Abstract
The aim of this study was to compare the phenotype of 40 strains of L. monocytogenes under food and host relevant stress conditions. The strains were chosen to represent food and clinical isolates and to be equally distributed between the most relevant clonal complexes for clinical and food isolates (CC1 and CC6 vs CC121 and CC9), plus one group of eight strains of rare clonal complexes. Human-associated CC1 had a faster maximal growth rate than the other major complexes, and the lag time of CC1 and CC6 was significantly less affected by the addition of 4% NaCl to the medium. Food-associated CC9 strains were hypohemolytic compared to other clonal complexes, and all strains found to be resistant to increased concentrations of benzalkonium chloride belonged to CC121 and were positive for Tn6188 carrying the qacH gene. Lactic acid affected the survival of L. monocytogenes more than HCl, and there was a distinct, strain specific pattern of acid tolerant and sensitive strains. Strains from CC6 and human clinical isolates are less resilient under acid stress than those from other complexes and from food. One strain isolated from a human patient exhibited significant growth defects across all conditions.
Similar content being viewed by others
Introduction
The severe clinical conditions resulting from food-borne infections with Listeria monocytogenes are a constant concern to public health agencies and food producers. The European Food Safety Authority reported 2‘161 confirmed cases of human listeriosis in 2014, which accounts for a notification rate of 0.52 cases per 100‘000 people1. Although the incidence level is low, the high hospitalization rate and a case fatality rate ranging from 15–30%2,3,4 rank listeriosis among the most threatening foodborne illnesses.
The ubiquitous nature and the wide genetic variability of L. monocytogenes have inspired efforts to identify markers of virulence by analyzing and comparing the compositions of strain populations found in clinical patients, the food production context and the natural environment5,6. The underlying aim across studies to determine the virulence potential of a given strain has been (i) to assess the level of risk associated with a strain found in the food chain and (ii) to gain deeper insight into the mechanisms of virulence7,8. L. monocytogenes are divided into four deeply separated evolutionary lineages9 and 13 serotypes10. Even though all L. monocytogenes are considered pathogenic, the vast majority of human clinical cases can be attributed to a narrow set of serotype 1/2a, 1/2b and 4b strains11,12. Further subtyping by multilocus sequence typing (MLST) identified up to six epidemic clones of genetically related strains, isolated from geographically and temporally distinct outbreaks13,14,15. Recently, Maury et al.16 analyzed the relative prevalence of different clonal complexes (CC) in a large dataset of 6633 L. monocytogenes strains isolated from human clinical cases and food. CC1, CC2, CC4 and CC6 strains showed the highest relative frequency in clinical cases, while CC121 and CC9 strains were mostly found in food. The same authors described that host factors played a role in infections with strains that are not typically associated with clinical cases. For example, CC121 or CC9 strains were found more frequently in patients with clinical comorbidities16. This may explain why an infrequent number of strains with mutations in important virulence factors are found in human patients6,17,18. Moura et al.15 identified groups of highly related strains responsible for multistate outbreaks and showed that sub lineages of L. monocytogenes were repeatedly spread internationally.
Based on the data provided by Maury et al.16, we hypothesized that the skew in relative frequencies of CC1 and CC6 towards clinical cases might be due to the fact that they are able to grow faster in food and that their defense mechanisms against host barriers such as hydrochloric acidity in the stomach might be superior to that of strains from CC121 or CC9.
We characterized a set of 40 L. monocytogenes isolates for their response to food associated stresses (preservatives like salt and lactic acid, and the antimicrobial benzalkonium chloride (BC)) as well as attributes relevant in the host environment (resistance to HCl, hemolytic activity as an approximation for the expression rate of virulence genes). The strain set was chosen to be equally distributed between food and clinical isolates as well as CC1, CC6, CC121 and CC9, plus an additional set of nine strains to represent rare isolates not commonly found in clinical cases or the food environment.
Results
Growth in BHI
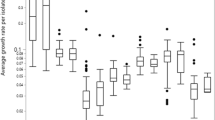
Growth characteristics of every strain were measured in BHI in order to create a baseline to which growth under stress can be compared to. All strains were grown in BHI at 37 °C and the maximal growth rate (Vmax) as well as the lag time were determined. The mean Vmax in BHI was 6.98 (SD = 0.8). There was no meaningful difference in Vmax between strains from different lineages or serotypes. CC1 showed a significantly higher Vmax (7.54, SD = 0.49) than CC6, CC121 and CC9 (combined mean Vmax 6.66, SD = 0.81; p < 0.001) (Fig. 1a). The highest Vmax was observed in strain N12-1996 (CC1, serotype 4b, food isolate; 8.1, SD = 0.5), the slowest Vmax was in strain N12-1387 (CC6, serotype 4b, human clinical isolate; 3.86, SD = 0.05).
