Abstract
Hepatitis E virus (HEV) is the causative agent of hepatitis E in humans worldwide. Although hepatitis E is self-limiting without chronic infection development, HEV infection often leads to severe liver diseases causing high mortality in pregnant women in addition to chronic hepatitis and cirrhosis in immunosuppressed patients. In this study, we investigated the effect of a Liriope platyphylla ethanol extract (LPE) on HEV replication. Interestingly, LPE suppressed replication of the genotype 3 HEV replicon. Sequential solvent fractionation revealed that the ethyl acetate (EA) fraction of LPE exerts the most potent inhibitory effects. With the aid of activity-guided fractionation and multi-step column chromatography, spicatoside A was subsequently isolated in the EA fraction of LPE and specifically shown to exert inhibitory effects on replication of the genotype 3 HEV replicon. In addition, spicatoside A interfered with replication of the HEV genotype 3 strain 47832c and expression of HEV ORF2 capsid proteins. Our findings clearly support the potential utility of spicatoside A as an effective anti-HEV agent.
Similar content being viewed by others
Introduction
Hepatitis E virus (HEV), a member of the Hepeviridae family transmitted via the fecal-oral route, is the causative agent of hepatitis E1. The virus has a single-stranded, positive-sense RNA genome 7.2 kb in size with a capped 5′ end and polyadenylated 3′ end2,3.
The HEV genome contains three open reading frames (ORFs) designated ORF1, 2 and 32. ORF1 encodes a non-structural replicase polyprotein with several functional domains including methyltransferase (Met), Y domain, papain-like cysteine protease (PCP), hypervariable region (HVR), X-domain, helicase domain (Hel) and RNA dependent RNA polymerase (RdRp)4. ORF2 encodes the capsid protein that binds cellular proteins, such as heparin sulfate proteoglycan (HSPG), heat-shock protein 90 (HSP90) and glucose-regulated protein 78 (Grp78) while ORF3 encodes a multifunctional phosphoprotein important for release of the HEV virion5,6,7,8. In addition, HEV ORF3 is reported to inhibit the host innate immune response by degrading tumor necrosis factor receptor 1-associated death domain (TRADD) protein, reducing ubiquitination of the receptor interacting protein 1 (RIP1) and suppressing NF-κB activation9.
After entry into the host cell, viral genomic RNA serves as an mRNA for ORF1 translation and a template for replication. The viral genomic RNA translates ORF1 protein, and the largest ORF1 domain, RdRp, synthesizes negative-sense RNA which is used as a template for subsequent synthesis of positive-sense genomic and subgenomic RNAs4. ORF2 and ORF3 are translated from subgenomic RNA, and viral genomic RNA is incorporated into the virion.
HEV isolates infecting humans belong to the Orthohepevirus A species10,11,12,13. Within Orthohepevirus A, four genotypes of HEV are implicated in human infection to date11,13. Genotypes 1 and 2 mainly exist in developing countries. Genotype 1 has been detected in Asia and Africa, and genotype 2 in Mexico and Africa with infections by both genotypes being limited to humans. In contrast, genotypes 3 and 4 are primarily zoonotic. Several animal species, including pigs, wild boar, deer and mongoose, serve as reservoirs of HEV. Genotype 3 has been detected not only in developing but also developed countries, such as Japan and the United States, and the European continent while genotype 4 is more prevalent in Asia12,13. In addition to genotypes 1–4, a human case of genotype 7 infection is reported14.
Approximately 20 million people worldwide are infected with HEV15. Despite its self-limiting nature, HEV infection can trigger a serious liver disease that is fatal in up to 3.3% cases15. More importantly, HEV infection can cause fulminant hepatic failure in pregnant women with an estimated mortality rate of ~30%16,17. In addition, HEV genotype 3 is reported to cause chronic viral hepatitis and liver cirrhosis in organ transplant recipients on immunosuppressive therapy18.
Currently, no specific treatments are available for HEV infection. Ribavirin and interferon α (IFN-α) are generally used to treat immunosuppressed or immunocompromised patients with chronic hepatitis E19. Both ribavirin and IFN-α efficiently suppress HEV but induce long-term side effects20. Attempts to develop novel antivirals against HEV infection have been reported21,22. Anang et al. showed that a cyclic peptide inhibitor blocks HEV release from the infected cells by preventing interactions between HEV ORF3 and the host tumor susceptibility gene 10123. Zinc salts have been demonstrated to inhibit HEV RdRP activity24. Another recent study indicated that natural compound silvestrol inhibits HEV replication25.
The plant species Liriope platyphylla is widely employed as a herbal medicine to treat various chronic diseases, such as diabetes and inflammation, in Korea and China and additionally is reported to possess antiviral activity against hepatitis B virus (HBV)26,27,28,29. In the present study, we investigated the potential antiviral effects of a 70% ethanol extract of L. platyphylla (LPE) and derived bioactive compound(s) on HEV genotype 3 replication.
