Abstract
Multidrug resistance (MDR) is the major cause, by which cancer cells expel the drugs out, developing a challenge against the current chemotherapeutic drugs regime. This mechanism is attributed to the over expression of ABC transporters like MRP1 on the surface of cells. Since nucleotide binding domains (NBD) of ABC transporters are the site of ATP binding and hydrolysis, thereby in this study we have targeted NBD1 of MRP1using molecular docking and molecular dynamic simulations (MDS). The compounds present in the FDA approved library were docked against NBD1 of the human multidrug resistance associated protein 1 (PDB ID: 2CBZ). For the docking studies, Standard Precision and Extra Precision methods were employed. After the EP docking studies, ligands showed an extremely low docking score that was indicative of very high binding affinity of the ligands to the NBD. Apart from the low docking score, another short listing criterion in simulation studies was the interaction of incoming ligand with the desired conserved residues of NDB involved in ATP binding and hydrolysis. Based on these measures, potassium citrate (DB09125) and technetium Tc-99m medronate (DB09138) were chosen and subjected to 100 ns simulation studies. From the MDS study we concluded that between these two compounds, potassium citrate is a better candidate for targeting MRP1.
Similar content being viewed by others
Introduction
The leading cause of chemotherapy failure is the multi drug resistance (MDR) exhibited by cancer cells due to which chemotherapeutic drugs are expelled out of the cells. The overexpressed efflux protein pumps belonging to ATP Binding Cassette (ABC) family1 are the key reason for MDR in cancer cells. The ABC superfamily can be divided into two major categories namely, ABC transporters which require ATP for substrate translocation and non-transporter proteins that are responsible for DNA maintenance and gene regulation2. In bacterial system, ABC transporters act as importers as well as exporters. They translocate molecules like ions, amino acids, sugar etc. inside and out of the bacterial cells. In contrary to this, only ABC family exporters are extant in a eukaryotic system. These drug pumps are an important part of a cell’s defense mechanism and this was first realized by DanØ who in 1973 demonstrated that Ehrlich ascites cells lower the concentration of intracellular daunorubicin by expelling it out3. After the discovery of P-glycoprotein by Ling & Juliano in 1976 as a 170 kDa transmembrane glycoprotein, a second type of drug transport protein i.e. multidrug resistance-associated protein (MRP) was later discovered by Chole in 19924. It was cloned from H69R lung cancer cell line. Both the P-gp and MRP belong to the family of ABC transporters and both are responsible for conferring multidrug resistance to cells. In spite of a very low structural identity between MRP1 and P-gp, there was a considerable similarity between the phenotype of H69AR cells and cells expressing P-gp5.
MRP1, a 190 kDa protein is capable of exporting hydrophobic drug molecules, folates, glutathione conjugated compounds, glucornoid conjugated steroids and organic anions to name a few6. It is a 1531 amino acid protein with two Nucleotide Binding Domains (NBD) and three Membrane Spanning Domains (MSDs) namely MSD0, MSD1 and MSD2 consisting of 17 transmembrane helices (TM) deviating from the structure of a typical ABC transporter that consists of two MSDs consisting of 6 TMs7. The MSD0 is not essential for function but it becomes essential for trafficking protein in which the missing COOH- terminal is either mutated or missing thus explaining the different trafficking requirement of MRP1 as compared to other ABC transporter proteins8. Nevertheless, despite the numbers of MSDs present in an ABC transporter their NBDs share many conserved features, especially NBD1. The two NBDs play a crucial role in the transport activity of ABCC proteins.
One feature common to all ABC family members is the dependence of their ATPase activity on the cooperative interactions between the two NBDs. It has already been proved that both of the NBDs are involved in the transport of substrates across the cell and the binding and release of substrate depends on the ordered binding of ATP to the two NBDs. In addition to this, ATP binding is essential for the export of substrates converting the protein from a high-affinity to low affinity substrate binding state9,10. Moreover, ATP binding to NBD1 is not very much dependent on the functional state of NBD2 but for ATP binding to NBD2 is highly dependent on the functional state of NBD19,10. The affinity of NBD1 for ATP is several times higher compared to NBD211. Therefore, it can be possible that NBD2 merely acts as a site of ATP hydrolysis.
Molecular docking provides a platform where using the crystal structure of a protein as a template novel ligands can be determined. A library of compounds and a protein template is required to perform docking studies12. Docking has been applied mainly to enzymes and has been proved useful in finding the novel ligands with good substrate affinities. In order to discover a promising ligand for a macromolecule, it is important to understand the interactions and principles by which the ligand will recognize and interact with it. This forms the basis of structure based drug designing (SBDD). For SBDD, method of molecular docking is employed because it is able to predict the conformation and binding of a ligand into the active site of the macromolecule with preciseness. Using molecular docking, the binding mode of ligand and macromolecule, types of interactions (electrostatic, hydrophobic, pi cation) and the binding affinities can be analyzed13. Although docking algorithms have developed and improved over years, some issues like involvement of water molecules in the protein structure, conformational changes that occur following ligand binding still needs to be resolved14.
Molecular docking and MD simulations approach has already been used by many research groups to target MRP1 as well as P-gp. Williamson et al. used molecular docking analysis to predict the potential binding site of quercetin glucuronides on ABCC2 and then examined these predictions experimentally15. From this study it was concluded that molecular model agreed with the experimentally determined interactions between ABCC2 and ligand. In 2008, a study conducted on MRP5 demonstrated a model for this protein. Further potential binding sites were determined and with docking of guanosine 3′–5′ cyclic monophosphate (cGMP) the importance and involvement of transmembrane helix (TMH) was determined16. Sirisha et al. used molecular docking to find the inhibitors of MRP1 among dihydropyridine derivatives17. In addition to this, potential substrates for other ABCC members have been determined using molecular dynamics simulations. Furthermore, the inhibitory effect of curcumin on MRP1 has been studied by in silico studies.