Maximal growth rate. The y axis represents the maximum growth rate (defined as the maximum slope calculated by a sliding window approach) in BHI (a) and BHI with 4% NaCl (b) plotted against clonal complex. The shaded area denotes the four larger CC included in the study. The white area contains CC which are represented by only one strain. Boxplots represent the mean, 25th and 75th quantile with 1.5*IQR whiskers. Dots represent outliers and *denotes a significant contribution of CC to the phenotype.
The average lag time in BHI was 124 min (SD = 15 min). There was no meaningful difference in lag time between strains from different lineages, serotypes or clonal complexes. Isolates from food had a marginally shorter lag time (120.5 min, SD = 18.7) than isolates from human clinical cases (129.6 min, SD = 19.7), p = 0.002. The shortest lag time was in strain N13-0703 (CC6, serotype 4b, food isolate; 108.8 min, SD = 0.005) and the longest lag time was observed in strain N12-1387 (CC6, serotype 4b, human clinical isolate; 166 min, SD = 0.0004).
Growth in BHI supplemented with 4% NaCl
When the BHI was supplemented with 4% w/v NaCl, the average Vmax (4.85, SD = 0.51, p < 0.0001) was significantly slower than without the added salt. At this slower growth rate, the pattern observed for Vmax across strains remained unchanged. There was no meaningful difference in Vmax between lineages and serotypes, while CC1 showed significantly faster growth (5.23, SD = 0.25) compared to CC6, CC121 and CC9 (combined mean Vmax 4.6, SD = 0.44, p < 0.001) (Fig. 1b). The fastest Vmax was observed in strain N13-0228 (CC31, serotype 1/2a, food isolate; 5.83, SD = 0.11), the slowest Vmax in strain N12-1387 (CC6, serotype 4b, human clinical isolate; 3.24, SD = 0.09).
When cells were grown in BHI supplemented with 4% NaCl compared to the growth in BHI, the average lag times significantly increased to 167 min (SD = 20.1), p < 0.001. Strains from lineage I showed a significantly shorter lag time (156 min, SD = 15.0) than strains from lineage II (176 min SD = 19.4), p < 0.001. No significant difference in lag times was observed between serotypes, and strains from CC1 had significantly shorter lag times (151.5 min, SD = 7.7) than CC9 and CC121 (combined mean lag time175.3 min, SD = 20.9) (p < 0.001). The shortest lag time in BHI with 4% NaCl was observed in strain N12-1339 (CC1, serotype 4b, food isolate; 143.7 min, SD = 0.005) and the longest lag time in BHI with 4% NaCl was for strain N11-1837 (CC9, serotype 1/2a, human clinical isolate; 237 min, SD = 0.01).
To differentiate strains that are generally fast growers (e.g. strain N13-0228 (CC31, serotype 1/2a, food isolate) which had a high Vmax in BHI as well as in BHI supplemented with NaCl) from strains that adapt faster than others to an increase in NaCl in the medium, relative values for Vmax (Vmax in 4% NaCl/Vmax in BHI) and lag time (lag time in BHI supplemented with 4% NaCl/lag time in BHI) were calculated.
Overall, Vmax decreased by a factor of 0.70 (SD = 0.05) in BHI with the added salt compared to BHI. No significant differences in relative Vmax was observed between lineages, serotypes or clonal complexes, indicating that Vmax was a property of the strain regardless of the two media tested here. The highest relative Vmax was observed for strain N12-1387 (CC6, serotype 4b, human clinical isolate; 0.84, SD = 0.02), the lowest for strain N12-0710 (CC9, serotype 1/2c, food isolate; 0.60, SD = 0.02).
The relative lag times indicate the factor by which the lag time increased upon the addition of 4% NaCl. The lag times increased less in strains that belong to lineage I (1.29, SD = 0.0.04) than in strains from lineage II (1.39, SD = 0.08, p < 0.001) (Fig. 2a). CC1 and CC6 had significantly shorter relative lag times than CC121 and CC9 (p < 0.01 for all contrasts) (Fig. 2b). Nested F-tests showed that while serotype explained a significant amount of variance in relative lag-time by itself, most of that variance was explained by clonal complex since adding serotype to the clonal complex model did not yield a significantly better model fit. The highest relative lag time was observed for strain N12-1107 (CC207, serotype 1/2a, human clinical isolate; 1.57, SD = 0.06), the lowest for strain N11-2542 (ST739, serotype 1/2a, food isolate; 1.03, SD 0.40).
Relative lag time. The y axis represents the relative lag time (lag time in BHI supplemented with 4% NaCl/lag time in BHI), plotted against lineage (a) and clonal complex (b). The shaded area denotes the four larger CC included in the study. The white area contains CC which are represented by only one strain. Boxplots represent the mean, 25th and 75th quantile with 1.5*IQR whiskers. Dots represent outliers and *denotes a significant contribution of the qualifier to the phenotype.
