Abstract
In this study we assessed the breeding population, or Management Unit (MU), origin of green turtles (Chelonia mydas) present at Yadua Island and Makogai Island foraging grounds in Fiji, central South Pacific. Based on analysis of mitochondrial (mt) DNA sequences from 150 immature green turtles caught during surveys carried out in 2015–2016, we identified a total of 18 haplotypes, the most common being CmP22.1 (44%) which is a primary haplotype characterizing the American Samoa breeding population. Results of a Bayesian mixed-stock analysis reveals that the two foraging grounds are used by green turtles from the American Samoa MU (72%, Credible Interval (CI): 56–87%), New Caledonia MU (17%, CI: 6–26%) and French Polynesia MU (7%, CI: 0–23%). The prominence of the contribution we found from the American Samoa MU compared to that of French Polynesia, both which have historic telemetry and tagging data showing connectivity with Fijian foraging areas, may reflect the current relative abundance of these two nesting populations and draws attention to a need to update population surveys and identify any significant nesting in Fiji that may have been overlooked.
Similar content being viewed by others
Introduction
The green turtle Chelonia mydas has a circumglobal distribution ranging from tropical to warm temperate waters1. In tropical waters, juvenile green turtles undergo an ontogenetic shift from an omnivorous diet in the pelagic environment to a primarily herbivorous diet in the shallow waters of coastal neritic environments2,3 when about 3–10 years old4,5. Green turtles reach maturity at 15–42 years5 and have a non-annual reproductive cycle; adult females exhibit natal homing and return to lay their eggs on the sandy beaches of their natal region1 with a remigration interval (non-nesting phase) of about 3 years6, while adult males show fidelity to courtship areas7 and have a non-breeding phase of about 1–2 years8. Green turtles of the same rookery may use different foraging grounds9, and a single individual can visit and exploit different foraging grounds10. Mixed aggregations from various rookeries have also been reported9.
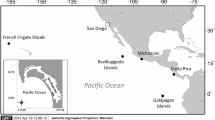
Green turtles nest and forage in Fiji, in the central South Pacific (Fig. 1). While there have not been comprehensive green turtle nesting surveys that adequately cover the entire region of Fiji, which is made up of more than 300 islands and 500 islets, effort to identify nesting sites over the last few decades have failed to reveal significant nesting, with a small number of nests recorded in a few localities11,12,13,14,15,16,17. The majority of the islands surveyed by Hirth18 and Guinea14 had no sign of green turtle nesting activity. However, four green turtles nesting sites, Makogai Island, Namena-lala Island, Heemskercq Reef and the Ringgold Reef systems have been identified and surveyed at least once every ten years in the last four decades11,14,15,16,17. The number of green turtle nests has been consistently low at Heemskercq Reef and the Ringgold Reef systems (average number of green turtle nests observed per survey <1011,14,16,17). Green turtle nests have no longer been found at Makogai Island and Namena-lala since the 1980s15. The green turtle nesting colony on Vatoa Island, in the south of Fiji, reportedly disappeared in the late 1970s13. The Fiji nesting population of green turtles has been estimated in about 50–75 adult females16 based on unpublished data from year 2000 and a market survey done in 199619. Despite the sparsity of observed nesting in Fiji20, the green turtle is considered to be the most abundant of the four sea turtles species regularly seen in Fiji waters14.
Green turtle foraging grounds surveyed in this study (Yadua Island 16.823S, 178.284W and Makogai Island 17.450S, 178.948W) and known green turtle nesting sites (Heemskercq Reef 16.283S, 179.467E and the Ringgold Reef 16.317S, 179.467E systems are currently active, while Makogai Island, Namena-lala Island 17.083 S, 179.050W and Vatoa Island 19.833S, 178.217E are no longer in use), Fiji, central South Pacific. Map created with R57 with packages “map” and “mapdata”.
Genetic characterization of mitochondrial (mt) DNA of the green turtle population nesting in Fiji is not known. Mitochondrial DNA has been extensively used to delineate breeding population structure into management units (MUs) which, in turn, can be used in mixed-stock analysis (MSA) to characterize the genetic composition and natal origins of sea turtle foraging aggregations21. These studies have provided insights into connectivity between foraging grounds used by resident green turtles and their natal breeding areas (rookeries) in the eastern, central North and Indo-Pacific22,23,24,25,26,27,28,29,30. However, there is a lack of information from genetic studies for the western and central South Pacific Ocean islands31.
