Abstract
The disruption of the Hippo pathway occurs in many cancer types and is associated with cancer progression. Herein, we investigated the impact of 32 Hippo genes on overall survival (OS) of cancer patients, by both analysing data from The Cancer Genome Atlas (TCGA) and reviewing the related literature. mRNA and protein expression data of all solid tumors except pure sarcomas were downloaded from TCGA database. Thirty-two Hippo genes were considered; for each gene, patients were dichotomized based on median expression value. Survival analyses were performed to identify independent predictors, taking into account the main clinical-pathological features affecting OS. Finally, independent predictors were correlated with YAP1 oncoprotein expression. At least one of the Hippo genes is an independent prognostic factor in 12 out of 13 considered tumor datasets. mRNA levels of the independent predictors coherently correlate with YAP1 in glioma, kidney renal clear cell, head and neck, and bladder cancer. Moreover, literature data revealed the association between YAP1 levels and OS in gastric, colorectal, hepatocellular, pancreatic, and lung cancer. Herein, we identified cancers in which Hippo pathway affects OS; these cancers should be candidates for YAP1 inhibitors development and testing.
Similar content being viewed by others
Introduction
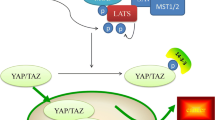
Since its discovery in Drosophila Melanogaster1, Hippo pathway has gained ever-increasing attention. Nowadays, the involvement of Hippo pathway in cancer development and progression is well recognised. However, the different and sometimes controversial roles that it may play rise the scientific interest about this pathway. The main example is the enhanced immune response against the tumor after depletion of the LATS1-2 oncosuppressors observed in immune-competent mice2. Nevertheless, the canonical oncosuppressor role is the widely accepted one3,4. In this view, the kinases axis, represented by STK3-4/LATS1-2, works as a brake, controlling cell cycle, apoptosis and cell patterning, thus avoiding uncontrolled proliferation and loss of epithelial-like features. LATS kinases can be activated by a great variety of stimuli through different groups of kinases, such as MAP4Ks and TAOKs3. The activity of these kinases depends on the presence of co-activators, among which SAV1, NF2 and FRMD6 represents the first to be discovered1,5.
The final outcome of Hippo pathway is the LATS-mediated phosphorylation of YAP1, mainly at the residue S127, leading to its cytoplasmic retention and eventually degradation6. Unphosphorylated YAP1, together with WWTR1, activates the TEAD1-4-mediated transcription in the nucleus, representing the cancer progression accelerator. Finally, VGLL4 is a peptide acting as an oncosuppressor by competing with YAP1-WWTR1 complex to TEADs binding3 (Fig. 1). The presence of natural YAP1 competitor uncovered a new scenario to counterbalance the insufficient Hippo pathway oncosuppressor activity. Several molecules are capable to interfere with YAP1 activity by both mimicking VGLL4 function and preventing YAP1-WWTR1 interaction7. Among YAP1 inhibitors, the photosensitizer verteporfin, already approved by the Food and Drug Administration for the macular degeneration treatment, showed excellent results both in vitro and in mice, with no or limited side effects8,9. Verteporfin is then one of the main candidate to move a step forward as a therapeutic agent for YAP1 inhibition. In the present study, we conducted a data analysis of all solid tumor datasets of The Cancer Genome Atlas (TCGA) except pure sarcomas, and a review of literature to investigate the impact of the Hippo pathway dysregulation on survival of cancer patients, providing food for thought and data-driven proposals for approaching future Hippo-directed therapies.
Hippo pathway. In orange are kinases, in green coactivators or scaffold proteins and in yellow transcription factors or proteins interacting with transcription factors. Green lines refer to active Hippo pathway, which leads to YAP1-WWTR1 inactivation; red lines relate the TEAD-mediated transcription, when the pathway is inactive.