Targeting ATP-binding site for drug discovery is not a new concept. The strategy has been explored against many proteins including tyrosine kinases. In 1999, Novartis Pharmaceuticals developed a pharmacophore model of the ATP-binding site present in Epidermal Growth Factor Receptor (EGFR)18. Since then this model has been used to find therapeutic molecules that can target the ATP binding site. In addition to this, with the help of docking and molecular dynamics studies on compounds like indirubins, it was predicted that these compounds bind at the active site where the adenine ring of the ATP binds19. Moreover, in order to target the ATP binding site of topoisomerase II a purine inhibitor scaffold was designed leading to identification of novel compounds20. In 2012, another research group targeted the ATP-binding site of type II topoisomerase by synthesizing 5-aryl-1, 3,4-thiadiazole coupled phthalimide derivatives21. These compounds were docked against the ATP-binding site of the protein. The compounds with best in silico results were further evaluated in vitro.
The ATP binding site for the docking studies was determined by previous studies. From the literature it was reviewed that NBD consists of some conserved motifs and loops that are essential for ATP binding and hydrolysis (Fig. 1). These include walker A which is a glycine rich motif responsible for ATP binding. Furthermore, walker B which consists of hydrophobic residues is also involved in ATP binding. The LSGGQ signature sequence also known as the C motif is placed opposite to the walker A and B of the other NBD and helps in completing the ATP binding sites22,23. The consensus D loop sequence (EATSALD) communicates with the other NBD24. In alpha-hemolysin translocation ATP binding protein HlyB, the H loop plays an essential role in ATP hydrolysis25 and in cystic fibrosis transmembrane conductance regulator it is required for the efficient closure of the channel26. Furthermore, the NBDs of the ABC family proteins share a high level of tertiary structure conservation. Therefore, an inhibitor that has been screened against the NBD of MRP1 can also be tried against the NBDs of other proteins from the same family. The transport cycle which involves the binding and consequently the hydrolysis of ATP is dependent on the walker A, walker B, D loop, H loop and Q loop of NBD. Walker A residues are responsible for H-bond formation between the α, β and γ phosphates and walker B motif is responsible for stabilization of ADP as well as for coordination of the magnesium ion. Residues in D and H loop (Asp792 and 793, and His827 respectively) are responsible for the stabilization of ATP. The Q loop has its role in the catalytic cycle in which the Gln713 residue makes contact with the magnesium ion and the attacking water molecule. Nucleotide binding is accompanied by structural changes in the protein involving walker A motif and the Q loop. Although the C signature sequence is not required by MRP1 for ATP binding, mutations in this sequence impedes the transition of protein from a high affinity binding state for substrate to a lower one. This can be attributed to the role of this sequence to form a correct interface for NBD dimer27. A similar phenomenon has been observed in case of P-gp28,29. Mutations in walker A and walker B of NBD2 did not affect the binding of ATP to NBD1 while on the other hand in case of NBD1, mutations in the Lys residue of walker can render the protein partially active or can eliminate the complete transport of leukotriene C4. Moreover, ATP is essential to change the high affinity substrate binding protein to a lower one. And thus it makes sense to target the NBD that is the site of ATP binding.
Within these conserved motifs and loops of NBDs, there are certain amino acid residues which are reported to be essential in ATP binding and hydrolysis. The conserved lysine residue located within the walker A is involved in ATP binding and when replaced by asparagine reduces the ATP hydrolysis activity in maltose/maltodextrin import (MaIK) as compared to the wild type30. The serine residue of the walker A forms H-bond with the Asp of Walker B and interacts with the Mg2+ and the β-phosphate of the bound ATP31. A conserved glutamate is present at the junction of walker B and D-loop is involved in co-ordination of magnesium ion32 and it also polarizes the incoming water molecules and acts as a catalytic base33. Mutation in this glutamate residue reduces the human Transporter Associated with Antigen Processing (TAP) protein transport activity as it is required for the abstraction of protons from the transition product formed34. However, this theory of glutamate acting as a catalytic base is disputed upon25. Study of the nucleotide binding domain of (TAP1) suggests that mutation in the D loop aspartate impedes the NBD dimerization35. In Q loop, glutamine residue is important as it is involved in co-ordination of catalytic metal and organizes the active site so that ATP hydrolysis can take place. The flexible nature of Q loop is utilized in separating the sub domains after the hydrolysis has occurred36,37. Additionally, the H loop histidine is involved in the transfer of protons in ATP hydrolysis25. Mutation in the conserved histidine of H loop reduces the ATPase activity in MaIK30. An interesting finding by Yang and research group suggests that a H-bond between H loop of NBD2 and ATP is essential for the ATP dependent transport of drugs by P-gp across the cell while mutations in H loop of NBD1 did not affect the protein conformation significantly38. Considering all this information about the NBDs of MRP1, it is likely to find an inhibitor that could make a complex with NBD1 thus hampering ATP binding.
The ATP switch and constant contact models are the two proposed models for ATP binding and hydrolysis in ABC transporters. In this model in the resting stage the two NBDs are wide apart without any ATP. As the ATP molecules start binding to the two NBDs sequentially, both the NBDs come together to form a dimer. After this one by one both the ATP molecules are hydrolyzed and converted to ADP and Pi. As the ADP and Pi are released, the protein reverts to the resting stage. On the other hand the constant contact model supports the alternating hydrolysis of ATP. In this the ATP-bound site is closed while the other one is empty and open ATP hydrolysis is followed by the binding of another ATP molecule and as a result of this binding the hydrolysis products of the first ATP bound are released. This cycle continues as long as ATP is available2.