Acid stress
The response of L. monocytogenes to inorganic (hydrochloric acid, HCl) and organic (lactic acid, LA) acid stress at 37 °C was determined with and without a prior adaptation step at an intermediate pH to determine the ability of the strains to survive an acid shock as well as their adaptive acid tolerance. The results are presented as the log reduction between before and after one hour incubation at the final pH (Fig. 3a).
(a) Log reduction of CFU/ml in individual strains between before and after 1 h acid exposure at pH3 for HCl, pH3.5 for LA. The upper panel represents data from strains that were pre-exposed to mild acid stress at pH5.5 for one hour prior to exposure the acid stress. The lower panel represents data from acid-shocked strains without prior adaptation to pH5.5. The horizontal line represents the delineation between “acid sensitive” or “acid resistant” phenotypes as determined by k-means. (b) Probabilities of belonging to the “acid sensitive” phenotype by source. (c) Probabilities of belonging to the “acid sensitive” phenotype by clonal complex. The shaded area denotes the four larger CC included in the study. The white area contains CC which are represented by only one strain. CCs with collapsed error bars had only acid-sensitive or acid-resistant observations, making a realistic estimate of the variance impossible. *Denotes a significant association of the qualifier with the phenotype.
Interestingly, there was a distinct pattern of acid-resistant and acid-sensitive strains that seemed mainly driven by properties of the individual strains. This is reflected in the large standard deviations within the dataset. The mean log reduction resulting from exposing L. monocytogenes to one hour of HCl stress at pH 3.0 was 3.84 (SD 2.99) with previous adaptation to pH 5.5 and 3.58 (SD 2.49) for acid shock. Initial experiments with LA showed that L. monocytogenes did not survive exposure to LA at pH 3.0 for an hour (data not shown). We therefore increased the pH to 3.5 in order to measure inter-strain differences in LA acid survival. In general, the results obtained with LA at pH 3.5 mirror the patterns obtained with HCl at pH 3.0 (Fig. 3a). The mean log reduction resulting from exposing L. monocytogenes to one hour of LA stress at pH 3.5 was 3.51 (SD 3.01) with previous adaptation to pH 5.5 and 3.77 (SD 3.00) for acid shock.
We used k-means to assign binary phenotypes of “acid resistant” and “acid sensitive” to each individual strain. Given that the large variance between strains overrode the effect of acid adaptation in individual strains, adaptation was not a good predictor of acid susceptibility. Neither lineage nor serotype were significantly associated with either the acid-susceptible or acid-resistant phenotype. Strains isolated from food had a significantly lower probability to be “acid sensitive” than strains isolated from human clinical cases (p < 0.001, Fig. 3b). Clonal complex was a significant predictor of acid susceptibility with CC6 having a slightly higher chance to fall into the “sensitive” category (p < 0.0001, Fig. 3c). For CCs with only one strain it was not possible to differentiate between the effect of CC and the strain itself to get a realistic estimate of the variance (e.g. CC54, CC224, ST226 and ST375 had only acid-resistant observations, while CC4, CC415 and CC207 had only acid-susceptible observations).
Resistance to benzalkonium chloride
The majority of strains included in this study (31/40) was resistant to 5 μg/mL BC or lower (Fig. 4). Strains were considered BC resistant if they were able to grow at double the concentration that inhibited growth in >50% of all strains, e.g. at 10 μg/mL19,20.
Strains from lineage II were more likely to be more highly resistant than strains from lineage I (p < 0.001). This was likely due to the underlying effect of the clonal complex, as CC121 was resistant to significantly higher concentrations of BC (median = 15 μg/mL BC), compared to the other clonal complexes (median = 5 μg/mL BC, p < 0.001) (Fig. 4). While all CC121 strains were resistant to high concentrations of BC; strain N12-1387 (CC6, serotype 4b, human clinical isolate) was the most sensitive (median = 0 μg/mL BC).
PCR revealed that all CC121 strains were positive for Tn6188 that carried qacH, an efflux pump for BC21. None of the strains carried any of the other known efflux pumps such as emrE22 or bcrABC23.
Hemolysis
We analyzed the hemolytic activity of the supernatants of all 40 strains. The hemolytic activity is a direct function of the amount of listeriolysin O (LLO) a strain secretes. LLO is encoded by hly under the control of the major transcriptional regulator for virulence genes, PrfA, and can be used as an approximation for the virulence of a given strain.
There was no significant difference in hemolytic activity between lineages. Strains belonging to serotype 1/2c had significantly lower hemolytic activity compared to 1/2a, 1/2b and 4b (p = 0.01). CC9 strains caused significantly less hemolysis compared to other CCs (Fig. 5). The highest hemolysis was observed for strain N13-0369 (CC121, serotype 1/2a, food isolate; 1.85, SD = 0.3), the lowest for strain N12–1387 (CC6, serotype 4b, human clinical isolate; 0.96, SD = 0.05).