Results
Power analysis and definitive datasets
Thirteen of the twenty-nine downloaded TCGA datasets had β above 0.8 with the set parameters and were selected for further analyses. Details and covariates for each dataset were reported in Table 1.
Survival analyses
Univariate and multivariate results were summarized in Table 2, p values of univariate and multivariate analyses were reported in Supplementary Tables S1 and S2 respectively. Briefly, univariate analyses showed that 12 out of 13 cancer models had at least one Hippo gene associated with patients prognosis and ten datasets had 3 or more significant genes. Brain lower grade glioma and kidney renal clear cell carcinoma had the higher number of Hippo genes associated with patients’ survival, 16 and 15 respectively, whereas liver hepatocellular carcinoma was the only dataset with no significant genes. With regard to genes, TEAD4 and LATS2 were the most frequently associated with patients’ survival, in 6 and 5 out of 13 datasets respectively. Genes and clinical-pathological parameters resulting associated with prognosis after univariate analyses were then used in the multivariate cox regression. Again, 12 out of 13 datasets had at least one Hippo gene as independent survival predictor, and TEAD4 resulted an independent prognostic factor in 3 different datasets. Survival curves of the independent predictors are reported in Fig. 2 and in Supplementary Figure S1.
Kaplan-Meier curves. In the panel are Kaplan-Meier curves of the four independent predictors that correlated with YAP1 protein, coherently with the canonical role of the Hippo pathway. In detail: (a) TEAD3 in Kidney Renal Clear Cell Carcinoma; (b) RASSF1 in Head and Neck Squamous Cell Carcinoma; (c) TEAD4 in Bladder Urothelial Carcinoma; (d) TEAD2 in Brain Lower Grade Glioma. The log-rank p values are also reported.
mRNA-protein correlation
Genes resulted as independent predictors were correlated with the expression of YAP1 and YAP1pS127 proteins. YAP1 and YAP1pS127 expression levels were always highly correlated, whereas a significant correlation between mRNA levels of Hippo genes and at least one of YAP1 or YAP1pS127 proteins was found in 7 datasets. Further details were reported in Table 3 and Supplementary Figure S2.
Review of literature
Seventy-two original articles associated 17 of the 32 Hippo genes with patients’ survival in more than 20 human cancers. Gastric and colorectal cancers were the most frequently tumors reporting association of Hippo genes with patients’ prognosis; whereas the most represented gene was YAP1, reported as prognostic factor in 29 different studies in 14 cancer models. The majority of these 29 studies were conducted on a protein level and, in all but 2, patients with a high expression level of YAP1 had a lower survival rate. In addition, more than 10 studies associated only nuclear and not cytoplasm staining with patients’ prognosis. Table 4 summarizes the review of literature, and Fig. 3 sums up the overall results.
Results summary. For each analysed TCGA datasets, grey circles indicate the presence of: an independent predictor among Hippo components (multivariate survival analysis); a correlation of the independent predictor with YAP1 protein; coherence between poor survival and canonical oncosuppressor role of the Hippo pathway; and the presence of at least 2 independent studies confirming our results.
Discussion
Genetic alterations affecting the Hippo pathway components are generally rare events in the cancer biology landscape, except for malignant pleural mesothelioma and some tumors of the nervous system, such as neurofibromas, meningiomas and shwannomas4,10,11. However, the disruption of this pathway was reported in several human cancers. Epigenetic events, post-transcriptional and post-translational modifications can all play a crucial effect on this pathway12, and simultaneously monitoring all these alterations is impracticable. If a positive aspect can exist in this scenario, it is the converging effect of a great variety of dysregulation on a single protein expression and/or phosphorylation, YAP1. Herein, we investigated the effect of mRNA and protein levels of the Hippo pathway components on survival of cancer patients by both analysing TCGA data and reviewing the literature.
In the large majority of analysed datasets, the mRNA levels of the Hippo pathway components were associated with patients’ survival, and most importantly, in almost all cancer models taken into account at least one of the considered genes was an independent predictor (Table 2). We then decided to move another step forward, on a protein level, to understand if the predictors correlated with the effector, YAP1 protein and its phosphorylation status.