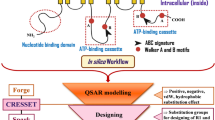
Based on this knowledge about the NBD domain of MRP1, the rational approach to screen for the inhibitors would be to check for the maximum number of interactions of the incoming ligand with the essential (target) residues of NBD. The target residues are those amino acids which are present in the loops and motifs of NBD and are essential for ATP binding and hydrolysis. In case of ABC transporters these residues are conserved. These include Gly681, Cys682, Lys684, Ser685 and Ser686 in Walker A motif, Glu713 in Q loop, Asp792 and Asp793 in D loop, and His827 in the H loop. The aim is to screen the molecules that will interact with maximum of these target residues as to obstruct ATP binding and hydrolysis. Considering this rationale, we have targeted the NBD 1 of the human multi drug resistance protein 1 to search for a compound that could halt the ATP binding and hydrolysis. For this, molecular docking studies were performed using FDA approved drugs as ligand to find the most suitable molecule. The FDA library was subjected to standard precision (SP) and extra precision (XP) mode of docking and the screened compounds demonstrated high binding affinity to the NBD domain that can be inferred by the low docking scores. Further the top hits were screened manually and the two prime ligands which were interacting with the maximum number of target residues of NBD were selected for molecular dynamic simulation studies. The flow chart for the methodology is shown in Fig. 2.
Results
As per the information about the active site and the residues of the NBDs involved in ATP hydrolysis, it is rational to look for the interaction of these target residues with the ligand. The aim was to block these residues so that ATP would not get hydrolyzed and consequently, there would be no efflux of drugs. Eight residues (Gly681, Cys682, Lys684, Cys685, Ser686, Gln713, His827) were short-listed and then the interaction of ligands with the target residues was assessed using docking. On the basis of the docking score and the number of interactions of the ligand with the target residues, the compounds for MD simulations were shortlisted.
Molecular docking studies
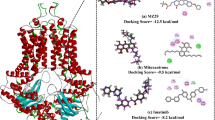
To assess the interactions between the incoming ligand and the active site of the protein docking was performed. First SP docking protocol was applied to shortlist the compounds. From the outcome of SP docking the compounds having lowest parameters were eliminated and a total of 68 compounds which were having a docking score less than −6 were chosen for XP docking. From the XP docking results (Supplementary Table S1), ligands having docking score less than −10 were selected and further manually screened for interactions with the target residues. Based on the highest binding affinity and the maximum number of interaction with the target residues (Supplementary Table S2), four compounds where selected from the XP docking result. Among the four compounds we identified that Potassium citrate (DB09125) has the highest binding affinity of −16.765 Kcal.mol−1 toward ATP binding site of NBD domain. The other three compounds i.e. Technetium Tc-99m medronate (DB09138), Polaprezinc (DB09221) and 4 Samarium (153 Sm) lexidronam (DB05273) showed the binding affinity of −13.059, −12.494 and −12.434 Kcal.mol−1respectively toward the active site (Supplementary Table S1).
Analysis of the four best docked compounds
All the four top hit compounds selected on the basis of XP docking score and number of interaction with the target residues, showed a perfect fit inside the ATP binding grove (Figs 3 and 4) and interacted with the active site residues. The interacting residues, interacting target residues and residues in vicinity of the four compounds in the active site were explained individually.
Potassium citrate (DB09125)
Potassium citrate showed the highest binding affinity toward ATP binding site of NBD domain with a docking score of −16.765. The compound interacted with four target amino acid residues with H-bonds (G681, S686, Q713) and salt bridge (K684). Additionally, one more H-bond interaction was observed with G683. Furthermore, the complex was stabilized by two salt bridge interactions with Mg2+ ion, a H-bond with surrounding water molecules and electrostatic interaction. Apart from this, other amino acid residues comprising the active site pocket consists of polar (Ser685, Gly679) and hydrophobic (Val680, Cys682) amino acids.
Technetium Tc-99m medronate (DB09138)
Technetium Tc-99m medronate showed interactions with the target residues as well as with the other residues present in the active sites. The ligand also showed a high binding affinity towards NBD1 with a docking score of −13.059. It interacted with three target amino acid residues i.e. Ser685, Gln713 (withH-bond) and Lys684 (with two salt bridges). Further, it formed a H-bond with Gly683 and four salt bridges with Mg2+ in the active site. Other amino acids comprising the active site of NBD1 include hydrophobic (Val680, Cys682) and polar amino acids (Ser686, Gly681).
Polaprezinc (DB09221)
Polaprezinc formed a salt bridge with Lys684, two H-bonds with Ser686, one H-bond with Ser 685 and one with Gly 683. Also it shared a pi cation interaction with Lys 684. Besides, it also formed two salt bridges with the Mg2+ion. The whole complex was stabilized by hydrophobic and electrostatic interactions. The other residues lining the active site include the polar amino acids His (827), Gln (713), Thr (660) and Gln (713), hydrophobic amino acids Val (680), Cys (682) and Trp (653) and a negatively charged Asp (793) residue.
Samarium (153Sm) lexidronam (DB05273)
Samarium (153Sm) lexidronam formed an energy complex of −12.434 with the active site of NBD1. The compound interacted with three target residues and formed one H-bond with Gly681, two H-bonds and a salt bridge with Lys684 and one H-bond with Ser685 while a salt bridge was formed between the ligand and the Mg2+ ion. Overall, the complex was stabilized by polar interactions. Apart from the target residues the active site was composed of polar (Ser686, Gln679), non-polar (Gly686) and hydrophobic (Cys682, Val680)amino acid residues.
From the results of molecular docking (Figs 3 and 4), we chose two compounds for MD simulations. The two compounds were selected on the basis of two criteria; the docking score (Supplementary Table S1) and the number of interactions between the ligand and the target residues (Supplementary Table S2). Therefore, potassium citrate and technetium Tc-99m medronate were chosen. Potassium citrate demonstrated a docking score of −16.795 while it interacted with four target residues forming four H-bonds and a salt bridge. Technetium Tc-99m medronate formed an energy complex of −13.059 and displayed interaction with three target residues forming two H-bonds and two salt bridges.