Hemolysis. The y-axis represents the amount of hemolysis caused by the supernatant of each strain, as determined by a mixed effects linear model, plotted against clonal complex. The dots represent the least square means, the whiskers the upper and lower 95% confidence level. CCs that share a number above the bars are not significantly different from each other, as determined by a false discovery rate of <0.05.
Discussion
We found that lineage I strains (comprising CC1 and CC6 and also serotype 4b) recover faster after exposure to 4% NaCl in the medium and CC1 had a higher Vmax compared to CC121 and CC9 in both BHI supplemented with NaCl and BHI. In combination, this might increase the chance of a CC1 strain to reach high numbers in food faster than strains from other clonal complexes and might in part explain why CC1 strains are among the most frequently isolated in human clinical cases16. This is in line with other authors who found that serotype 4b (lineage I) strains had a shorter lag time than serotype 1/2a (lineage II) strains in 11% NaCl grown at 10 °C24. However, in the data provided in Magalhães et al.25, lineage seems not to be a significant predictor of lag time in TSBYE with 4% NaCl as determined by a t-test on the data in Table 2 of their data (t = −0.13138, p-value = 0.9).
Resistance to acid is a relevant factor in the ability of a given L. monocytogenes strain to reach a human host and result in an infection. Lactic acid is an important preservative in the food industry, and is often added to meat or fish in the form of potassium or sodium lactate. In addition, lactic acid is naturally present in dairy products through fermentation. The relatively common use of a low pH as a preservative measure in food may exert a selective pressure that explains the higher likelihood of food isolates to be acid tolerant. Interestingly, human clinical isolates were significantly less resilient under acid stress. The low pH in the mammalian stomach is achieved by the secretion of HCl and is an important “first line of defense” mechanism against pathogens entering the body. L. monocytogenes is capable of surviving acidic conditions as low as pH 2.5 for 2 h26. In our initial experiments, BHI at pH 3 was used for the HCl as well as the LA assays, resulting in less than 100 CFU/ml (limit of detection) surviving bacteria after LA stress. We therefore concluded that L. monocytogenes was more sensitive to LA than to HCl, which is in line with literature on the adaptive acid tolerance in L. monocytogenes which was more effectively induced by LA than HCl27,28. We found a distinct pattern of acid tolerant and acid sensitive strains that did not consistently align with any of the classification systems according to lineage, serotype, or CC of the strains. Throughout the literature on acid tolerance of L. monocytogenes, a strain-dependent survival pattern is discernable. For instance, Lianou et al.29 challenged different strains of L. monocytogenes with LA at pH 3.5, and a strain specific pattern that did not follow serotype or origin designation is visible in the data. Strains grown on food matrices for different time intervals were challenged with pH 1.5 (HCl) and the survival pattern was strain dependent30. After 2h of exposure to pH 2.4 achieved with HCl, Lundén et al.31 observed a variability in log reductions between strains that exceeds 6 log. A strain specific pattern of acid resistance is also visible in the datasets presented by Ramalheira et al.32 and Cheng et al.28. Mobile genetic elements such as plasmids, transposons or insertion sequences are likely candidates to explain phenotypic patterns that refuse to align with any classification systems derived from the genome such as lineage or CC. It is tempting to speculate their involvement in the pattern observed here and in other studies. Bacteria faced with the challenge of an increased concentration of H+ ions have the options of (i) consuming H+ ions through decarboxylation reactions, (ii) exporting H+ ions through pumps and (iii) repairing the damage resulting from low pH, for instance mediated by heat shock proteins. All these mechanisms are growth-phase and temperature dependent27, have been described for L. monocytogenes and were reviewed by Ryan, Hill & Gahan33. Any of the above mechanisms could be encoded on mobile genetic elements.
We found that all strains with an increased resistance to BC belonged to CC121 and harbored the Tn1688 transposon that carries the qacH gene encoding an efflux pump21, which goes along with previous findings34,35. While strains coding for bcrABC are commonly found in Switzerland34,35, we found no strains carrying these genes in the panel of strains used in this study. Not surprisingly, we did not find any strains carrying the emrE gene; this efflux pump for BC has so far only been demonstrated in strains from Canada22 and in one strain originating from Finland34.
The hemolytic activity was used as an indicator of virulence for a given strain. Differences in hemolytic activity between strains may indicate differences in PrfA activity as the main transcriptional activator of hly, or in the post-transcriptional regulation of LLO. hly mutants are unable to escape phagocytic vacuoles36, are avirulent in vivo37, and the controlled regulation of LLO production is directly linked to virulence in vivo38. However, while LLO encoded by hly is the main hemolysin of L. monocytogenes, a variety of genes is known to influence the hemolytic activity of a strain, which should caution against using LLO activity as the only explanation for phenotypic differences in hemolysis39,40. Hypohemolytic mutants occur at low frequencies in L. monocytogenes belonging to CC9 and CC121 as well as CC1 and CC6, as was shown in a screening of 57820 strains40. All 60 hypohemolytic strains found in that study carried mutations in the prfa-hly regulation pathway. In this context, it is interesting that strains from CC9, which are mostly isolated from the food environment, showed the lowest hemolytic activity and might partially explain why strains from CC9 are less frequently isolated from human clinical cases. On the other hand, CC121 that is also mainly associated with the food environment had significantly higher hemolytic activity compared to CC9.