Results
LPE inhibits replication of the HEV genotype 3 replicon
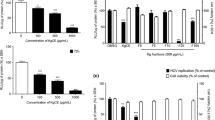
Huh7.5 cells were transfected with transcripts from the pSHEV3-luc replicon and subsequently treated with 10 μg/ml LPE. Four days after transfection, luciferase activity of the pSHEV3-luc replicon was increased 234.7-fold in control (DMSO-treated) cells (Fig. 1). The increase in luciferase activity in LPE-treated cells was significantly lower, up to a value of 67% that in control cells (Fig. 1). To identify the specific solvent fraction of LPE responsible for inhibiting replication of the pSHEV3-luc replicon, LPE was subjected to sequential extraction with ethyl acetate (EA), butanol (n-BuOH) and ddH2O. Huh7.5 cells transfected with transcripts from the pSHEV3-luc replicon were treated with the EA, n-BuOH or ddH2O fraction of LPE (10 μg/ml), and luciferase activity measured 1, 2, 3 and 4 d after transfection. Interestingly, solvent fractions of LPE inhibited replication more effectively than LPE (Fig. 1). Specifically, treatment of cells with the EA, n-BuOH and ddH2O fractions reduced luciferase activity of the pSHEV3-luc replicon to 33.8%, 47.2% and 40.8%, respectively, relative to that in control DMSO-treated cells (Fig. 1).
LPE inhibits replication of the HEV luciferase replicon. Huh7.5 cells were mock-transfected or transfected with capped RNA transcripts from the pSHEV3-luc replicon and luc-pcDNA3. After incubation at 37 °C for 5 h, cells were treated with DMSO (vehicle control), LPE or solvent fractions of LPE (EA, n-BuOH or ddH2O) at a concentration of 10 μg/ml. Cells were re-treated with DMSO, LPE or LPE fractions at 3 d after transfection. At 1, 2, 3 and 4 days after treatment, luciferase activity was determined using a dual luciferase assay system. Luciferase activity of the pSHEV3-luc replicon was expressed as relative light units (RLU) by normalizing Renilla luciferase activity to the constitutive firefly luciferase activity of luc-pcDNA3 transcripts. Relative luciferase activity was calculated by reference to luciferase activity of the pSHEV3-luc replicon in the presence of DMSO 1 d after transfection and defined as 1. P-values were determined by unpaired two-tailed Student’s t-tests (significant at *P < 0.05; **P < 0.01, ***P < 0.001). The data shown here represent four independent experiments.
The EA fraction of LPE inhibits replication of HEV genotype 3 strain 47832c in vitro
To ascertain the inhibitory effects of the EA fraction of LPE against HEV replication, we inoculated A549 cells with HEV genotype 3 strain 47832c, followed by treatment with the EA fraction. At 1, 3, 7 and 14 d after inoculation, HEV copy numbers in cells (cell-associated HEV RNA) and culture media (cell-free HEV RNA) were estimated using qRT-PCR (Fig. 2). At 14 d after inoculation, the amount of HEV RNA in cells was increased 28.3-fold in DMSO-treated control cells (Fig. 2A). In cells treated with the EA fraction, the increase in HEV RNA was 70% lower than that in the DMSO treatment group (Fig. 2A). In parallel, the increase in HEV RNA in the culture media of cells treated with the EA fraction was 52.8% lower than that in the DMSO treatment group (Fig. 2B).
The EA fraction of LPE inhibits replication of the HEV genotype 3 strain 47832c. A549 cells were inoculated with cell culture supernatants containing HEV genotype 3 strain 47832c and incubated at room temperature for 1 h. Cells were treated with either DMSO or the EA fraction of LPE and re-treated again with either DMSO or the EA fraction every 3 d after the initial treatment. At 1, 3, 7 and 14 d after inoculation, (A) the relative amounts of HEV RNA in cells and (B) the amounts of HEV RNA copies in culture media were determined via qRT-PCR using primers specific for HEV ORF1. P-values were determined by unpaired two-tailed Student’s t-tests (significant at *P < 0.05). Relative amounts of cell-associated HEV RNA in the presence of DMSO 1 d after inoculation was defined as 1. The data shown here represent three independent experiments.
The EA fraction of LPE exerts no significant cytotoxic effects against Huh7.5 or A549 cells
To examine the possibility that the inhibitory effects of the EA fraction of LPE on HEV replication are mediated via cytotoxic effects, Huh7.5 and A549 cells were treated with different concentrations of 0, 10, 25, 50 and 100 µg/ml EA and cell viability assessed. During the incubation period, cells were re-treated with the EA fraction of LPE every 3 d after the initial treatment. Notably, after a 4-d treatment period, the EA fraction of LPE exerted no significant cytotoxic effects against Huh7.5 cells, except at the highest concentration (100 µg/ml) where Huh7.5 cell viability was only slightly reduced (87%) (Fig. 3A). In A549 cells, the EA fraction had no cytotoxic effects up to the maximum concentration tested (100 µg/ml) after 14 d treatment (Fig. 3B). As previously reported, HEV genotype 3 strain 47832c infection had no cytotoxic effects (data not shown)30. Furthermore, HEV genotype 3 strain 47832c infection exerted no additional or synergistic cytotoxic effects against A549 cells with the EA fraction of LPE (data not shown). These findings indicate that the antiviral effect of the EA fraction of LPE against HEV is not attributable to cytotoxicity.