The protein levels from TCGA were obtained by standard reverse phase protein lysate microarray, a technique that allows to reliably estimate protein levels and post-translational modifications, without considering the initial compartmentalization13. As a consequence, we always found a very high direct correlation between YAP1 and YAP1pS127 that theoretically should determine a very different output: TEAD-mediated transcription and YAP1 inactivation respectively. Considering that this incongruence should be overcome by other techniques such as immunohistochemistry (IHC), we found that 7 of the 19 predictors were correlated with high levels of YAP1 protein (Table 3). Interestingly, MAP4Ks never correlated with YAP1 protein, and, when they were independent predictors, very often the expression levels associated with a worse prognosis were not justified by their theoretical role within Hippo pathway. Nevertheless, this is in agreement with other well-known functions of MAP4Ks14 and with 8 out of 9 previous studies that associated high MAP4Ks levels with a worse prognosis (Table 4). Assuming that MAP4Ks should not play a pivotal role in the regulation of Hippo pathway, more than half (7 out of 12) of the other independent predictors were correlated with YAP1. In addition, because of mRNA levels were compared with survival of patients, some incongruence should be accounted for feedback mechanisms such as in the case of LATS2. In fact, LATS2 is a direct transcriptional target of activated YAP1-WWTR1-TEADs15, thus explaining high LATS2 mRNA levels associated with poor prognosis.
Yet, more than half of Hippo genes were already associated with patients’ prognosis in different independent studies in several human cancers (Table 4). High expression levels of YAP1 were repeatedly reported as a poor prognostic factor, especially in gastric, colorectal, hepatocellular, pancreatic and lung cancer. These cancer types should then really benefit from treatment with YAP1 inhibitors, as well as kidney renal clear cell carcinoma, head and neck carcinoma, bladder cancer and lower grade glioma, in which we found not only at least one Hippo gene as an independent prognostic factor, but also a correlation between the predictors and YAP1 protein levels, coherently with their role within Hippo pathway.
In conclusion, the independent impact of YAP1 activation on patients’ survival was repeatedly proven by several independent studies and in a large variety of human cancers. Several molecules can disrupt YAP1 activation, and showed very promising results both in vitro and in mice. Some of these molecules directly bind to YAP1 thus allowing to use its expression levels as a potential predictive biomarker. Moreover, YAP1 evaluation by IHC would provide not only the direct quantification of the protein levels, but also the visualization of its compartmentalization: this is a relevant point because nuclear YAP1 is the real biological effector and strongly correlated with patients prognosis. Indeed, YAP1 quantification by IHC needs to be uniformly assessed because of the wide interpretation criteria that still exist.
Finally, Kary Mullis truly said that the majority of the scientific studies are correlation and not cause-effect, but when a great number of independent studies point in the same direction, maybe the time is ripe to move a step forward.
Methods
Selection of genes and datasets
Thirty-two genes belonging to the core Hippo pathway were considered in the present study (Table 5). Level 3 RNA Seq, level 3 reverse phase protein lysate microarray and clinical data of all solid tumor datasets of TCGA except pure sarcomas were downloaded from cBioPortal (www.cbioportal.org). In order to select datasets for further investigation, power analysis for survival data was performed with the powerSurvEpi R package version 0.0.9. In detail, two hypothetical groups with the same number of patients and the same probability of death were considered. Moreover, postulated risk ratio of 2.3 and alpha of 0.05 were set to assess the statistical power of each dataset. Datasets with β above 0.8 were selected for further analyses.