Across the board, we found little association of the phenotype with the source a strain was isolated from (clinical vs food). This indicates that the genotype rather than the environment a strain is isolated from dictates its ability to grow under the tested conditions, a finding that is confirmed by other authors as well24.
Among the panel of 40 strains used in this study, strain N12-1387 (CC6, serotype 4b, human clinical isolate) stood out in several assays: it was the strain with the lowest tolerance to BC (<2.5 mg/ml), had the lowest hemolytic activity, by far the lowest Vmax in BHI with and without 4% added NaCl, and belonged to the group of acid sensitive strains. Interestingly, this strain was isolated from blood in a human clinical case. No further information is available on the patient or his treatment, but it is conceivable that there were considerable comorbidities involved to result in invasive listeriosis by a strain with such substantial growth impairments.
In conclusion, CC1 had a high maximal growth rate, CC1 and CC6 strains recovered faster after the addition of 4% NaCl to the medium and CC1 and CC6 showed higher hemolytic activity than CC9 strains, which may provide them with a growth advantage in food and increased virulence in human hosts. The molecular mechanism of the distinct pattern of acid resistance is subject to further investigation.
Materials and Methods
Bacterial Strains and Growth Conditions
Forty L. monocytogenes strains were selected out of two existing strain collections described by Ebner et al.35 and Althaus et al.6 (Table 1). The strains were isolated either from food or human patients and collected at the Swiss National Reference Laboratory (NENT) between 2011 and 2014. The selection comprised eight strains of each CC1, CC9, CC121, and seven strains of CC6. An additional group consisted of nine rare strains35 with less than nine isolates recorded at the Listeria MLST database hosted by the Institute Pasteur (http://bigsdb.web.pasteur.fr/listeria/). If available, equal numbers of strains isolated from food and clinical sources were included. Where the strain collection contained more than the desired number of strains with given parameters, Google’s random number generator (accessed in March 2017) was used to facilitate the strain selection.
Bacterial strains were stored in brain heart infusion (BHI) (Oxoid, Pratteln, Switzerland) broth and 15% glycerol (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) at −80 °C. Fresh cultures were obtained for each experiment by streaking frozen stock on BHI agar followed by overnight incubation at 37 °C. Single colonies were inoculated into five ml BHI broth and incubated at 37 °C shaking (200 rpm) for 15–18 h. These overnight cultures were then sub cultured (1:100) into fresh BHI broth and grown for six hours at 37 °C with shaking to an early stationary phase (corresponding to a culture grown to OD590 1.0 plus 1 h, 1–3 × 109 colony forming units (CFU)/ml). Where indicated, strains were adjusted to equal an OD590 of 1.5 in fresh BHI. All experiments were carried out in triplicates.
Growth rate in rich medium
Early stationary phase cultures were prepared and adjusted to the same OD590. Five μl of each of the adjusted cultures were inoculated into 200 μl BHI in a 96 well plate in triplicate. The plates were incubated for 12 h at 37 °C with continuous shaking in an EON microplate reader (BioTek, Lucerne, Switzerland). OD600 was measured every 15 min. Data acquisition (lag phase, maximal growth rate and OD600 max) and visualization was done using the proprietary software Gen5 All-In-One Microplate Reader Software (Version 2.01.14, BioTek, Lucerne, Switzerland).
Growth under 4% salt stress
Early stationary phase cultures were prepared and adjusted to the same OD590. Triplicate 5 μl aliquots of each strain were transferred into 200 μl of BHI broth with 4% (w/v) in a 96 well plate. The plates were incubated for 12 h at 37 °C with continuous shaking in an EON microplate reader (BioTek, Lucerne, Switzerland). Data acquisition was the same as described above for the growth rate in rich medium.