MDS
From the docking results the above two compounds were selected whichfulfilled both the criteria. The selected two complexes (potassium citrate and technetium Tc-99m medronate) with apo-NBD1 were employed for MDS study. MDS is a widely usable technique for understanding the dynamics of the system as well as stability of protein ligand complexes39,40,41,42. A total of three systems (apo-NBD1, NBD1-potassium citrate and NBD-technetium Tc-99m medronate) were created. After that all the systems were employed for 100 ns MDS study. All the systems got the equilibrated state after 40 ns hence last 60 ns stable trajectories were considered for further analysis. In this study, RMSD, RMSF, radius of gyration (Rg), H-bond, PCA, and binding free energy werecalculated.
RMSD
The RMSD was used to measure the scalar distance between the atoms and for prediction of stability of the systems. The backbone RMSD indicated that all the systems are stable and well equilibrated after 40 ns and they produced stable trajectories for further analysis. The apo-NBD1 showed very stable RMSD average value of 0.28 while NBD1-potassium citrate and NBD1-technetium Tc-99m medronate complexes showed 0.28 and 0.30 nm RMSD respectively (Fig. 5A). The result indicated that NBD1-potassium citrate complex showed almost same RMSD as apo-NBD1 and formed much more stable complex as compared to NBD1-technetium Tc-99m medronate complex. From the RMSD result we concluded that all the systems are stable and NBD1-potassium citrate complex is more stable as compared to NBD1-technetium Tc-99m medronate complex.
Molecular dynamics simulations. (A) RMSD of the Cα backbone of NBD1 (black) and the ligand complexes, NBD1-potassium citrate (red), NBD1-technetium Tc-99m medronate (cyan) over the 100 ns MDS at 300 K. (B) RMSF of residues during MDS for NBD1 (black) and the ligand complexes NBD1-potassium citrate (red) and NBD1-technetium Tc-99mmedronate (cyan).
RMSF
The conformational changes during ligand binding were observed by the calculation of RMSF values. The RMSF describes the conformational changes of whole protein or each residue after ligand binding. The apo-NBD1 and both complexes (NBD1-potassium citrate and NBD1-technetium Tc-99m medronate) were used for RMSF calculation. The last 60 ns trajectories were considered for the calculation of RMSF. The average RMSF value for apo-NBD1, NBD1-potassium citrate and NBD1-technetium Tc-99m medronate complexes were 0.12, 0.12 and 0.13 nm respectively. It was observed that the NBD1-potassium citrate complex does not cause much fluctuation after binding and showed RMSF value like apo-NBD1 while the NBD1-technetium Tc-99m medronate complex showed higher fluctuation compared to this ligand. From the Fig. 5B we observed that apo-NBD1 and NBD1-technetium Tc-99m medronate complex showed higher fluctuation in the C-terminal domain while NBD1-potassium citrate does not show this fluctuation. From the RMSF result we concluded that NBD1-potassium citrate complex is novel as compared to NBD-technetium Tc-99m medronate complex.
Radius of gyration
For prediction of the conformational changes and the compactness of the protein-ligand complexes, radius of gyration (Rg) was calculated for the last 60 ns (Fig. 6A). The Rg is defined as the mass weighted root mean square distance of a collection of atoms from their common center of mass. The average Rg value for apo-NBD1, NBD1-potassium citrate and NBD1-technetium Tc-99m medronate complexes were 1.86, 1.88 and 1.84 nm respectively. The Rg value clearly suggests that the NBD1-technetium Tc-99m medronate complex show least Rg value and forms a well compact complex than the NBD1-potassium citrate complex. It also show that after binding of the ligand, the protein attain a compact conformation in the case of NBD1-technetium Tc-99m medronate complex. The NDB1-2 complex showed higher Rg peak in the Fig. 6A. The result represents that both complexes are compact while NBD1-technetium Tc-99m medronate complex is more compact.
Structural stability analysis. (A) Plot of Radius of gyration vs time for NBD1 (black) and the ligands potassium citrate (red), NBD1-technetium Tc-99m medronate (cyan). (B) Number of hydrogen bond interactions formed between NBD1 and the ligands NBD1-potassium citrate (red), NBD1-technetium Tc-99m medronate (cyan) complexes during the last 60 ns simulation time period.
H-bonds
The interaction between protein and ligand is transient and H-bonds play key role in the stability of protein-ligand complex. We have analyzed the number of H-bonds for last 60 ns trajectories (Fig. 6B). The average number of H-bonds for NBD1-potassium citrate and NBD1-technetium Tc-99m medronate complexes was 0–2 and 0–1 respectively. From the Figure we can clearly see that NBD1-potassium citrate complex show more number of H-bonds as compared to NBD1-technetium Tc-99m medronate complex. So the result clearly suggests that the NBD1-potassium citrate complex is more stable. To predict the key residues that play major role in H-bonding between the protein and ligand complex. The percent occupancy of H-bonds was calculated with the interacting residues from last 60 ns trajectory and shown in Supplementary Table S3.
PCA
The correlated motions during ligand binding were predicted by PCA. The first few eigenvectors play key role in the overall significant motions of protein. The first fifty eigenvectors were considered for the analysis. Covariance matrix of atomic fluctuations was diagonalized for predicting the eigenvalues. Figure 7A represents the eigenvalue in decreasing order versus corresponding eigenvector for apo-NBD1, NBD1-potassium citrate and NBD1-technetium Tc-99m medronate complexes. From this result we have analyzed ten eigenvectors that accounted for 74.76%, 78.89% and 79.45% motions for apo-NBD1, NBD1-POTASSIUM CITRATE and NBD1-technetium Tc-99m medronate complexes respectively. The correlated motions showed that apo-NBD1 showed lesser correlated motions as compared to the complexes. While in the case of only two complex the NBD1-technetium Tc-99m medronate showed higher motion and NBD1-POTASSIUM CITRATE showed lower motions. All these calculations were performed on the last 60 ns equilibrated trajectories. The results suggested that NBD1-potassium citrate complex could act as a potential inhibitor than NBD1-technetium Tc-99m medronate complex.
