reports. 6, 22958 (2016).
Weintraub, P. G. Integrated control of pests in tropical and subtropical sweet pepper production. Pest Management Science. 63, 753–760 (2007).
Carletto, J. et al. Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Molecular Ecology. 18, 2198–2212 (2009).
Barbagallo, S., Cocuzza, G., Cravedi, P. & Komazaki, S. IPM case studies: deciduous fruit trees. In: Aphids as crop pests. ed. by Emden, H. F. v. & Harrington, R. Wallingford. UK: CABI, 651–661, http://www.cabi.org/cabebooks/ebook/20073204012 (2007).
Black, L. L., Green, S. K., Hartman, G. L. & Poulos, J. M. Pepper diseases: A field guide. Asian Vegetable Research and Development Centre publication. 91–101 (1991).
Corporación colombiana internacional. AJÍ COLOMBIANO: Grandes oportunidades de mercado. Portafolio 2007. pp. 7.
Frago, E., Dicke, M. & Godfray, H. C. J. Insect symbionts as hidden players in insect-plant interactions. Trends in Ecology &. Evolution. 27, 705–711 (2012).
Moran, N. A., Dunbar, H. E. & Wilcox, J. L. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. Journal of Bacteriology. 187, 4229–4237 (2005).
Moran, N. A. & Degnan, P. H. Functional genomics of Buchnera and the ecology of aphid hosts. Molecular Ecology. 5, 1251–1261 (2006).
Hansen, A. K. & Moran, N. A. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proceedings of the National Academy of Sciences USA 108, 2849–2854 (2011).
Russell, J., Latorre, A., Sabater-Muñoz, B., Moya, A. & Moran, N. A. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Molecular Ecology. 12, 1061–1075 (2003).
Carletto, J., Gueguen, G., Fleury, F. & Vanlerberghe-Masutti, F. Screening the bacterial endosymbiotic community of sap-feeding insects by terminal-restriction fragment length polymorphism analysis. Entomologia Experimentalis et Applicata. 129, 228–234 (2008).
Desneux, N., Barta, R. J., Hoelmer, K. A., Hopper, K. R. & Heimpel, G. E. Multifaceted determinants of host specificity in an aphid parasitoid. Oecologia 160, 387–398 (2009).
Najar-Rodríguez, A. J., McGraw, E. A., Mensah, R. K., Pittman, G. W. & Walter, G. H. The microbial flora of Aphis gossypii: Patterns across host plants and geographical space. Journal of invertebrate pathology. 100, 123–126 (2009).
Augustinos, A. A. et al. Detection and characterization of Wolbachia infections in natural populations of aphids: Is the hidden diversity fully unraveled? Plos One. 6, e28695 (2011).
Jones, R., Bressan, A., Geenwell, A. M. & Fierer, N. Bacterial Communities of Two Parthenogenetic Aphid Species Cocolonizing Two Host Plants across the Hawaiian Islands. Applied and environmental microbiology. 77(23), 8345–8349 (2011).
Jousselin, E., Coeur d’acier, A., Vanlerberghe-Masutti, F. & Duron, O. Evolution and diversity of Arsenophonus endosymbionts in aphids. Molecular Ecology. 22, 260–270 (2013).
Vorburger, C. & Gouskov, A. Only helpful when required: a longevity cost of harbouring defensive symbionts. Journal of Evolutionary Biology. 24, 1611–1617 (2011).
Oliver, K. M., Smith, A. H. & Russell, J. A. Defensive symbiosis in the real world -advancing ecological studies of heritable, protective bacteria in aphids and beyond. Functional Ecology. 28, 341–355 (2014).
Fukatsu, T., Nikoh, N., Kawai, R. & Koga, R. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera). Applied and Environmental Microbiology. 66, 2748–2758 (2000).
Wille, B. D. & Hartman, G. L. Two species of symbiotic bacteria present in the soybean aphid (Hemiptera: Aphididae). Environmental Entomology. 38, 110–115 (2009).
Duron, O., Wilkes, T. E. & Hurst, G. D. Interspecific transmission of a male-killing bacterium on an ecological timescale. Ecology Letters. 13, 1139–1148 (2010).
Dale, C., Beeton, M., Harbison, C., Jones, T. & Pontes, M. Isolation, pure culture, and characterization of “Candidatus Arsenophonus arthropodicus,” an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Applied. Environmental Microbiology. 72, 2997–3004 (2006).
Raina, H. S. et al. Elimination of Arsenophonus and decrease in the bacterial symbionts diversity by antibiotic treatment leads to increase in fitness of whitefly, Bemisia tabaci. Infection, Genetics and Evolution. 32, 224–230 (2015).
Hansen, A., Jeong, G., Paine, T. & Stouthamer, R. Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California. Applied. Environmental Microbiology. 73, 7531–7535 (2007).
Wagner, S. M. et al. Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Functional Ecology. 29, 1402–1410 (2015).
Wulff, J. A. & White, J. A. The Endosymbiont Arsenophonus Provides a General Benefit to Soybean Aphid (Hemiptera: Aphididae) Regardless of Host Plant Resistance (Rag). Environmental Entomology. 44(3), 574–81 (2015).
Ceja-Navarro, J. A. et al. Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nature comunications. 6, 7618 (2015).