HCl Acid Stress Assay
Acid resistance at pH3 with and without prior acid adaptation was tested as previously described by Ferreira et al.41 with minor modifications. The pH of the BHI was adjusted with hydrochloric acid solution (1 M HCL) (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) immediately prior to the experiment. Aliquots of 1 ml of early stationary phase cultures were transferred to Eppendorf tubes and centrifuged for 5 min at 6000 × g. For acid shock experiments, the pellets were resuspended in 1 ml BHI pH 3 and incubated for 1 h at 37 °C without shaking. For acid adaptation experiments, 1 ml of the same culture were pelleted, resuspended in 1 ml BHI adjusted to pH 5.5 and incubated for 1 h at 37 °C without shaking. Then, the culture was centrifuged again, resuspended in BHI pH 3 and incubated for 1 h at 37 °C without shaking. Bacteria were enumerated by direct colony count from serial dilutions before and after the one-hour incubation at pH 3. Serial dilutions were performed in 96-well plates in BHI (pH 7.4). 10 μl of each well were spotted onto the edge of a BHI agar plate and run over the plate by tilting as described by Jordan et al.10. Plates were incubated at 37 °C for 18 h prior to performing colony counts.
Organic Acid Stress Assay
DL-Lactic acid ~90% (LA, Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) was used as a second acid stress. The experimental procedure was carried out in the same way as described above, apart from the final pH being 3.5 instead of 3. This change in final pH was determined after preliminary experiments showed no growth after incubation at pH 3 in LA in an effort to expose the strains to the most selective possible acid stress.
Benzalkonium Chloride Resistance
BC susceptibility was tested as previously described by Meier et al.34. Early stationary phase cultures were prepared. All cultures were adjusted to the same OD590 and 10 μl of the adjusted cultures were spotted onto BHI agar plates containing increasing BC (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) concentrations (0, 2.5, 5, 7.5, 10, 15, 20, 25 and 30 μg/ml). The strains could either show (i) no growth, (ii) growth of single colonies or (iii) confluent growth in the spotted area after incubation at 37 °C for 48 h. Strains were considered resistant to the concentration of BC where confluent growth was observed. To further characterize resistant strains PCR analysis for resistance genes (bcrABC, Tn6188, emrE) was carried out with primers and conditions described by Meier et al.34.
Hemolysis Assay
The hemolytic activity in the supernatant of all L. monocytogenes strains was measured similarly to the method previously described by Schärer et al.42. Human blood (blood type A+) was obtained from Blutspende Zürich, Zürich, Switzerland. The erythrocytes were washed twice by centrifugation (5 min, 500 × g) and resuspension in phosphate buffered saline (PBS, pH 7.4, Sigma, Buchs, Switzerland). Early stationary phase cultures of L. monocytogenes were obtained as described above. The OD590 of the cultures was measured and 4 ml were centrifuged for 10 min at 10 000 × g, 4 °C. The supernatants were diluted in BHI to account for differences in OD590 of the cultures. The adjusted supernatants were sterile filtered and 5 μl dithiotreitol (DTT Sigma, Buchs, Switzerland) were added to 995 μl of the supernatant. After one hour of incubation at 37 °C, 100 μl of the supernatants were mixed with an equal amount of 0.05% washed human erythrocytes in a 96-well round bottom plate in triplicate technical replicates. After 40 minutes of incubation at 37 °C, the plate was centrifuged for 5 min at 4300 × g and 100 μl of the supernatants were transferred into a new 96-well plate. The OD at 540 nm was measured with a Microplate reader (Synergy HT, BioTek, Lucerne, Switzerland). Supernatant from washed erythrocytes mixed with DTT reduced BHI served as a blank. In every run, L. monocytogenes 10403S prfA* (G145S43, and ΔprfA mutants44 served as positive and negative controls, respectively.
Statistical analysis
All statistical analysis was performed in R (Version 3.4.2)45 using Studio (Version 1.1.383)46. All figures were produced using the ggplot2 package47.
Medians of Vmax and lag times in BHI were compared using Kruskal-Wallis with Dunn’s post hoc tests as the data did not satisfy the assumptions of a linear model. For the relative growth rates and relative lag times in BHI with 4% NaCl linear models were calculated. Contrasts were calculated using lsmeans48. Model selection was done by nested F-tests.
The acid data did not satisfy the assumptions of a linear model, and exploratory graphs made a clear clustering into two groups apparent. We therefore used k-means49 clustering to group the observations into two clusters and then modelled the probabilities of an observation falling into a cluster using logistic regressions.
BC resistance was modelled using an ordinal regression with polr in MASS50. The six categories of BC concentration were considered in order (0 < 2.5 < 5 < 7.5 < 10 < 15) and an effect for the probability of a subject to be in the next higher category was fitted.
The hemolysis data was analyzed with linear mixed effect models (lmer in LmerTest51) using the OD540 in the supernatant as the dependent variable and lineage, serotype, clonal complex, and source of the strain as fixed effects. To hedge for differences in different batches of human blood obtained at different days, “date” was used as random effect. Lsmeans48 was used to create contrasts and p-values were corrected for multiple comparisons using a false discovery rate of 0.05. Since there were generally no differences in phenotype between strains isolated from food vs clinical sources, they are only discussed in the results section where source was identified as a predictor of the phenotype.
All statistical analysis can be obtained as supplementary materials.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
References
Food, E., Authority, S. & Centre, E. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2014. EFSA J. 13 (2015).