Principal component analysis. (A) Plot of eigenvalues was plotted vs eigenvector index. First 50 eigenvectors were considered. NBD1 (black) and the ligands, NBD1-potassium citrate (red), NBD1-technetium Tc-99m medronate (cyan) indicate the eigenvalues. (B) Projection of the motion of the protein in phase space along the PC1 and PC2 for NBD1 (black) and the ligands NBD1-potassium citrate (red), NBD1-technetium Tc-99m medronate (cyan) at 300 K.
The 2D projection plot is a better way to clearly achieve the essential dynamics of the systems. We observed from the first 50 eigenvectors PCA results that the first two eigenvectors play crucial role in overall motions of the protein. So for predicting the conformational changes induced after ligand binding, we have drawn a 2D projection plot from the last 60 ns MDS trajectories (Fig. 7B). The figure clearly represents that apo-NBD1 showed less stable cluster as compared to both the complex but in the case of the two predicted ligand the NBD1-potassium citrate complex show more stable cluster than NBD1-technetium Tc-99m medronate complex. Thus we can conclude that NBD1-potassium citrate complex is more stable and that the 2D PCA result is in corroboration with the PCA result.
Binding free energy analysis
The binding free energy was calculated for prediction of binding energy of the complexes using MM-PBSA tool. The results are shown in Supplementary Table S4. The last 10 ns trajectories were used for the calculation of binding free energy. The binding free energy also reveals that NBD1-potassium citrate complex has more favorable binding affinity than NBD1-technetium Tc-99m medronate complex. The NBD1-potassium citrate showed a binding energy of −85.31 kJ.mol−1 while NBD1-technetium Tc-99m medronate complex showed −51.53 kJ.mol−1 binding affinity. Furthermore, it was observed that the electrostatic interactions, non-polar solvation energy and Van der Waals interactions negatively complimented the overall interaction energy while the polar solvation energy had positively enriched the binding energy. So from the binding energy result we have concluded that NBD1-potassium citrate complex is more stable as compared to NBD2-12 complex.
Discussion
From the results of molecular docking and MD simulations, we conclude that potassium citrate has a better binding affinity towards the active site of NBD 1 as compared to Tc-99m medronate and the NBD1- Potassium citrate complex was found to be more stable than NBD1-technetium Tc-99m medronate complex. From the docking analysis it was observed that the binding affinity of Potassium citrate (−16.795) was better compared to that of technetium Tc-99m medronate (−13.059). From the 2D ligand interaction diagram it was observed that potassium citrate interacted with maximum number of target residues (Gly681, Lys684, Ser686, Gln713) as compared to technetium Tc-99m medronate (Lys684, Ser685, Gln713). The walker A of NBD is responsible for stabilizing the incoming ATP by forming H-bond with its phosphates. Three of the four target residues of NBD1 that are showing interaction with the potassium citrate are conserved residues of walker A and are essential for ATP binding. All these three residues form H-bond with the phosphates of bound ATP30,31. Additionally, the fourth target residue that is showing interaction with potassium citrate is Gln of Q loop. This residue is crucial as it is involved in co-ordination of catalytic metal and organizes the active site so that ATP hydrolysis can take place36,43. The docking result shows that the ligand potassium citrate occupies the ATP binding domain of NBD and interacts with the crucial amino acid residues that are necessary for ATP binding and active site organization. Therefore, potassium citrate binding will obstruct the binding of ATP and subsequent hydrolysis. As already discussed in the introduction, ATP binding is essential to change the high affinity substrate binding protein to high affinity substrate binding protein. Thus, obstruction in ATP binding will lead to the inhibition of MRP1 and reversal of MDR in cancer cell. Moreover from the overall RMSD, RMSF, PCA, H-bonding, Rg and binding free energy results, it was shown that potassium citrate is a better ligand for the active site of NBD as compared to technetium Tc-99m medronate.
The high expression level of MRP1 leads to multidrug resistance in cancer cells. This poses significant problem as the chemotherapeutic drugs employed to kill cancer cells are expelled out. Therefore it becomes quite evident that MRP1 is an ideal target to tackle MDR in cancer cells. Compounds like verapamil have been used to tackle this problem but its use has been limited because of its cardiotoxicity and the search for inhibitors of drug pumps like P-gp and MRP1 is still going on. In the current scenario, researchers are exploring different approaches to target MRP1 including increased GSH flux mediated by MRP1 that increases the oxidative stress in cells, interactions of MRP1 with different compounds that could obstruct the substrate transport and targeting expression level of MRP1 at mRNA level44.ABC transporters work by binding and hydrolyzing ATP. As already discussed above many other ATPase proteins targeting their ATP binding domain is a well-known approach especially for tyrosinase kinases and topo isomerases. As the transport activity of ABC transporters is powered by ATP binding and hydrolysis targeting its NBD that contains the ATP binding site is worth trying.
With the help of molecular docking and MD simulations we have proposed potassium citrate, an FDA approved compound, for inhibiting the NBD of MRP1.Potassium citrate is not new to be used as a drug. It is an FDA approved compound that has been used in preventing reoccurrence of kidney stones. Additionally, it is also used to deter the age dependent bone density loss45. Researchers have shown that potassium citrate supplementation along with thiazide prevented stone formation in patients with hypocitraturia46. We have hereby proposed for the first time that potassium citrate can bind to the active site of NBD1. Therefore, potassium citrate can act as an inhibitor of MRP1 and thereby it has the potential to hamper the efflux of drug from the cancer cells. Although, further in-vitro studies are needed to ensure the inhibiting potential of potassium citrate. This present study only provides the docking and simulation confidence on potassium citrate to be the best candidate for targeting MRP1 among the FDA approved compounds library.