Effects of the EA fraction of LPE on cell viability. (A) Huh7.5 or (B) A549 cells were treated with varying concentrations of the EA fraction of LPE (0, 10, 25, 50 and 100 µg/ml). Cells were re-treated with the EA fraction every 3 d after the initial treatment. Huh7.5 and A549 cell viability was determined at 4 and 14 d, respectively, after treatment using the CellTiter-Glo Luminescent Cell Viability Assay. Relative luciferase activity was calculated by reference to RLU of cells treated with 0 µg/ml LPE and defined as 100%. P-values were determined by paired two-tailed Student’s t-tests (significant at *P < 0.05). The data shown here represent four independent experiments.
Identification of Spicatoside A
To determine the chemical constituent(s) in the EA fraction of LPE that inhibit HEV replication, multi-step column chromatography was employed to yield 100 mg pure compound (spicatoside A) (Fig. 4). The complete NMR spectral data for spicatoside A are presented as follows: 1H-NMR (pyridine-d5, 600 MHz): δ 0.82 (3 H, s, 18-CH3), 1.02 (3 H, d, J = 7.02 Hz, 27-CH3), 1.07 (3 H, d, J = 6.96 Hz, 21-CH3), 1.31 (3 H, s, 19-CH3), 1.49 (3 H, d, J = 6.36 Hz, fucose-CH3), 4.82 (1 H, d, J = 7.6 Hz, anomeric H), 5.25 (1 H, d, J = 7.6 Hz, anomeric H), 5.45 (1 H, d, J = 7.82 Hz, anomeric H), 5.53 (1 H, d, J = 5.58, H-6). 13C-NMR ((pyridine-d5, 600 MHz): δ 82.74 (C-1), 37.11 (C-2), 67.69 (C-3), 43.46 (C-4), 139.5 (C-5), 124.28 (C-6), 32.15 (C-7), 32.73 (C-8), 50.13 (C-9), 42.63 (C-10), 23.39 (C-11), 40.13 (C-12), 39.90 (C-13), 56.70 (C-14), 31.78 (C-15), 82.65 (C-16), 62.58 (C-17), 16.5 (C-18), 14.6 (C-19), 42.21 (C-20), 14.86 (C-21), 109.37 (C-22), 26.12 (C-23), 25.95 (C-24), 27.36 (C-25), 64.76 (C-26), 16.05 (C-27), 100.3 (fuc-1), 80.96 (fuc-2), 83.5 (fuc-3), 74.87 (fuc-4), 71.73 (fuc-5), 16.95 (fuc-6), 106.02 (xyl-1), 76.32 (xyl-2), 78.16 (xyl-3), 70.90 (xyl-4), 67.06 (xyl-5), 104.7 (glc-1), 80.96 (glc-2), 78.68 (glc-3), 72.05 (glc-4), 78.41 (glc-5), 63.08 (glc-6) (Fig. 5A).
Determination of the anti-HEV effect of spicatoside A. (A) Spicatoside A structure. (B) Concentration-dependent inhibitory effects of spicatoside A on luciferase activity of the pSHEV3-luc replicon. Huh7.5 cells were mock-transfected or transfected with capped RNA transcripts from the pSHEV3-luc replicon and luc-pcDNA3. After incubation at 37 °C for 5 h, cells were treated with either DMSO or spicatoside A at concentrations of 0, 0.5, 1, and 2 µg/ml. Cells were re-treated with spicatoside A every 3 d after the initial treatment. At 4 d after treatment, luciferase activity was determined using a dual luciferase assay system. The luciferase activity of the pSHEV3-luc replicon was expressed in RLU by normalizing Renilla luciferase activity to constitutive firefly luciferase activity of luc-pcDNA3 transcripts. Relative luciferase activity was calculated by reference to luciferase activity of the pSHEV3-luc replicon in the presence of DMSO 4 d after transfection and defined as 100. P-values were determined by unpaired two-tailed Student’s t-tests (significant at ***P < 0.001). The data shown here represent six independent experiments.