Crim, S. M. et al. Incidence and Trends of Infection with Pathogens Transmitted Commonly Through Food — Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2006–2013. MMWR. Morb. Mortal. Wkly. Rep. 63, 328–332 (2014).
de Valk, H. et al. Surveillance of Listeria infections in Europe. Euro Surveill. Bull. Eur. sur les Mal. Transm. = Eur. Commun. Dis. Bull. 10, 251–255 (2005).
Popovic, I., Heron, B. & Covacin, C. Listeria: an Australian perspective (2001–2010). Foodborne Pathog. Dis. 11, 425–32 (2014).
Kovacevic, J., Arguedas-Villa, C., Wozniak, A., Tasara, T. & Allen, K. J. Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures. Appl. Environ. Microbiol. 79, 1915–1922 (2013).
Althaus, D. et al. Characterization of Listeria monocytogenes Strains Isolated During 2011-2013 from Human Infections in Switzerland. Foodborne Pathog. Dis. 11, 753–758 (2014).
Mulay, S. R. et al. Listeria Pathogenesis and Molecular Virulence Determinants. Nature. Communications 7, 10274 (2016).
Jacquet, C., Gouin, E., Jeannel, D., Cossart, P. & Rocourt, J. Expression of ActA, Ami, InlB, and Listeriolysin O in Listeria monocytogenes of human and food origin. Appl. Environ. Microbiol. 68, 616–622 (2002).
Orsi, R. H., Bakker, H. Cde & Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96 (2011).
Jordan, K., Fox, E. M. & Walker, J. M. Listeria monocytogenes: Methods and Protocols, https://doi.org/10.1007/978-1-4939-0703-8 (2014).
McLauchlin, J. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur. J. Clin. Microbiol. Infect. Dis. 9, 210–213 (1990).
Gray, M. J. et al. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70, 5833–5841 (2004).
Chen, Y., Zhang, W. & Knabel, S. J. Multi-virulence-locus sequence typing identifies single nucleotide polymorphisms which differentiate epidemic clones and outbreak strains of Listeria monocytogenes. J. Clin. Microbiol. 45, 835–846 (2007).
Bergholz, T. M. et al. Determination of Evolutionary Relationships of Outbreak-Associated Listeria monocytogenes Strains of Serotypes 1/2a and 1/2b by Whole Genome Sequencing. Appl. Environ. Microbiol. 82, 928–938 (2016).
Moura, A. et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol., 1–10, https://doi.org/10.1038/nmicrobiol.2016.185 (2016).
Maury, M. M. et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–13 (2016).
Gelbíčová, T., Koláčková, I., Pantůček, R. & Karpíšková, R. A novel mutation leading to a premature stop codon in inlA of Listeria monocytogenes isolated from neonatal listeriosis. New Microbiol. 38, 293–296 (2015).
Jacquet, C. et al. A Molecular Marker for Evaluating the Pathogenic Potential of Foodborne Listeria monocytogenes. J Infect Dis. 189, 2094–2100 (2004).
Langsrud, S., Sidhu, M. S., Heir, E. & Holck, A. L. Bacterial disinfectant resistance—a challenge for the food industry. Int. Biodeterior. Biodegradation 51, 283–290 (2003).
Xu, D. et al. Benzalkonium chloride and heavy-metal tolerance in Listeria monocytogenes from retail foods. Int. J. Food Microbiol. 190, 24–30 (2014).
Müller, A. et al. Tn6188 - A Novel Transposon in Listeria monocytogenes Responsible for Tolerance to Benzalkonium Chloride. Plos One 8, 1–11 (2013).
Kovacevic, J. et al. Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 82, 939–953 (2015).
Elhanafi, D., Utta, V. & Kathariou, S. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998–1999 outbreak. Appl. Environ. Microbiol. 76, 8231–8238 (2010).
Ribeiro, V. B. & Destro, M. T. Listeria monocytogenes Serotype 1/2b and 4b Isolates from Human Clinical Cases and Foods Show Differences in Tolerance to Refrigeration and Salt Stress. J. Food Prot. 77, 1519–1526 (2014).
Magalhães, R. et al. Persistent and non-persistent strains of Listeria monocytogenes: A focus on growth kinetics under different temperature, salt, and pH conditions and their sensitivity to sanitizers. Food Microbiol. 57, 103–108 (2016).
Smith, J. L., Liu, Y. & Paoli, G. C. How does Listeria monocytogenes combat acid conditions? Can. J. Microbiol. 59, 141–52 (2013).
O’Driscoll, B., Gahan, C. G. & Hill, C. Adaptive acid tolerance response in Listeria monocytogenes: isolation of an acid-tolerant mutant which demonstrates increased virulence. Appl. Environ. Microbiol. 62, 1693–8 (1996).