Materials and Methods
Protein preparation
The crystal structure of human Multi drug resistance protein 1 Nucleotide Binding Domain 1 (PDB ID: 2CBZ) with a resolution of 1.5 Å was taken from RCSB Protein Data Bank (PDB) (http://www.rcsb.org/pdb/). Protein was prepared using the “protein preparation” wizard of Schrödinger Maestro suite. The missing loops and missing side chains were filled and the water molecules, except for those present in the vicinity of the active site, were removed. In this way the protein structure was refined and finally optimized and minimized.
Receptor grid generation
The prepared protein was used to generate the grid upon which the ligands were screened for their affinity towards the active site of protein. Receptor grid was generated using “Glide” module (Schrödinger), around the active site at which ATP along with Mg2+ was bound. Grid was generated in such a way that all the conserved motifs and loops involved in the binding and hydrolysis of ATP were included. The co-ordinates of the generated grid were x = −17.43, y = 47.21, z = 0.6.
Ligand preparation
FDA approved compounds were used for ligand preparation (1996) and were prepared using “LigPrep” module (Schrödinger). LigPrep suite of Schrödinger can generate different states for a single ligand while looking for tautomers, isomers and ionization states. Using LigPrep, the ligands were optimized to possess lowest energy, right chirality and right ionization state. The pH was set to the physiological pH of 7.0. The ligands were optimized using OPLS-2005 force field. Once all the ligands were optimized, they were further considered for docking studies.
Docking
To determine the interactions between the ligands and the active site, docking experiments were performed using the “Ligand Docking” module of Schrödinger. The prepared ligands were docked into receptor grid that was generated on the prepared protein. First the Standard Precision (SP) docking protocol was followed in which the ligands were sorted based on the docking score and glide score. Glide module of Schrödinger uses the Emodel scoring function to compare between the ligand-protein complex poses and the Glide score. It chooses the best pose and then compares the poses with the Glide score. From the result of SP docking, the compounds having lowest parameters were eliminated and then extra precision (XP) docking was performed.
Molecular dynamics simulation (MDS)
MD simulations were performed in an in-house supercomputer using GROMACS 4.6.547,48 with GROMOS 9653a6 force field and SPC216 water model as previously mentioned49,50,51,52. A total of three systems (one for apo-protein and two for protein-ligand complex) were created for 100 ns MDS. The ligand topology was generated by using PRODRUG server53. A cubic simulation box was created and filled with water molecules and then allowed for padding around the protein; Four Na+ ions were included to neutralize the system. Energy minimization was carried out using the steepest descent algorithm for removing the steric clashes of the systems. Then, all the systems were equilibrated under NVT and NPT for 1 ns for position restraint. The electrostatic interactions were computed with particle mesh Ewald (PME) method54. To compute Lennard-Jones and Coulomb interactions, 1.0 nm radius cut-off distance was used. The LINCS algorithm55 was used to constrain the H-bond lengths. The time step was maintained at 2 fs for the simulation. To predict the short-range non-bonded interaction, 10 Å cut-off distance was used. 1.6 Å Fourier grid spacing was used for the PME method for long-range electrostatics. All bonds including H-bonds were fixed by Shake algorithm56. The MD simulations were performed at 300 K temperature with the integration time step of 2 fs.
Analysis of MD data
The analyses on the obtained trajectories were performed using GROMACS utilities. To understand the conformational changes, root mean square deviation (RMSD) and radius of gyration (Rg) of all backbone Cα atoms were computed with respect to the native structure at 300 K. The structural stability of the proteins was defined in terms of root mean square fluctuations (RMSF) and H-bond interaction to provide direct evidence of protein ligand stability. The RMSD and RMSF were calculated using g_rms and g_rmsf tools as mentioned earlier52 while the H-bonds were and principal component analysis (PCA) were carried out by using g_hbond, g_covar and g_anaeig tool of GROMACS package. The trajectories were analyzed using Chimera 1.10.2, and the graphs were plotted by Origin 6.0 software.
Binding free energy calculation
The binding free energy of protein-ligand complexes was calculated using the molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method54,57. Free energy of solvation (polar + non-polar solvation energies) and molecular mechanics potential energy (electrostatic + Van der Waals interaction) were calculated by this tool. In this study, the last 10 ns of the MD trajectories were taken for the calculation of MM-PBSA.
Conclusion
In this study we have described two compounds, potassium citrate and technetium Tc-99m medronate that showed the prospective of reversing MDR in cancer cells. These compounds were screened through rigorous molecular docking methods and subjected to simulations study. We conclude that NBD1-potassium citrate is more stable as compared to NBD1-technetium Tc-99m medronate and thus can be the lead compound against NBD1 of MRP1. Moreover, as the NBD domain is conserved among the members of ABC transporter family the ligand could prove its mettle against other drug pumps also. The experimental studies based on our findings would lead to a better understanding of working with potassium citrate to tackle MDR in cancer cells.
References
Choi, C.-H. ABC transporters as multidrug resistance mechanisms and the development of chemosensitizers for their reversal. Cancer cell international 5, 30 (2005).
George, A. M. & Jones, P. M. Perspectives on the structure–function of ABC transporters: the switch and constant contact models. Progress in biophysics and molecular biology 109, 95–107 (2012).
Danø, K. Active outward transport of daunomycin in resistant Ehrlich ascites tumor cells. Biochimica et Biophysica Acta (BBA)-Biomembranes 323, 466–483 (1973).
Cole, S. et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 258, 1650–1654 (1992).
Conseil, G., Deeley, R. G. & Cole, S. P. Role of two adjacent cytoplasmic tyrosine residues in MRP1 (ABCC1) transport activity and sensitivity to sulfonylureas. Biochemical pharmacology 69, 451–461 (2005).