Both adult and immature green turtles forage in Fiji. Use of Fijian foraging grounds by adult males and by females nesting in other Pacific countries has been demonstrated by tagging studies and satellite telemetry carried out between 1972 and 20101,13,20. A total of 19 adult females were found (mark-recapture) or tracked (satellite telemetry) to Fiji during post-reproductive migration. Of these, eight individuals were nesters from American Samoa1,32, five from French Polynesia1,14,20,33, three from Australia (one reported by Jit34, the other two are from Colin Limpus pers. comm.), two from Tonga35, and one in the Cook Islands1. Connectivity with adult males from French Polynesia waters was also suggested by the migration to Fiji of three individuals released from pens and tagged on Scilly Atoll1,36. Additional insights on the origin of immature and adult green turtles foraging in Fiji were provided by a market survey carried out in Suva, Fiji’s capital, in 1996. Results from a pilot study using genetic analysis of green turtles sampled from market stalls suggested additional possible links to rookeries in the northern Great Barrier Reef and Micronesia19. In general, origin of immature green turtles and the contribution of each regional rookery to Fijian foraging aggregations remain unknown.
The aim of this study was to assess the stock composition of green turtle aggregations at Yadua Island and Makogai Island foraging grounds by using molecular markers (mtDNA) on samples from wild ranging individuals captured and released in situ.
Results
A total of 150 immature green turtles were sampled during six surveys in Yadua (n = 91, CCL: mean 52.1 cm, range: 25.5–74.5 cm) and four surveys in Makogai (n = 59, CCL: mean 57.9 cm, range: 43.5–97.0 cm).
There was a total of 18 haplotypes identified within the two study areas (Table 1). Seventeen haplotypes were observed in Yadua and nine in Makogai. CmP33.1 was the only haplotype found in Makogai that was not found in Yadua. CmP22.1 was the most common haplotype at both sites, identified in 44% of the samples analysed. CmP65.1 was the second most common haplotype observed in 22%, followed by CmP44.1 in 8.6% of the sample set. CmP20.1 was found in 5.3% of the samples in equal proportions between the two sites. There were five newly described haplotypes, in 4.0% of the samples, which have not been identified in any MU to date (known as orphan haplotypes) (CmP33.1, CmP239.1-CmP242.1; Genbank IDs: MK471335-MK471339). A sixth orphan haplotype (CmP192.1), found in 2.7% of the samples (Table 1) has been previously described at a foraging ground in Australia, but to date not identified at any rookery25. The remaining haplotypes were found in low frequencies among both sites (Table 1). There was no difference in haplotype frequencies between the Yadua and Makogai (P > 0.05), so the samples were combined for the MSA.
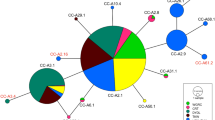
The MSA with uniform priors indicated that the majority of green turtles foraging in the two foraging grounds were composed of animals from the American Samoa MU (72%; Credible Interval (CI): 56–87%). New Caledonia was estimated to contribute 17% (CI: 6–26%) and French Polynesia 7% (CI: 0–23%) (Fig. 2). The combined contributions from the Commonwealth of the Northern Mariana Islands (CNMI)/Guam and Marshall Islands MUs were minimal with an estimated mean of 1%, and CI spanning zero. In general, results were similar for weighted priors, with distance having a slightly less effect than population size (Fig. 2). Taking population size into account reduced the contribution from French Polynesia and eliminated Marshall Islands and CNMI/Guam, allocating a greater contribution from American Samoa (Fig. 2), however all point estimates had overlapping CIs. Potential contributions from other Pacific locations were inconclusive and therefore unlikely sources.
Discussion
Our findings that the Fijian foraging aggregations in our study comprised immature green turtles from American Samoa and French Polynesia, taken together with previous telemetry studies showing post-nesting migration of nesters from American Samoa and French Polynesia to Fiji1,37, highlight the relevance of Fijian foraging grounds for these two MUs. The contribution we detected from New Caledonia MU represents a new finding that adds to the complex connectivity among green turtle rookeries and foraging areas in the Pacific Islands, which extends outside the central South Pacific to the western South Pacific. The similarity of results for the MSA weighted by population size and distance and those using uniform priors suggest that the estimates based on our genetic data are relatively robust, as indicated also by the similar CI for all three models. Even though the New Caledonia population is thought to be orders of magnitude larger than the American Samoa38, the weighted model estimates barely change for New Caledonia, and increase slightly for American Samoa.