Cheng, C. et al. Listeria monocytogenes varies among strains to maintain intracellular pH homeostasis under stresses by different acids as analyzed by a high-throughput microplate-based fluorometry. Front. Microbiol. 6 (2015).
Lianou, A., Stopforth, J. D., Yoon, Y., Wiedmann, M. & Sofos, J. N. Growth and stress resistance variation in culture broth among Listeria monocytogenes strains of various serotypes and origins. J. Food Prot. 69, 2640–2647 (2006).
Skandamis, P. N., Gounadaki, A. S., Geornaras, I. & Sofos, J. N. Adaptive acid tolerance response of Listeria monocytogenes strains under planktonic and immobilized growth conditions. Int. J. Food Microbiol. 159, 160–166 (2012).
Lundén, J., Tolvanen, R. & Korkeala, H. Acid and heat tolerance of persistent and nonpersistent Listeria monocytogenes food plant strains. Lett. Appl. Microbiol. 46, 276–280 (2008).
Ramalheira, R. et al. Survival of clinical and food isolates of Listeria monocytogenes through simulated gastrointestinal tract conditions. Foodborne Pathog. Dis. 7, 121–128 (2010).
Ryan, S., Hill, C. & Gahan, C. G. M. Chapter 3: Acid Stress Responses in Listeria monocytogenes. Advances in Applied Microbiology 65, 67–91 (2008).
Meier, A. B. et al. Comparative phenotypic and genotypic analysis of swiss and finnish Listeria monocytogenes isolates with respect to benzalkonium chloride resistance. Front. Microbiol. 8, 1–9 (2017).
Ebner, R. et al. Phenotypic and genotypic characteristics of Listeria monocytogenes strains isolated during 2011–2014 from different food matrices in Switzerland. Food Control 57, 321–326 (2015).
Gaillard, J. L., Berche, P., Mounier, J., Richard, S. & Sansonetti, P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55, 2822–2829 (1987).
Portnoy, Da, Jacks, P. S. & Hinrichs, D. J. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167, 1459–1471 (1988).
Schnupf, P. et al. Regulated translation of listeriolysin O controls virulence of Listeria monocytogenes. Mol. Microbiol. 61, 999–1012 (2006).
Zemansky, J. et al. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J. Bacteriol. 191, 3950–3964 (2009).
Maury, M. M. et al. Spontaneous Loss of Virulence in Natural Populations of Listeria monocytogenes. Infect. Immun. 85, e00541–17 (2017).
Ferreira, A., Sue, D., O’Byrne, C. P. & Boor, K. J. Role of Listeria monocytogenes sigma(B) in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl Env. Microbiol 69, 2692–2698 (2003).
Scharer, K., Stephan, R. & Tasara, T. Cold shock proteins contribute to the regulation of listeriolysin O production in Listeria monocytogenes. Foodborne Pathog Dis 10, 1023–1029 (2013).
Vega, Y. et al. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Mol. Microbiol. 52, 1553–1565 (2004).
Wong, K. K. Y. & Freitag, N. E. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J. Bacteriol. 186, 6265–6276 (2004).
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing Vienna Austria 0, {ISBN} 3-900051-07-0 (2015).
R Studio, T. R Studio. Integrated Development for R. R Studio, Inc., Boston, MA, http//www.rstudio.com R Studio, Inc., Boston, MA, https://doi.org/10.1007/978-81-322-2340-5 (2016).
Wickham, H. Ggplot2 Elegant Graphics for Data Analysis. Media 35 (2009).
Lenth, R. V. Least-Squares Means: The R Package lsmeans. J. Stat. Softw. 69, 1–33 (2016).
Hartigan, J. A. & Wong, M. A. A K-Means Clustering Algorithm. Appl. Stat. 28, 100–108 (1979).
Venables, W. N. & Ripley, B. D. MASS: modern applied statistics with S. Issues of Accuracy and Scale 868, https://doi.org/10.1198/tech.2003.s33 (2002).
Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. Lmer Test: Tests in linear mixed effects models. R package version 2.0-33 (2016).
Acknowledgements
We are grateful to Prof. Paul Torgerson at the University of Zurich for his statistical insights on the outset of this project.
Author information
Authors and Affiliations
Contributions
C.G., R.S. and J.H. designed the study. J.H. performed all laboratory work. J.H. and C.G. wrote the manuscript. D.K. performed all statistical analyses.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Horlbog, J.A., Kent, D., Stephan, R. et al. Surviving host - and food relevant stresses: phenotype of L. monocytogenes strains isolated from food and clinical sources. Sci Rep 8, 12931 (2018). https://doi.org/10.1038/s41598-018-30723-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-30723-z
- Springer Nature Limited
This article is cited by
-
Persistent and sporadic Listeria monocytogenes strains do not differ when growing at 37 °C, in planktonic state, under different food associated stresses or energy sources
BMC Microbiology (2019)
-
Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products
Nature Communications (2019)