Deeley, R. G. & Cole, S. P. Substrate recognition and transport by multidrug resistance protein 1 (ABCC1). FEBS letters 580, 1103–1111 (2006).
Bakos, E. et al. Characterization of the amino-terminal regions in the human multidrug resistance protein (MRP1). Journal of cell science 113, 4451–4461 (2000).
Westlake, C. J., Cole, S. P. & Deeley, R. G. Role of the NH2-terminal membrane spanning domain of multidrug resistance protein 1/ABCC1 in protein processing and trafficking. Molecular biology of the cell 16, 2483–2492 (2005).
Gao, M. et al. Comparison of the functional characteristics of the nucleotide binding domains of multidrug resistance protein 1. Journal of Biological Chemistry 275, 13098–13108 (2000).
Hou, Y.-X., Cui, L., Riordan, J. R. & Chang, X.-B. Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. Journal of Biological Chemistry 275, 20280–20287 (2000).
Yang, R., Cui, L., Hou, Y.-X., Riordan, J. R. & Chang, X.-B. ATP binding to the first nucleotide binding domain of multidrug resistance-associated protein plays a regulatory role at low nucleotide concentration, whereas ATP hydrolysis at the second plays a dominant role in ATP-dependent leukotriene C4 transport. Journal of Biological Chemistry 278, 30764–30771 (2003).
Shoichet, B. K., McGovern, S. L., Wei, B. & Irwin, J. J. Lead discovery using molecular docking. Current Opinion in Chemical Biology 6, 439–446 (2002).
Ferreira, L. G., dos Santos, R. N., Oliva, G. & Andricopulo, A. D. Molecular docking and structure-based drug design strategies. Molecules 20, 13384–13421 (2015).
Ghosh, S., Nie, A., An, J. & Huang, Z. Structure-based virtual screening of chemical libraries for drug discovery. Current opinion in chemical biology 10, 194–202 (2006).
Williamson, G. et al. Interaction of positional isomers of quercetin glucuronides with the transporter ABCC2 (cMOAT, MRP2). Drug metabolism and disposition (2007).
Ravna, A. W., Sylte, I. & Sager, G. A molecular model of a putative substrate releasing conformation of multidrug resistance protein 5 (MRP5). European journal of medicinal chemistry 43, 2557–2567 (2008).
Sirisha, K. et al. Molecular docking studies and in vitro screening of new dihydropyridine derivatives as human MRP1 inhibitors. Bioorganic & medicinal chemistry 19, 3249–3254 (2011).
Traxler, P. & Furet, P. Strategies toward the design of novel and selective protein tyrosine kinase inhibitors. Pharmacology & therapeutics 82, 195–206 (1999).
Hayes, J. M. et al. Kinetics, in silico docking, molecular dynamics, and MM‐GBSA binding studies on prototype indirubins, KT5720, and staurosporine as phosphorylase kinase ATP‐binding site inhibitors: The role of water molecules examined. Proteins: Structure, Function, and Bioinformatics 79, 703–719 (2011).
Stahura, F. L., Xue, L., Godden, J. W. & Bajorath, J. Molecular scaffold-based design and comparison of combinatorial libraries focused on the ATP-binding site of proteinkinases1. Journal of Molecular Graphics and Modelling 17, 1–52 (1999).
Gupta, A., Singh, P., Kamble, B., Kulkarni, A. & Joghee Nanjan Chandrasekar, M. Synthesis, Docking and Biological Evaluation of Some Novel 5-bromo-2-(5-aryl-1, 3, 4-thiadiazol-2-yl) isoindoline-1, 3-dione Derivatives Targeting ATP-binding Site of Topoisomerase II. Letters in Drug Design & Discovery 9, 668–675 (2012).
Buchaklian, A. H. & Klug, C. S. Characterization of the LSGGQ and H motifs from the Escherichia coli lipid A transporter MsbA. Biochemistry 45, 12539–12546 (2006).
Jones, P. M. & George, A. M. Subunit interactions in ABC transporters: towards a functional architecture. FEMS microbiology letters 179, 187–202 (1999).
Walker, J. E., Saraste, M., Runswick, M. J. & Gay, N. J. Distantly related sequences in the alpha‐and beta‐subunits of ATP synthase, myosin, kinases and other ATP‐requiring enzymes and a common nucleotide binding fold. The EMBO journal 1, 945–951 (1982).
Zhou, Y., Ojeda-May, P. & Pu, J. H-loop histidine catalyzes ATP hydrolysis in the E. coli ABC-transporter HlyB. Physical Chemistry Chemical Physics 15, 15811–15815 (2013).
Kloch, M. et al. The H-loop in the second nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator is required for efficient chloride channel closing. Cellular Physiology and Biochemistry 25, 169–180 (2010).
Payen, L. et al. Functional interactions between nucleotide binding domains and leukotriene C4 binding sites of multidrug resistance protein 1 (ABCC1). Molecular pharmacology 67, 1944–1953 (2005).
Tombline, G., Bartholomew, L., Gimi, K., Tyndall, G. A. & Senior, A. E. Synergy between conserved ABC signature Ser residues in P-glycoprotein catalysis. Journal of Biological Chemistry 279, 5363–5373 (2004).
Szakács, G., Özvegy, C., Bakos, É., Sarkadi, B. & Váradi, A. Role of glycine-534 and glycine-1179 of human multidrug resistance protein (MDR1) in drug-mediated control of ATP hydrolysis. Biochemical Journal 356, 71–75 (2001).
Davidson, A. L. & Sharma, S. Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. Journal of bacteriology 179, 5458–5464 (1997).
Yang, R., Scavetta, R. & Chang, X. B. Interaction between the bound Mg.ATP and the Walker A serine residue in NBD2 of multidrug resistance-associated protein MRP1 plays a crucial role for the ATP-dependent leukotriene C4 transport. Biochemistry 47, 8456–8464, https://doi.org/10.1021/bi8007643 (2008).