Spicatoside A inhibits replication of the HEV genotype 3 replicon in a concentration-dependent manner
The effects of spicatoside A, a major chemical constituent of the EA fraction of LPE (Fig. 5A), on luciferase activity of the pSHEV3-luc replicon were investigated. Huh7.5 cells transfected with transcripts from the pSHEV3-luc replicon were treated with 0, 0.5, 1 and 2 µg/ml spicatoside A, and luciferase activity of the replicon determined at four days after transfection. At doses of 0.5, 1 and 2 µg/ml, spicatoside A significantly reduced the luciferase activity of the pSHEV3-luc replicon (Fig. 5B). Our analyses yielded an estimated 50% inhibitory concentration (IC50) value of spicatoside A against pSHEV3-luc replicon of 0.469 ± 0.06 µg/ml (Fig. 5B). In addition, the effects of spicatoside A on replication of HEV genotype 3 strain 47832 were investigated using qRT-PCR (data not shown). Similar to data obtained with the pSHEV3-luc replicon, spicatoside A interfered with replication of HEV genotype 3 strain 47832c with an estimated IC50 of 0.727 ± 0.15 µg/ml.
Spicatoside A inhibits replication of HEV genotype 3 strain 47832c and expression of HEV ORF2 in vitro
The inhibitory effects of spicatoside A against HEV replication were further established by inoculating A549 cells with HEV genotype 3 strain 47832c and treating with 2 µg/ml spicatoside A. At 1, 3, 7 and 14 d after inoculation, HEV copy numbers in cells and culture media were estimated using qRT-PCR (Fig. 6A,B). Our results showed a 32.3-fold increase in the amount of HEV RNA in DMSO-treated control cells at 14 d after inoculation (Fig. 6A). Interestingly, in cells treated with spicatoside A, the increase in HEV RNA was 15.4% lower than that in the DMSO treatment group (Fig. 6A). In addition, the increased in HEV RNA in the culture media of cells treated with spicatoside A was 23.3% lower than that in the DMSO treatment group (Fig. 6B).
Spicatoside A inhibits replication of the HEV genotype 3 strain 47832c. A549 cells were inoculated with cell culture supernatants containing HEV genotype 3 strain 47832c and incubated at room temperature for 1 h, followed by treatment with either DMSO or spicatoside A. Cells were re-treated with either DMSO or spicatoside A every 3 d after the initial treatment. At 1, 3, 7 and 14 d after inoculation, (A) the relative amounts of HEV RNA in cells and (B) the amounts of HEV RNA copies in culture media were determined via qRT-PCR using primers specific for HEV ORF1. Relative amounts of cell-associated HEV RNA in the presence of DMSO 1 d after inoculation was defined as 1. The data shown here represent three independent experiments. At 14 d after inoculation, expression of HEV ORF2 capsid protein was determined via (C) western blot and (D) immunofluorescence microscopy. BF, bright-field microscopic image; FL, fluorescence microscopic image. P-values were determined by unpaired two-tailed Student’s t-tests (significant at *P < 0.05; **P < 0.01).
To further confirm the effects of spicatoside A on replication of HEV genotype 3 strain 47832c, western blot and immunofluorescence microscopy were applied to analyze expression of the HEV ORF2 capsid protein. A549 cells inoculated with HEV genotype 3 strain 47832c were treated with different concentrations of 0, 0.2, 0.5, 1 and 2 µg/ml spicatoside A. At 14 d after inoculation, ORF2 was robustly expressed in DMSO-treated cells inoculated with HEV genotype 3 strain 47832c (Fig. 6C, compare lane 2 with 1). Notably, treatment with spicatoside A significantly reduced ORF2 expression in inoculated cells compared with vehicle controls in a dose-dependent manner (Fig. 6C, compare lane 2 with lanes 3 to 6). Consistent with western blot, down-regulation of HEV ORF2 protein was observed by immunofluorescence microscopy (Fig. 6D, compare lane 3 with lanes 4 and 5). The collective data clearly demonstrate that spicatoside A effectively inhibits HEV replication and ORF2 expression.
Effects of Spicatoside A on viability of Huh7.5 and A549 cells
To determine whether spicatoside A exerts cytotoxic effects, we treated Huh7.5 and A549 cells with the compound at concentrations of 0, 1, 2, 5 and 10 µg/ml and assessed cell viability. After a 4 d treatment period, spicatoside A at 5 and 10 µg/ml doses exerted significant cytotoxic effects against Huh7.5 cells inducing reduced viability to 55.9% and 31%, respectively (Fig. 7A). In A549 cells, spicatoside A had no cytotoxic effects up to 5 µg/ml after 14 d treatment (Fig. 7B) while, at 10 µg/ml spicatoside A, viability was significantly reduced to 28.6% (Fig. 7B). The IC50 values of spicatoside A for inhibition of Huh 7.5 and A549 cell viability were determined as 7.169 ± 9.64 and 7.486 ± 0.28 µg/mL, respectively. HEV genotype 3 strain 47832c infection exerted no additional or synergistic cytotoxic effects against A549 cells with spicatoside A (data not shown). In view of the finding that spicatoside A inhibits HEV replication and ORF2 expression at concentrations that exert no cytotoxic effects (<2 µg/ml), we propose that the antiviral activity of spicatoside A is not attributable to cytotoxicity.