Overall, both immature (this study) and mature1,14,20,32,35 green turtles from the central South Pacific and western South Pacific use seagrass meadows and algal beds in Fiji. However, the lack of large individuals in our survey suggests that they may do so in separate aggregations. For example, none of the adult females that travelled to Fiji after being equipped with a satellite transmitter at their nesting sites reached waters around Yadua Island or Makogai Island32,34. Seagrass meadows are present in the shallow coastal waters around the two main islands of Fiji (Viti Levu and Vanua Levu)39 and several of the small island groups (Yasawa, Mamanuca, Lomaiviti and Lau)16,39, however the extent of seagrass area coverage is unknown40,41. Information on sea turtles foraging at some of these other seagrass areas has been limited to occasional reports11,14,18 as well as single-individual tracking32 so it is not possible to assess the overall size and distribution of the Fijian foraging aggregations of green turtles. Mapping presence and area coverage of seagrass meadows together with their use by green turtles, would be a useful baseline to understand green turtle populations’ ecology and dynamics in this largely unexplored area of the central South Pacific.
We found Yadua had twice the number of haplotypes detected at Makogai, however this is likely a consequence of the larger sample size obtained in the first site, rather than an indication of greater haplotype diversity, especially given that there was no significant difference in haplotype frequencies. Additional sampling is warranted to explore this further.
The majority of the green turtles we sampled were from the American Samoa MU. Rose Atoll, the major nesting area in American Samoa38, is about 1400 km north-east from the study sites and it is an uninhabited National Wildlife Refuge administered jointly by the U.S. Fish and Wildlife Service and the American Samoa Government. The site is also included in Rose Atoll Marine National Monument and part of the American Samoa’s Whale and Turtle Sanctuary. The size of the breeding population has been estimated at 105 breeding females38, however this may be an underestimation due to the nesting sites being remote, largely unsurveyed and the breeding population might be potentially recovering (Mark MacDonald pers. comm.).
Our estimates of 6–26% (mean 17%) contribution from the New Caledonia MU, located about 1700 km west of Fiji is of interest given the lack of connectivity evident from previous tagging and telemetry studies in adult nesters. A recent review of 50 years of tagging data at the main nesting site, D’Entrecasteaux atolls, shows that the majority of the New Caledonian green turtle nesters forage in the Great Barrier Reef in Australia, while a smaller percentage remain in New Caledonia’s waters42. However, it is worth noting that a recent genetic MSA study found that the immature green turtles at one of New Caledonia’s largest foraging ground, Gran Lagon Sud, primarily belonged to the New Caledonia MU24. The reviewed studies however, do not necessarily track dispersal of hatchlings from nesting beaches. Our results suggest that a portion of the population of immature green turtles from New Caledonia may be transported eastward periodically by eddies that feed into the South Pacific Gyre, despite the prevailing westward flow of the South Equatorial current that passes New Caledonia (see Read et al.24). Fine-scale ocean current modelling would be useful to test this hypothesis (see Naro-Maciel et al.28).