Survival and correlation analyses
For each dataset, clinical-pathological features mainly affecting patients’ survival according to the eighth edition of the American Joint Committee on Cancer16 were taken into account as covariates. In order to directly compare the effect of genes and covariates, patients with missing values for any of the selected clinical-pathological parameters were removed from the analyses. For each gene, patients were divided into two groups, high and low expression levels, based on the median value. Also for age, the median was used to dichotomize patients. Survival curves were estimated with the Kaplan-Meier method and compared using the log-rank test. Multivariate Cox proportional hazard modelling of genes and covariates identified as potential prognostic factors in the univariate analyses was then used to determine their independent impact on patients’ survival, and to estimate the corresponding hazard ratio, setting high expression as reference group. All survival analyses were performed with the survival R package version 2.41-3. All p values below 0.05 were considered to be statistically significant.
All genes identified as independent prognostic factors were correlated with YAP1 and YAP1pS127 protein expression levels using Pearson’s correlation, following the procedures of Hmisc R package version 4.1-1. The flow chart of data analyses is reported in Fig. 4.
Review of literature
PubMed database (www.ncbi.nlm.nih.gov/pubmed) was used to search papers investigating Hippo genes and survival of cancer patients. All aliases provided by HUGO nomenclature (www.genenames.org) were used. Only English-written original articles were selected, and only papers containing original data and concerning protein or mRNA levels were considered.
Data availability
The datasets analysed during the current study are available at www.cbioportal.org.
References
Wu, S., Huang, J., Dong, J. & Pan, D. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 (2003).
Moroishi, T. et al. The Hippo Pathway Kinases LATS1/2 Suppress Cancer Immunity. Cell 167, 1525–1539.e17 (2016).
Meng, Z., Moroishi, T. & Guan, K.-L. Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016).
Moroishi, T., Hansen, C. G. & Guan, K.-L. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79 (2015).
Hamaratoglu, F. et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27–36 (2006).
Zhao, B. et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007).
Nakatani, K. et al. Targeting the Hippo signalling pathway for cancer treatment. J. Biochem. (Tokyo) 161, 237–244 (2017).
Kang, M.-H. et al. Verteporfin inhibits gastric cancer cell growth by suppressing adhesion molecule FAT1. Oncotarget 8, 98887–98897 (2017).
Liu-Chittenden, Y. et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26, 1300–1305 (2012).
Zygulska, A. L., Krzemieniecki, K. & Pierzchalski, P. Hippo pathway - brief overview of its relevance in cancer. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 68, 311–335 (2017).
Bueno, R. et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 48, 407–416 (2016).
Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 (2010).
Akbani, R. et al. Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report: the RPPA (Reverse Phase Protein Array) society. Mol. Cell. Proteomics MCP 13, 1625–1643 (2014).
Chuang, H.-C., Wang, X. & Tan, T.-H. MAP4K Family Kinases in Immunity and Inflammation. Adv. Immunol. 129, 277–314 (2016).
Moroishi, T. et al. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 29, 1271–1284 (2015).
AJCC Cancer Staging Manual. https://doi.org/10.1007/978-3-319-40618-3 (Springer International Publishing, 2017).
Zhang, J. et al. Loss of large tumor suppressor 1 promotes growth and metastasis of gastric cancer cells through upregulation of the YAP signaling. Oncotarget 7, 16180–16193 (2016).
Ji, T. et al. Decreased expression of LATS1 is correlated with the progression and prognosis of glioma. J. Exp. Clin. Cancer Res. CR 31, 67 (2012).
Lin, X.-Y., Zhang, X.-P., Wu, J.-H., Qiu, X.-S. & Wang, E.-H. Expression of LATS1 contributes to good prognosis and can negatively regulate YAP oncoprotein in non-small-cell lung cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 35, 6435–6443 (2014).
Xu, B. et al. Expression of LATS family proteins in ovarian tumors and its significance. Hum. Pathol. 46, 858–867 (2015).
Zhang, Y. et al. LATS2 is de-methylated and overexpressed in nasopharyngeal carcinoma and predicts poor prognosis. BMC Cancer 10, 538 (2010).