Effects of spicatoside A on cell viability. (A) Huh7.5 or (B) A549 cells were treated with varying concentrations of spicatoside A (0, 1, 2, 5 and 10 µg/ml). Cells were re-treated with spicatoside A every 3 d after the initial treatment. Huh7.5 and A549 cell viability was determined at 4 and 14 d, respectively, after treatment using the CellTiter-Glo Luminescent Cell Viability Assay. Relative luciferase activity was calculated by reference to RLU of the 0 µg/ml LPE treatment group and defined as 100%. P-values were determined by unpaired two-tailed Student’s t-tests (significant at **P < 0.01; ***P < 0.001). The data shown here represent five independent experiments.
Discussion
L. platyphylla, a perennial plant distributed throughout Korea, Japan, and China, is reported to exert various biological activities against chronic human diseases26,29,31,32. In the present study, we determined the effects of LPE and its solvent fractions on HEV infection based on experiments with the HEV genotype 3 pSHEV3-luc replicon. In each case, EA, n-BuOH and ddH2O fractions of LPE were more effective inhibitors of the pSHEV3-luc replicon than LPE itself. These individual solvent fractions may contain higher concentrations of bioactive compounds against HEV than the total extract. Additionally, it is possible that bioactive compounds with anti-HEV activity in LPE counteract each other. The EA fraction showed greater efficacy against HEV infection relative to other solvent fractions and inhibited replication of both the pSHEV3-luc replicon and HEV strain 47832c without adversely affecting host cell viability. Further analysis of the EA fraction via anti-HEV activity-guided fractionation and multi-column chromatography led to the isolation of spicatoside A.
Spicatoside A is a steroidal saponin isolated from the tubers of liriopogons, cultivated genera of Liriope and Ophiopogon33. Various biological activities of spicatoside A have been identified to date34. For example, spicatoside A interferes with lipopolysaccharide (LPS)-induced activation of extracellular signal-regulated kinases (ERK1/2), p38 mitogen-activated protein kinase (MAPK), c-Jun N-terminal kinase (JNK) and NF-κB35. In addition to anti-inflammatory properties, spicatoside A exhibits anti-asthma, anti-osteoclastogenesis and anti-cancer activities34. The possibility that other bioactive compounds in LPE additionally interfere with HEV replication cannot be discounted. Considering that the n-BuOH and ddH2O fractions of LPE also inhibited replication of the pSHEV3-luc replicon, the potential bioactive compounds in these fractions warrant further investigation.
L. platyphylla roots are suggested to contain a bioactive compound that inhibits HBV viral promoter activity by interfering with NF-κB, but not AP-1 activity27. Since spicatoside A inhibits nuclear translocation of NF-κB in LPS-treated RAW264.7 macrophages35, one possibility is that NF-κB inhibitory activity affects replication of HEV. However, HEV utilizes ORF2 and ORF3 to inhibit NF-κB activity and thereby evade the host innate immune response9,36. Thus, it is likely that spicatoside A employs mechanism(s) other than NF-κB inhibition to suppress HEV replication, which will be the focus of future studies.
Materials and Methods
Cells, viruses and plant materials
The human hepatocellular carcinoma cell line Huh 7.5 was kindly provided by Dr. Charles M. Rice (Rockefeller University, New York, NY, USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM; Hyclone, Logan, UT) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). The human adenocarcinoma cell line A549 was purchased from the Korean Cell Line Bank. A549 cells were cultured in Minimum Essential Medium (MEM; Hyclone) supplemented with 10% FBS and 1% P/S. HEV strain 47832c was a kind gift from Dr. Reimar Johne (Federal Institute for Risk Assessment, Berlin, Germany)30. For inoculation with HEV 47832c, A549 cells were washed three times with PBS, treated with a culture supernatant containing HEV (5 genome copies/cell) and incubated at room temperature for 1 h. The supernatant was removed and replaced with medium supplemented with 5% FBS and 0.5% P/S, followed by cell incubation for 14 d. Dried root of Liriope platyphylla was purchased from herbal markets in Seoul, South Korea. The sample was identified by Dr. Sun Yeou Kim (College of Pharmacy (COP), Gachon University, Incheon, Korea). The plant material was preserved and a voucher specimen (KSY-SYJ1001) deposited in the herbarium of COP.
Extraction, partition and isolation of spicatoside A from Liriope platyphylla root
The dried material (20 kg) was crushed to powder, extracted with 60 L of 70% ethanol at room temperature for 72 h and concentrated using a rotary evaporator resulting in 290 g ethanol extract. The ethanol extract was suspended in ddH2O and partitioned using the liquid partitioning technique, resulting in the following fractions: ethyl acetate (EA, 20 g), butanol (BuOH, 30 g) and ddH2O (238 g). Dried fractions were maintained at 4 °C before experimental use. Based on anti-HEV activity, the EA fraction was selected for further isolation using a silica gel column with a mobile phase chloroform and methanol gradient (9:1 → 0:10). Thirteen sub-fractions were obtained, and based on anti-HEV activity, sub-fraction 10 was selected for further fractionation leading to isolation of a compound (100 mg). Thin-layer chromatography (TLC) was conducted using EA: MeOH: water (100:13.5:10). To identify the isolated compound, nuclear magnetic resonance (NMR) spectroscopy was performed.