Hung, L.-W. et al. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature 396, 703 (1998).
Davidson, A. L., Dassa, E., Orelle, C. & Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiology and Molecular Biology Reviews 72, 317–364 (2008).
Procko, E., Ferrin-O’Connell, I., Ng, S.-L. & Gaudet, R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Molecular cell 24, 51–62 (2006).
Vakkasoglu, A. S., Srikant, S. & Gaudet, R. D-helix influences dimerization of the ATP-binding cassette (ABC) transporter associated with antigen processing 1 (TAP1) nucleotide-binding domain. PLoS ONE 12, e0178238 (2017).
Campbell, J. D., Deol, S. S., Ashcroft, F. M., Kerr, I. D. & Sansom, M. S. Nucleotide-dependent conformational changes in HisP: molecular dynamics simulations of an ABC transporter nucleotide-binding domain. Biophysical journal 87, 3703–3715 (2004).
Orelle, C. et al. Dynamics of α-helical subdomain rotation in the intact maltose ATP-binding cassette transporter. Proceedings of the National Academy of Sciences 107, 20293–20298 (2010).
Yang, R. & Chang, X.-B. Hydrogen-bond formation of the residue in H-loop of the nucleotide binding domain 2 with the ATP in this site and/or other residues of multidrug resistance protein MRP1 plays a crucial role during ATP-dependent solute transport. Biochimica et Biophysica Acta (BBA)-Biomembranes 1768, 324–335 (2007).
Sonkar, A. et al. UDP-N-Acetylglucosamine enolpyruvyl transferase (MurA) of Acinetobacter baumannii (AbMurA): Structural and functional properties. International journal of biological macromolecules 97, 106–114 (2017).
Pandey, T. et al. A combined biochemical and computational studies of the rho-class glutathione s-transferase sll1545 of Synechocystis PCC 6803. International journal of biological macromolecules 94, 378–385 (2017).
Pathak, R. K. et al. In Silico Identification of mimicking molecules as defense inducers triggering jasmonic acid mediated immunity against alternaria blight disease in Brassica species. Frontiers in plant science 8, 609 (2017).
Shukla, R., Shukla, H. & Tripathi, T. Activity loss by H46A mutation in Mycobacterium tuberculosis isocitrate lyase is due to decrease in structural plasticity and collective motions of the active site. Tuberculosis 108, 143–150 (2018).
Orelle, C. et al. Dynamics of alpha-helical subdomain rotation in the intact maltose ATP-binding cassette transporter. Proc Natl Acad Sci USA 107, 20293–20298, https://doi.org/10.1073/pnas.1006544107 (2010).
Cole, S. P. Targeting multidrug resistance protein 1 (MRP1, ABCC1): past, present, and future. Annual review of pharmacology and toxicology 54 (2014).
Pak, C. Y., Peterson, R. D. & Poindexter, J. Prevention of spinal bone loss by potassium citrate in cases of calcium urolithiasis. The Journal of urology 168, 31–34 (2002).
Pak, C. Y. et al. Correction of hypocitraturia and prevention of stone formation by combined thiazide and potassium citrate therapy in thiazide-unresponsive hypercalciuric nephrolithiasis. The American journal of medicine 79, 284–288 (1985).
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013).
Hess, B., Kutzner, C., Van Der Spoel, D. & Lindahl, E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of chemical theory and computation 4, 435–447 (2008).
Shukla, R. et al. Identification of potential inhibitors of Fasciola giganticathioredoxin1: computational screening, molecular dynamics simulation, and binding free energy studies. Journal of Biomolecular Structure and Dynamics, 1–16 (2017).
Shukla, R., Shukla, H., Kalita, P. & Tripathi, T. Structural insights into natural compounds as inhibitors of Fasciola gigantica thioredoxin glutathione reductase. Journal of cellular biochemistry 119, 3067–3080 (2018).
Shukla, R., Shukla, H., Sonkar, A., Pandey, T. & Tripathi, T. Structure-based screening and molecular dynamics simulations offer novel natural compounds as potential inhibitors of Mycobacterium tuberculosis isocitrate lyase. Journal of Biomolecular Structure and Dynamics, 1–13 (2017).
Kalita, J. et al. Comprehensive analysis of the catalytic and structural properties of a mu-class glutathione s-transferase from Fasciola gigantica. Scientific reports 7, 17547 (2017).
SchuÈttelkopf, A. W. & Van Aalten, D. M. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallographica Section D: Biological Crystallography 60, 1355–1363 (2004).
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98, 10037–10041 (2001).
Hess, B., Bekker, H., Berendsen, H. J. & Fraaije, J. G. LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18, 1463–1472 (1997).
Ryckaert, J., Ciccotti, G. & Berendsen, H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23, 327–341 (1977).
Kumari, R., Kumar, R., Consortium, O. S. D. D. & Lynn, A. g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. Journal of chemical information and modeling 54, 1951–1962 (2014).
Acknowledgements
IIT Mandi is thanked for providing laboratory facilities. NG is supported by DST Ramanujan grant SB/S2/RJN-072/2015. Financial assistance from MHRD for DD and research fellowship to AS from UGC is gratefully acknowledged. Authors acknowledge the funding from IIT Mandi and SERB, grant number SB/S2/RJN-072/2015.
Author information
Authors and Affiliations
Contributions
D.D. and A.S. designed, performed the experiments, analysed the data and did manuscript writing. R.S. performed simulation experiments and assisted in manuscript writing. T.T. helped in planning the experiments. N.G. supervised the study and implemented final approval of manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dhasmana, D., Singh, A., Shukla, R. et al. Targeting Nucleotide Binding Domain of Multidrug Resistance-associated Protein-1 (MRP1) for the Reversal of Multi Drug Resistance in Cancer. Sci Rep 8, 11973 (2018). https://doi.org/10.1038/s41598-018-30420-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-30420-x
- Springer Nature Limited