Scilly Atoll in French Polynesia, located about 3000 km east of Fiji, was historically the largest green turtle nesting site of the central South Pacific, with an estimated 1050 breeding females38 (based on 1991 data), but contributed only a small percentage (7%) to the aggregations at Yadua Island and Makogai Island foraging grounds. There is relatively high uncertainty around the mean estimated contribution from French Polynesia from our MSA as indicated by a CI of 0–23%. This likely stems from the low sample size for this MU in the baseline dataset31 and the presence of CmP65.1 in both American Samoa and French Polynesia which would have resulted in uncertainty in assignments between these MUs, possibly underestimating the relative contribution of French Polynesia. Nevertheless, even at the upper bound of the CI (23%) the estimate for French Polynesia is well below that of the lower bound of the CI for American Samoa (56%) (Fig. 2). Although our sample was limited to immature individuals, based on data from nesting females’ movements from early 1970s to early 2010s14,33,36, a higher contribution from the French Polynesia MU was expected. Based on satellite telemetry, the average home range of adult females resident in Fiji was about 27 km232, so the low estimated number of immature green turtles from this MU might result from a different distribution of the French Polynesian turtles in other Fijian seagrass meadows. However, these results might also reflect a decline for the French Polynesia population43. With average age at maturity estimated at approximately 29 years for green turtles (range 15–42 years in Avens and Snover5 for different populations and aging methods), none of the immature individuals we sampled were born when data for the currently available estimates of nesting female population size were collected. The relatively low proportion we estimated as a contribution from the French Polynesia MU might be the result of a lower relative abundance from the dramatic population decline that appears to have occurred in recent decades coupled with the closer proximity of American Samoa to Fiji. It is worth noting that green turtles from French Polynesia made up an estimated 37% of the green turtles sampled at Suva fish markets in 1996–199719. While the genetic pilot study referred to in Boyle19 did not include baseline data from several key rookeries, and the turtle harvest reported by Boyle19 did not come from our study sites, this intense and sustained harvest of immature green turtles in Fiji may have played a part in reducing the number of individuals reaching adulthood and migrating back to French Polynesia to reproduce. Exploration of other Fiji foraging grounds is necessary to ascertain whether there are spatial or temporal patterns that may reflect historic patterns of recruitment. At present, we can only make inferences based upon the two foraging areas that were investigated in this study, therefore additional information from other coastal waters may change the picture.
Having representative samples from all the potential nesting populations is required for a more accurate MSA (see Komoroske et al.21), which underscores the need for extended surveys of Fiji’s remote islands to assess presence of green turtle nesting and collect samples for genetic analysis. Larger representative sample sizes from nesting sites in French Polynesia and American Samoa will also help improve accuracy of future MSA. The presence of orphan haplotypes in our Fiji sample set illustrates the need for larger sample sizes, and may represent haplotypes that are present in Fiji nesting populations that have been overlooked. The relatively low frequencies of each of the orphan haplotypes generally indicates presence of rare haplotypes in populations that have been under-sampled, and that are detected once larger sample sizes are obtained (see Komoroske et al.21). Our results do however suggest that the American Samoa nesting population is much larger than current knowledge, or alternatively, that there is significant nesting somewhere in Fiji that has been overlooked.
We did not find any haplotypes from the eastern Pacific rookeries in Fiji which, while rare, have been detected at foraging areas off New Zealand44 and in waters around the Oceanic Islands (Hawaii and Palmyra Atoll) in the North Pacific22,28. Also lacking from the mixed stock aggregations were individuals from northern Great Barrier Reef and Micronesia, whose presence in Fiji was suggested in the 20 year old pilot study reported by Boyle19 from the market survey but for which no size range of harvested turtles was provided. Finally, the elimination of CNMI/Guam as a source based on the population size weighted MSA model combined with the CI spanning zero in both weighted and uniform prior MSA results is consistent with the small nesting population.
Based on our findings, we suggest monitoring of Yadua Island and Makogai Island foraging grounds to ascertain temporal variation in stock composition and demographic (size-classes) structure, and exploration of other foraging grounds to determine presence of mature green turtles. This type of analysis would greatly benefit from the mtDNA characterization of the local green turtle rookery. The prominence of the contribution we found from the American Samoa MU compared to that of French Polynesia, both which have historic telemetry and tagging data showing connectivity with Fijian foraging areas, may reflect the current relative abundance of these two nesting populations and draws attention to a need to update population surveys for the French Polynesia MU, whose relatively high population estimates are based on old information from a 1991–1995 survey38.
This MSA study is the first to be carried out in the waters of Fiji and provides insight into the foraging habits of immature green sea turtles in the region. The patterns found in this study are restricted to two sites (that together represent less than 1% of total Fiji coastline) and should not be extended to foraging aggregations throughout the country’s coastal waters. Nevertheless, the contributions of the American Samoa, New Caledonia and French Polynesia MUs that we have found, further emphasize the connection between these four Pacific Island Countries and highlight the importance of looking beyond territorial waters when planning population management of green turtles in the region.