HEV replicon, in vitro transcription, transfection and luciferase-reporter assay
A genotype 3 swine HEV wild-type (pSHEV3) and Renilla luciferase (pSHEV3-luc) replicons were kindly provided by Dr. Xiang-Jin Meng (Virginia Polytechnic Institute and State University, Blacksburg, VA, USA)37. The transfection control firefly luciferase-pcDNA3 (luc-pcDNA3) plasmid was a gift from William Kaelin (Addgene plasmid # 18964)38. pSHEV3-luc plasmids were linearized by treatment with XbaI (New England BioLabs, Ipswich, MA) and firefly luciferase plasmid linearized with BglII (New England BioLabs). Capped RNA transcripts from pSHEV3-luc and luc-pcDNA3 plasmids were generated using the mMESSAGE mMACHINE T7 kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Huh 7.5 cells were transfected with capped RNA transcripts produced from pSHEV3-luc (0.34 μg/104 cells) and luc-pcDNA3 (0.17 μg/104 cells) plasmids using DMRIE-C reagent (Invitrogen, Carlsbad, CA) in keeping with the manufacturer’s instructions. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer’s protocol.
Quantitative RT-PCR (qRT-PCR)
Quantitative analysis of cell-free and cell-associated HEV RNA was performed using qRT-PCR as previously described24,30,39. HEV RNA was isolated using TRI reagent (Molecular Research Center, Inc., Cincinnati, OH) and reverse-transcribed to cDNA using a TOPscript™ cDNA Synthesis kit (Enzynomics, Daejeon, Korea) as described by the manufacturer. Real-time quantitative polymerase chain reactions (PCRs) were performed using 1X HOT FIREPol EvaGreenq PCR Mix Plus (Solis BioDyne, Tartu, Estonia) according to previous reports24,30. The primer sequences used for amplification were as follows: HEV ORF1, 5′-CCTGTTAGTGACATTTGGGTGT-3′ (forward) and 5′-AGACCTTTGCCCCATCCGGATA-3′ (reverse); GAPDH, 5′-CATGAGAAGTATGACAACAGCCT-3′ (forward) and 5′-AGTCCTTCCACGATACCAAAGT-3′ (reverse). The amounts of HEV RNA in culture media were determined according to the method previously described39,40. An external standard curve was generated by using in vitro transcribed RNA from pSHEV3.
Western blot analysis
Cells were harvested, fractionated and transferred onto nitrocellulose membrane, as described previously41. Antibodies to HEV ORF2 and tubulin were purchased from Millipore (Billerica, MA) and Sigma-Aldrich (St. Louis, MO), respectively. Secondary peroxidase-labeled anti-mouse immunoglobulin G antibodies and enhanced chemiluminescence detection reagent were purchased from (Amersham Biosciences (Piscataway, NJ) and Pierce (Rockford, IL), respectively.
Immunofluorescence assay
Fourteen days after inoculation, A549 cells were washed three times with phosphate-buffered saline (PBS) for 5 min. After washing, cells were fixed with 80% acetone at −20 °C for 10 min and blocked with 1X PBS containing 0.5% BSA for 30 min. Subsequently, cells were rinsed with PBS and stained with mouse-HEV ORF2 antibody (MAB8002) (1:200 dilution; Millipore, Billerica, MA). After overnight incubation, cells were washed with PBS and stained with fluorochrome-conjugated anti-mouse IgG secondary antibody (4408) (1:200 dilution; Cell Signaling, Beverly, MA) at 37 °C for 1 h. Finally, cells were washed and counterstained with 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Fluorescence was examined and images analyzed using an inverted Nikon TS100-F fluorescence microscope (Tokyo, Japan) equipped with a digital camera and Nikon NIS-Elements microscope imaging software.
Cell viability assay
Cell viability was analyzed using the Cell Titer-Glo Luminescent Cell Viability Assay (Promega, Madison, WI) as described by the manufacturer.
Statistical analysis
Data are presented as means ± standard deviations (SD). The significance of differences between two means was determined with the Student’s t-test. P-values were determined by unpaired two-tailed Student’s t-tests. P-values < 0.05 were considered statistically significant. Half-maximal inhibitory concentration (IC50) values were calculated using GraphPad Prism 7 (Graphpad Software, San Diego, CA).
Data Availability
All data and materials supporting the conclusions are described and included in this manuscript.
References
Kamar, N., Dalton, H. R., Abravanel, F. & Izopet, J. Hepatitis E virus infection. Clin Microbiol Rev 27, 116–138, https://doi.org/10.1128/CMR.00057-13 (2014).