Zahner, V., Lucarotti, C. J. & McIntosh, D. Application of 16S rDNA-DGGE and plate culture to characterization of bacterial communities associated with the sawfly, Acantholyda erythrocephala (Hymenoptera, Pamphiliidae). Current Microbioly. 57(6), 564–569 (2008).
Clark, E. L., Daniell, T. J., Wishart, J., Hubbard, S. F. & Karley, A. J. How conserved are the bacterial communities associated with aphids? A detailed assessment of the Brevicoryne brassicae (Hemiptera: Aphididae) using 16S rDNA. Environmental Entomology. 41, 1386–1397 (2012).
Singh, S. T. et al. Diversity and phylogenetic analysis of endosymbiotic bacteria from field caught Bemisia tabaci from different locations of North India based on 16S rDNA library screening. Infection, Genetics and Evolution. 12, 411–419 (2012).
Budachetri, K. et al. An Insight Into the Microbiome of the Amblyomma maculatum (Acari: Ixodidae). Journal of Medical Entomology. 51(1), 119–129 (2014).
Guidolin, A. S. & Cônsoli, F. L. Symbiont Diversity of Aphis (Toxoptera) citricidus (Hemiptera: Aphididae) as Influenced by Host Plants. Microbial Ecology. 73, 201–210 (2017).
Jousselin, E. et al. Assessment of a 16S rRNA amplicon Illumina sequencing procedure for studying the microbiome of a symbiont-rich aphid genus. Molecular ecology. 16, 628–640 (2016).
Staubach, F., Baines, J. F., Kunzel, S., Bik, E. M. & Petrov, D. A. Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. Plos one. 8(8), e70749 (2013).
Weiss, B. & Aksoy, S. Microbiome influences on insect host vector competence. Trends in Parasitology. 27(11), 514–522 (2011).
Oliver, K. M., Campos, J., Moran, N. A. & Hunter, M. S. Population dynamics of defensive symbionts in aphids. Proceedings of the Royal Society of London B. 275, 293–299 (2008).
Hodgson, F., Obertová, Z., Brown, C. & Lawrenson, R. PSA testing in general practice. Journal of Primary Health Care. 4(3), 199–204 (2012).
Melo, C.I & Manzano, M.R. Influencia de insectos depredadores en poblaciones de Aphis gossypii (Glover) (Hemiptera: Aphididae) en Capsicum frutescens L. Poster presentado en el 44 congreso de la sociedad colombiana de entomología. Bogotá, Colombia (2017, Julio).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biology. 12, 87 (2014).
Glassing, A., Dowd, S. E., Galandiuk, S., Davis, B. & Chiodini, R. J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut. Pathogens. 8, 24 (2016).
Herlemann, D. P. R. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME Journal. 5, 1571–1579 (2011).
Schloss, P. D. et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Applied. Environmental Microbiology. 75(23), 7537–7541 (2009).
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and Environmental Microbiology. 79(17), 5112–5120 (2013).
Kozich, J., Westcott, S., Baxter, N. & Highlander, S. MiSeq SOP. University of Michigan, United States. Visitado en noviembre de 2017, https://mothur.org/wiki/MiSeq_SOP (2013).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 27, 2194–2200 (2011).
Edgar, R. C. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 32(5), 1792–1797 (2004).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 33, 1870–1874 (2016).
Stamatakis, A. RAxML-VI-HPC: Maximum Likelihood-based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics 22(21), 2688–2690 (2006).
Miller, M. A., Pfeiffer, W., & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees in Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA. Pag. 1–8 14 Nov. 2010.
Lozupone, C. & Knight, R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Applied. Environmental Microbiology. 71(12), 8228–8235 (2005).
Excoffier, L., Smouse, P. E. & Quattro, J. M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 131, 479–491 (1992).
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 26, 32–46 (2001).
Martin, A. P. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Applied Environmental Microbiology. 68, 3673–3682 (2002).
Acknowledgements
We acknowledge financial support from the Colombian Science, Technology and Innovation Fund-General Royalties System (Fondo CTeI-Sistema General de Regalías, contract BPIN 2013000100007) and Centre for Bioinformatics and Photonics—CIBioFi. We thank the research group Interacción Planta - Microorganismo - Ambiente (IPMA) from the Universidad Nacional-Palmira for their support in the collection and identification of samples. We thank to Hugo Restrepo y Cía. S.A for the support during the field phase. We thank to the Network and Distributed Systems Laboratory, from Universidad del Valle for their support in the bioinformatic analysis and to the Posgrado en Ciencias-Biología from Universidad del Valle for their academic assistance. We are grateful to N. Rivera, R. Viáfara for his technical assistance and contributions to the data analysis. We are grateful to M. Peñuela for a critical reading of the manuscript and contributions to the data analysis.
Author information
Authors and Affiliations
Contributions
J.J.G. and D.N.D. designed the experiments and collected the samples. J.J.G. and N.T. performed the experiments and analyzed the data. J.J.G. wrote the manuscript. All of the authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gallo-Franco, J.J., Duque-Gamboa, D.N. & Toro-Perea, N. Bacterial communities of Aphis gossypii and Myzus persicae (Hemiptera: Aphididae) from pepper crops (Capsicum sp.). Sci Rep 9, 5766 (2019). https://doi.org/10.1038/s41598-019-42232-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-42232-8
- Springer Nature Limited