Methods
This study was carried out at two green turtle foraging grounds: Yadua Island (16.823S, 178.284W) on the West of Vanua Levu and Makogai Island (17.450S, 178.948W) in the Lomaiviti group. The survey extended along a total of 54 km of coastline; 38 km at Yadua Island and 16 km at Makogai Island. The total length of coastline in Fiji is estimated at 6123 km (all coastline lengths estimated from Fiji Ministry of Lands and Mineral Resources, Department of Lands, 1:50,000 scale GIS data of Fiji coastline). Seagrass meadows at the two study sites have a patchy distribution and occurred from the intertidal to the subtidal zones, to a maximum depth of 4 m. The two sites, totalling 10 km2 (surveyed surface was 7 km2 at Yadua Island and 3 km2 at Makogai Island), are located about 100 km apart at the opposite entrances of the Vatu-i-Ra Channel, where the Bligh Water Current flows from South-East to North-West (Fig. 1).
Green turtles were surveyed using capture, mark (flipper-tagging) and recapture, during a pilot study in July and August 2015, and then from November 2015 to July 2016. Each survey lasted four consecutive days, during which sea turtles were hand captured from a small boat45 during daylight at high tide. At first capture of each individual, biological data (species ID, curved carapace length (CCL), weight and sex when possible) were recorded and two skin samples collected by using a biopsy punch (2 mm diameter on small immatures and 6 mm diameter on large immatures), as described in Dutton et al.46. With the exception of three individuals previously tagged by Fiji Fisheries Department, all turtles were marked on the right front flipper20 with a titanium tag for long term identification.
Genetic analysis
Genomic DNA was extracted from epidermal tissue samples using a sodium chloride protocol described by Miller et al.47. Primers LCM15382 and H950g48 were used to amplify an ~889 bp fragment at the 5′ end of the control region of the mitochondrial genome using polymerase chain reaction (PCR) methodologies49. PCR products were sequenced with an Applied Biosystems® model 3730 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA). Haplotypes were assigned by comparing aligned sequences against a local reference library of published and unpublished green turtle haplotype sequences using Geneious version 8 (Biomatters) as well as searching the database on GenBank (http://www.ncbi.nlm.nih.gov). The haplotype composition and frequency of our sampled Fiji foraging aggregation (as the stock mix) was compared with those published for MUs (as potential sources) across the Pacific23,24,25,31,50,51,52,53,54. We estimated stock composition of the Fiji foraging index site by conducting MSA using the program BAYES55. Only 25 MUs that had any haplotypes also present in our Fiji foraging study were included as potential source MUs for the MSA and haplotypes not reported at any MU were excluded in the MSA. A total of 100,000 Markov Chain Monte Carlo steps were run for four independent chains, each with different starting points. A burn-in of 50,000 runs was used to calculate the posterior distribution. The Gelman and Rubin shrink factor diagnostic was calculated as part of the BAYES analysis to test for convergence of all chains55. The MSA was run using uniform priors, where potential source MUs were weighted equally, and two models that weighted priors by relative population size and distance from the foraging ground respectively. The weighted models assume that larger MUs are more likely to contribute to the mixed stock than smaller ones (population size priors) or alternatively that MUs in closer proximity to the foraging grounds are more likely to contribute than distant ones, and can help address uncertainty arising from haplotypes that are shared among multiple MUs, as long as the assumption is correct and biologically meaningful25. The size of each nesting population was based on numbers summarized in Ng et al.27 and FitzSimmons and Limpus56 (Supplementary Table S1). Distances between the midpoint of our two foraging sites and each MU, or its centroid in case of multiple rookeries, were measured as straight lines with ImageJ (http://rsb.info.nih.gov/ij) on the map from Jensen et al.25 (Supplementary Table S1).
This research was performed in accordance with relevant guidelines and regulations, and in compliance with the 2004–2018 Fiji “Moratorium on molesting, taking or killing of turtles”. Sampling procedures were approved by the “Animal Ethics Committee” section of The University of the South Pacific (USP) Research Committee.
References
Hirth, H. F. Synopsis of the Biological Data on the Green Turtle (Chelonia mydas) (Linnaeus 1758). (Fish and Wildlife Service, U.S. Department of Interior, 1997).
Hawkes, L. A. et al. Phenotypically linked dichotomy in sea turtle foraging requires multiple conservation approaches. Current Biology 16, 990–995 (2006).
Arthur, K., Boyle, M. & Limpus, C. Ontogenetic changes in diet and habitat use in green sea turtle (Chelonia mydas) life history. Marine Ecology Progress Series 362, 303–311 (2008).