Ahmad, I., Holla, R. P. & Jameel, S. Molecular virology of hepatitis E virus. Virus Res 161, 47–58, https://doi.org/10.1016/j.virusres.2011.02.011 (2011).
Emerson, S. U. & Purcell, R. H. Hepatitis E virus. Reviews in medical virology 13, 145–154 (2003).
Nan, Y. & Zhang, Y. J. Molecular Biology and Infection of Hepatitis E Virus. Front Microbiol 7, 1419, https://doi.org/10.3389/fmicb.2016.01419 (2016).
Cao, D. & Meng, X. J. Molecular biology and replication of hepatitis E virus. Emerg Microbes Infect 1, e17, https://doi.org/10.1038/emi.2012.7 (2012).
Kalia, M., Chandra, V., Rahman, S. A., Sehgal, D. & Jameel, S. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J Virol 83, 12714–12724, https://doi.org/10.1128/JVI.00717-09 (2009).
Yu, H. et al. Homology model and potential virus-capsid binding site of a putative HEV receptor Grp78. J Mol Model 17, 987–995, https://doi.org/10.1007/s00894-010-0794-5 (2011).
Zheng, Z. Z. et al. Role of heat-shock protein 90 in hepatitis E virus capsid trafficking. J Gen Virol 91, 1728–1736, https://doi.org/10.1099/vir.0.019323-0 (2010).
He, M. et al. The ORF3 Protein of Genotype 1 Hepatitis E Virus Suppresses TLR3-induced NF-kappaB Signaling via TRADD and RIP1. Sci Rep 6, 27597, https://doi.org/10.1038/srep27597 (2016).
Purdy, M. A. et al. ICTV Virus Taxonomy Profile: Hepeviridae. J Gen Virol 98, 2645–2646, https://doi.org/10.1099/jgv.0.000940 (2017).
Smith, D. B. et al. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95, 2223–2232, https://doi.org/10.1099/vir.0.068429-0 (2014).
Song, Y. J. Studies of hepatitis E virus genotypes. Indian J Med Res 132, 487–488 (2010).
Sridhar, S., Teng, J. L. L., Chiu, T. H., Lau, S. K. P. & Woo, P. C. Y. Hepatitis E Virus Genotypes and Evolution: Emergence of Camel Hepatitis E Variants. Int J Mol Sci 18, https://doi.org/10.3390/ijms18040869 (2017).
Lee, G. H. et al. Chronic Infection With Camelid Hepatitis E Virus in a Liver Transplant Recipient Who Regularly Consumes Camel Meat and Milk. Gastroenterology 150, 355–357 e353, https://doi.org/10.1053/j.gastro.2015.10.048 (2016).
Rein, D. B., Stevens, G. A., Theaker, J., Wittenborn, J. S. & Wiersma, S. T. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 55, 988–997, https://doi.org/10.1002/hep.25505 (2012).
Kumar, A., Beniwal, M., Kar, P., Sharma, J. & Murthy, N. Hepatitis E in pregnancy. International Journal of Gynecology & Obstetrics 85, 240–244 (2004).
Navaneethan, U., Al Mohajer, M. & Shata, M. T. Hepatitis E and pregnancy: understanding the pathogenesis. Liver Int 28, 1190–1199, https://doi.org/10.1111/j.1478-3231.2008.01840.x (2008).
Kamar, N. et al. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358, 811–817, https://doi.org/10.1056/NEJMoa0706992 (2008).
Debing, Y. et al. Ribavirin inhibits in vitro hepatitis E virus replication through depletion of cellular GTP pools and is moderately synergistic with alpha interferon. Antimicrob Agents Chemother 58, 267–273, https://doi.org/10.1128/AAC.01795-13 (2014).
Pischke, S. et al. Ribavirin treatment of acute and chronic hepatitis E: a single-centre experience. Liver Int 33, 722–726, https://doi.org/10.1111/liv.12114 (2013).
Debing, Y. & Neyts, J. Antiviral strategies for hepatitis E virus. Antiviral Res 102, 106–118, https://doi.org/10.1016/j.antiviral.2013.12.005 (2014).
Kenney, S. P. & Meng, X. J. Therapeutic targets for the treatment of hepatitis E virus infection. Expert Opin Ther Targets 19, 1245–1260, https://doi.org/10.1517/14728222.2015.1056155 (2015).
Anang, S. et al. Potent inhibition of hepatitis E virus release by a cyclic peptide inhibitor of the interaction between viral open reading frame 3 protein and host tumor susceptibility gene 101. J Virol, https://doi.org/10.1128/JVI.00684-18 (2018).
Kaushik, N. et al. Zinc Salts Block Hepatitis E Virus Replication by Inhibiting the Activity of Viral RNA-Dependent RNA Polymerase. J Virol 91, https://doi.org/10.1128/JVI.00754-17 (2017).
Todt, D. et al. The natural compound silvestrol inhibits hepatitis E virus (HEV) replication in vitro and in vivo. Antiviral Res 157, 151–158, https://doi.org/10.1016/j.antiviral.2018.07.010 (2018).