Musick, J. & Limpus, C. J. Habitat utilization and migration in juvenile sea turtles. In The Biology of Sea Turtles (eds Lutz, P. L. & Musick, J.) I, 137–163 (CRC Press) (1997).
Avens, L. & Snover, M. Age and age estimation in sea turtles. in The Biology of Sea Turtles (eds Wyneken, J., Lohmann, K. J. & Musick, J.) III, 97–134 (CRC Press) 2013).
Broderick, A. C., Glen, F., Godley, B. J. & Hays, G. C. Variation in reproductive output of marine turtles. Journal of Experimental Marine Biology and Ecology 288, 95–109 (2003).
FitzSimmons, N. N. et al. Philopatry of male marine turtles inferred from mitochondrial DNA markers. Proceedings of the National Academy of Sciences of the United States of America 94, 8912–8917 (1997).
Limpus, C. The green turtle, Chelonia mydas, in Queensland: breeding males in the southern Great Barrier Reef. Wildlife Research 20, 513 (1993).
Bowen, B. W. & Karl, S. A. Population genetics and phylogeography of sea turtles. Molecular Ecology 16, 4886–4907 (2007).
Senko, J., López-Castro, M. C., Koch, V. & Nichols, W. J. Immature east Pacific green turtles (Chelonia mydas) use multiple foraging areas off the Pacific coast of Baja California Sur, Mexico: first evidence from mark-recapture data. Pacific Science 64, 125–130 (2010).
Bustard, H. R. Turtles and an iguana in Fiji. Oryx 10, 317 (1970).
Hirth, H. F. Synopsis of biological data on green turtle Chelonia mydas (Linnaeus) 1758. (FAO Fisheries Synopsis) (1971).
Pritchard, P. Marine turtles of the south Pacific. In Biology and Conservation of Sea Turtles (ed. Bjorndal, K. A.) 253–262 (Smithsonian Institution Press 1982).
Guinea, M. Sea Turtles of Fiji. (South Pacific Regional Environmental Programme, 1993).
Batibasaga, A. Sea turtles status and conservation initiatives in Fiji. In Proceedings of the Western Pacific Sea Turtle Cooperative Research and Management Workshop (ed. Kinan, I.) 115–118 (2002).
Batibasaga, A., Waqainabete, S. & Qauqau, A. Notes on Fijian Sea Turtles: Estimates on Population Status. Information provided for Sea Turtle Working Group Meeting Nadave/CATD, 31st May– 1st June. (Fiji Fisheries Department, 2006).
Sharma-Gounder, S. & Veeran, R. Ringgold Isles Green Turtle Nesting and Tagging Survey (03rd –09th December, 2010). (Fiji Fisheries Department, 2010).
Hirth, H. F. South Pacific Islands - Marine Turtle Resources. (FAO, 1971).
Boyle, M. Sea turtles of Fiji: aspects of population biology and conservation implications of harvesting. (University of Otago, 1998).
Hirth, H. F. Chapter 10 - Marine turtles. In Nearshore Marine Resources of the south Pacific - Information for Fisheries Development and Management (eds Wright, A. & Hill, L.) 329–370 (Forum Fisheries Agency [FFA], Institute of Pacific Studies [IPS], 1993).
Komoroske, L. M., Jensen, M. P., Stewart, K. R., Shamblin, B. M. & Dutton, P. H. Advances in the application of genetics in marine turtle biology and conservation. Frontiers in Marine Science 4, 156 (2017).
Dutton, P. H. et al. Composition of Hawaiian green turtle foraging aggregations: mtDNA evidence for a distinct regional population. Endangered Species Research 5, 37–44 (2008).
Nishizawa, H. et al. Composition of green turtle feeding aggregations along the Japanese archipelago: implications for changes in composition with current flow. Marine Biology 160, 2671–2685 (2013).
Read, T. C. et al. Mixed stock analysis of a resident green turtle, Chelonia mydas, population in New Caledonia links rookeries in the South Pacific. Wildlife Research 42, 488 (2015).
Jensen, M. P. et al. Spatial and temporal genetic variation among size classes of green turtles (Chelonia mydas) provides information on oceanic dispersal and population dynamics. Marine Ecology Progress Series 543, 241–256 (2016).
Jensen, M. P., Pilcher, N. & FitzSimmons, N. Genetic markers provide insight on origins of immature green turtles Chelonia mydas with biased sex ratios at foraging grounds in Sabah, Malaysia. Endangered Species. Research 31, 191–201 (2016).
Ng, C. K. Y. et al. Regional conservation implications of green turtle (Chelonia mydas) genetic stock composition in China. Chelonian Conservation and Biology 16, 139–150 (2017).
Naro-Maciel, E. et al. Predicting connectivity of green turtles at Palmyra Atoll, central Pacific: a focus on mtDNA and dispersal modelling. Journal of The Royal Society Interface 11, 20130888–20130888 (2014).
Amorocho, D. F., Abreu-Grobois, F. A., Dutton, P. H. & Reina, R. D. Multiple Distant Origins for Green Sea Turtles Aggregating off Gorgona Island in the Colombian Eastern Pacific. PLoS ONE 7, e31486 (2012).
Dutton, P. H. et al. Genetic analysis and satellite tracking reveal origin of the green turtles in San Diego Bay. Marine Biology 166 (2019).
Dutton, P. H. et al. Genetic stock structure of green turtle (Chelonia mydas) nesting populations across the Pacific Islands. Pacific Science 68, 451–464 (2014).
Craig, P., Parker, D., Brainard, R., Rice, M. & Balazs, G. Migrations of green turtles in the central South Pacific. Biological Conservation 116, 433–438 (2004).
Petit, M. Double Programme de Recherche sur les Tortues Marines de l’Archipel de la Société, Polynésie française [Dual Research Program on Sea Turtles of the Society Archipelago - French Polynesia]. (Critical Ecosystem Partnership Fund (CEPF) & Conservation International, 2013).
Jit, J. N. Status of sea turtle conservation in Fiji: assessment of the international, regional and national focus. (University of the South Pacific, 2007).
Craig, P. Fiji’s strategic role as a turtle foraging area in the central South Pacific region. In SPREP 6th Regional Marine Turtle Conservation Programm, Feb. 2003, Apia, Samoa (2003).
Balazs, G. H., Siu, P. & Landret, J.-P. Ecological aspects of green turtles nesting at Scilly Atoll in French Polynesia. In Proceedings of the Twelfth Annual Workshop on Sea Turtle Biology and Conservation (eds. Richardson, J. I. & Richardson, T. H.) 361, 7–10 (NOAA-NMFS-SEFSC, 1995).
Craig, R. K. Protecting international marine biodiversity: international treaties and national systems of marine protected areas. Journal of Land Use & Environmental Law 20, 333–369 (2005).
Seminoff, J. A. et al. Status review of the green turtle (Chelonia mydas) under the Endangered Species Act. 539, (NOAA-NMFS-SWFSC, 2015).
Skelton, P. A. & South, G. R. Seagrass biodiversity of the Fiji and Samoa islands, South Pacific. New Zealand Journal of Marine and Freshwater Research 40, 345–356 (2006).
Prasad, B. C. Natural resource inventory report of the Fiji Islands. Volume 2: Marine resources inventory of the Fiji Islands. 94 (University of the South Pacific, 2010).
Brodie, G. & N’Yeurt, D. R. A. Effects of climate change on seagrasses and seagrass habitats relevant to the Pacific Islands. 112–131 (Pacific Marine Climate Change Report Card: Science Review, 2018).
Read, T. C. et al. Migrations of green turtles (Chelonia mydas) between nesting and foraging grounds across the Coral Sea. PLoS ONE 9, e100083 (2014).
Lesage, P. Double Programme de Recherche sur les Tortues Marines de l’Archipel de la Société, Polynésie française [Dual Research Program on Sea Turtles of the Society Archipelago – French Polynesia]. (Conservation International Pacific Islands Program, 2013).
Godoy, D. A. The ecology and conservation of green turtles (Chelonia mydas) in New Zealand. (Massey University, 2016).
Ehrhart, L. M. & Ogren, L. H. Studies in foraging habitats: capturing and handling turtles. In Research and management techniques for the conservation of sea turtles (eds Eckert, K. L., Bjorndal, K. A., Abreu-Grobois, F. A. & Donnelly, M.) (1999).