Choi, S. B., Wha, J. D. & Park, S. The insulin sensitizing effect of homoisoflavone-enriched fraction in Liriope platyphylla Wang et Tang via PI3-kinase pathway. Life Sci 75, 2653–2664, https://doi.org/10.1016/j.lfs.2004.04.039 (2004).
Huang, T. J. et al. Anti-viral effect of a compound isolated from Liriope platyphylla against hepatitis B virus in vitro. Virus Res 192, 16–24, https://doi.org/10.1016/j.virusres.2014.07.015 (2014).
Kim, H. J. et al. Effects of the roots of Liriope Platyphylla Wang Et tang on gastrointestinal motility function. Journal of ethnopharmacology 184, 144–153 (2016).
Lee, Y. C., Lee, J. C., Seo, Y. B. & Kook, Y. B. Liriopis tuber inhibit OVA-induced airway inflammation and bronchial hyperresponsiveness in murine model of asthma. Journal of Ethnopharmacology 101, 144–152, https://doi.org/10.1016/j.jep.2005.04.030 (2005).
Johne, R. et al. An ORF1-rearranged hepatitis E virus derived from a chronically infected patient efficiently replicates in cell culture. J Viral Hepat 21, 447–456, https://doi.org/10.1111/jvh.12157 (2014).
Kim, J. E. et al. Aqueous extract of Liriope platyphylla, a traditional Chinese medicine, significantly inhibits abdominal fat accumulation and improves glucose regulation in OLETF type II diabetes model rats. Lab Anim Res 28, 181–191, https://doi.org/10.5625/lar.2012.28.3.181 (2012).
Kim, J. E. et al. Aqueous extracts of Liriope platyphylla induced significant laxative effects on loperamide-induced constipation of SD rats. BMC Complement Altern Med 13, 333, https://doi.org/10.1186/1472-6882-13-333 (2013).
Fantz, P. R. Macrophytography of cultivated liriopogons and genera delineation. Horttechnology 18, 334–342 (2008).
Ramalingam, M. & Kim, S. J. Pharmacological Activities and Applications of Spicatoside A. Biomol Ther (Seoul) 24, 469–474, https://doi.org/10.4062/biomolther.2015.214 (2016).
Han, Y. et al. Anti-inflammatory effects of prosapogenin III from the dried roots of Liriope platyphylla in LPS-stimulated RAW264.7 cells. J Asian Nat Prod Res 15, 1038–1049, https://doi.org/10.1080/10286020.2013.825253 (2013).
Surjit, M., Varshney, B. & Lal, S. K. The ORF2 glycoprotein of hepatitis E virus inhibits cellular NF-kappaB activity by blocking ubiquitination mediated proteasomal degradation of IkappaBalpha in human hepatoma cells. BMC Biochem 13, 7, https://doi.org/10.1186/1471-2091-13-7 (2012).
Pudupakam, R. S. et al. Mutational analysis of the hypervariable region of hepatitis e virus reveals its involvement in the efficiency of viral RNA replication. J Virol 85, 10031–10040, https://doi.org/10.1128/JVI.00763-11 (2011).
Safran, M. et al. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci USA 103, 105–110, https://doi.org/10.1073/pnas.0509459103 (2006).
Schielke, A., Filter, M., Appel, B. & Johne, R. Thermal stability of hepatitis E virus assessed by a molecular biological approach. Virol J 8, 487, https://doi.org/10.1186/1743-422X-8-487 (2011).
Schemmerer, M. et al. Enhanced Replication of Hepatitis E Virus Strain 47832c in an A549-Derived Subclonal Cell Line. Viruses 8, https://doi.org/10.3390/v8100267 (2016).
Kim, J. E., Kim, S. Y., Lim, S. Y., Kieff, E. & Song, Y. J. Role of Ca2+/calmodulin-dependent kinase II-IRAK1 interaction in LMP1-induced NF-kappaB activation. Mol Cell Biol 34, 325–334, https://doi.org/10.1128/MCB.00912-13 (2014).
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2017R1D1A1B03032494). This work was also supported by the KIST Institutional Program (2E27832-18-106). Jung-Eun Kim was supported by the National Junior Research Fellowship through the NRF funded by the Ministry of Education in Korea (2014H1A8A1021537).
Author information
Authors and Affiliations
Contributions
G.P., A.P., J.E.K. and K.H.C. performed research and analyzed data. G.P., A.P., S.Y.K. and Y.J.S. drafted the manuscript. B.J.P. contributed the reagents. Y.J.S. designed the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, G., Parveen, A., Kim, JE. et al. Spicatoside A derived from Liriope platyphylla root ethanol extract inhibits hepatitis E virus genotype 3 replication in vitro. Sci Rep 9, 4397 (2019). https://doi.org/10.1038/s41598-019-39488-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-39488-5
- Springer Nature Limited