Dutton, P. H., Davis, S. K., Guerra, T. & Owens, D. Molecular phylogeny for marine turtles based on sequences of the ND4-Leucine tRNA and control regions of mitochondrial DNA. Molecular Phylogenetics and Evolution 5, 511–521 (1996).
Miller, S. A., Dykes, D. D. & Polesky, H. F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16, 1215 (1988).
Abreu-Grobois, F. A. et al. New mtDNA D-loop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analysis. In Proceedings of the 26th Annual Symposium onSea Turtle Biology (eds Frick, M., Panagopoulou, A., Rees, A. F. & Williams, K.) 179 (International Sea Turtle Society, 2006).
Leroux, R. A. et al. Re-examination of population structure and phylogeography of hawksbill turtles in the wider Caribbean using longer mtDNA sequences. Journal of Heredity 103, 806–820 (2012).
Cheng, I.-J. et al. Comparison of the genetics and nesting ecology of two green turtle rookeries. Journal of Zoology 276, 375–384 (2008).
Guo, Y., Wang, Z. & Liu, C. MtDNA sequence analysis of green sea turtle (Chelonia mydas) of the South China Sea. Journal of Guangdong Ocean University 29, 6–9 (2009).
Hamabata, T., Kamezaki, S., Kamezaki, N. & Koike, H. Genetic structure of populations of the green turtle (Chelonia mydas) in Japan using mtDNA control region sequences. Bulletin of the Graduate School of Social and Cultural Studies, Kyushu University 15, 35–50 (2009).
Hamabata, T., Kamezaki, N. & Hikida, T. Genetic structure of green turtle (Chelonia mydas) peripheral populations nesting in the northwestern Pacific rookeries: evidence for northern refugia and postglacial colonization. Marine Biology 161, 495–507 (2014).
Dutton, P. H. et al. Population structure and phylogeography reveal pathways of colonization by a migratory marine reptile (Chelonia mydas) in the central and eastern Pacific. Ecology and Evolution 4, 4317–4331 (2014).
Pella, J. & Masuda, M. Bayesian methods for analysis of stock mixtures from genetic characters. Fish. Bull. 99, 151–167 (2001).
FitzSimmons, N. N. & Limpus, C. J. Marine turtle genetic stocks of the Indo-Pacific: identifying boundaries and knowledge gaps. Indian Ocean Newsletter 20, 2–18 (2014).
R Core Team. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2017).
Acknowledgements
We express our most sincere gratitude to Turaga ni Yavusa Ratu Jone Cakau for allowing this research project to be carried out in Denimanu village’s iqoliqoli, Dau ni Vonu Pita Qarau for his commitment and hospitality, and Shritika Prakash for the accurate field work. We thank the large number of Dau ni Vonu, volunteers from Yadua and USP, and Fiji Fisheries officers, in particular Waisea Delainimati and Suliasi Takape, that helped during the surveys. Many thanks to Amy Lanci and Vicki Pease for their help with laboratory analysis at the NOAA-SWFSC in La Jolla, CA, USA. We also thank Michael Jensen for valuable input on data analysis and manuscript review. This work was supported by US NOAA PIRO (grant: NA15NMF4540131), US NOAA SWFSC and Fiji Fisheries Department. SPREP kindly provided tags and applicator.
Author information
Authors and Affiliations
Contributions
S.P. conceived the project. S.P. and P.H.D. designed the project. A.B. helped with planning, logistics and permits. A.C. sampled all specimens. E.L.L. and P.H.D. performed the genetic analysis. S.P., E.L.L. and P.H.D. wrote the first draft of the manuscript, which was reviewed by all authors.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Piovano, S., Batibasaga, A., Ciriyawa, A. et al. Mixed stock analysis of juvenile green turtles aggregating at two foraging grounds in Fiji reveals major contribution from the American Samoa Management Unit. Sci Rep 9, 3150 (2019). https://doi.org/10.1038/s41598-019-39475-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-39475-w
- Springer Nature Limited
This article is cited by
-
“Draw the sea…”: Children’s representations of ocean connectivity in Fiji and New Caledonia
Ambio (2022)
-
Conservation status and cultural values of sea turtles leading to (un)written parallel management systems in Fiji
Ambio (2022)
-
A global review of green turtle diet: sea surface temperature as a potential driver of omnivory levels
Marine Biology (2020)