Luo, S. Y. et al. Aberrant large tumor suppressor 2 (LATS2) gene expression correlates with EGFR mutation and survival in lung adenocarcinomas. Lung Cancer Amst. Neth. 85, 282–292 (2014).
Wu, A. et al. LATS2 as a poor prognostic marker regulates non-small cell lung cancer invasion by modulating MMPs expression. Biomed. Pharmacother. Biomedecine Pharmacother. 82, 290–297 (2016).
Zhang, X. et al. Expression of NF-κB-inducing kinase in breast carcinoma tissue and its clinical significance. Int. J. Clin. Exp. Pathol. 8, 14824–14829 (2015).
Hao, J.-M. et al. A five-gene signature as a potential predictor of metastasis and survival in colorectal cancer. J. Pathol. 220, 475–489 (2010).
Liu, A.-W. et al. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 17, 710–720 (2011).
Qiu, M.-H. et al. Expression and prognostic significance of MAP4K4 in lung adenocarcinoma. Pathol. Res. Pract. 208, 541–548 (2012).
Liang, J. J. et al. Expression of MAP4K4 is associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 14, 7043–7049 (2008).
Wang, O. H. et al. Prognostic and Functional Significance of MAP4K5 in Pancreatic Cancer. PloS One 11, e0152300 (2016).
Sugimachi, K. et al. Altered Expression of Hippo Signaling Pathway Molecules in Intrahepatic Cholangiocarcinoma. Oncology 93, 67–74 (2017).
Luo, Z.-L. et al. A splicing variant of Merlin promotes metastasis in hepatocellular carcinoma. Nat. Commun. 6, 8457 (2015).
Meerang, M. et al. Low Merlin expression and high Survivin labeling index are indicators for poor prognosis in patients with malignant pleural mesothelioma. Mol. Oncol. 10, 1255–1265 (2016).
Klacz, J. et al. Decreased expression of RASSF1A tumor suppressor gene is associated with worse prognosis in clear cell renal cell carcinoma. Int. J. Oncol. 48, 55–66 (2016).
Guo, W. et al. Decreased expression of RASSF1A and up-regulation of RASSF1C is associated with esophageal squamous cell carcinoma. Clin. Exp. Metastasis 31, 521–533 (2014).
Zhang, Y. et al. Prognostic and predictive role of COX-2, XRCC1 and RASSF1 expression in patients with esophageal squamous cell carcinoma receiving radiotherapy. Oncol. Lett. 13, 2549–2556 (2017).
Zhou, R. et al. RASSF6 downregulation promotes the epithelial-mesenchymal transition and predicts poor prognosis in colorectal cancer. Oncotarget 8, 55162–55175 (2017).
Wen, Y. et al. Decreased expression of RASSF6 is a novel independent prognostic marker of a worse outcome in gastric cancer patients after curative surgery. Ann. Surg. Oncol. 18, 3858–3867 (2011).
Guo, W. et al. Decreased expression and frequent promoter hypermethylation of RASSF2 and RASSF6 correlate with malignant progression and poor prognosis of gastric cardia adenocarcinoma. Mol. Carcinog. 55, 1655–1666 (2016).
Ye, H.-L. et al. Low RASSF6 expression in pancreatic ductal adenocarcinoma is associated with poor survival. World J. Gastroenterol. 21, 6621–6630 (2015).
Wang, L. et al. Expression profile and prognostic value of SAV1 in patients with pancreatic ductal adenocarcinoma. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. https://doi.org/10.1007/s13277-016-5457-4 (2016).
Lin, X. et al. Prognostic significance of mammalian sterile 20-like kinase 1 in breast cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 34, 3239–3243 (2013).
Lin, X.-Y. et al. Mammalian sterile 20-like kinase 1 expression and its prognostic significance in patients with breast cancer. Oncol. Lett. 14, 5457–5463 (2017).
Yu, J. et al. Identification of MST1 as a potential early detection biomarker for colorectal cancer through a proteomic approach. Sci. Rep. 7, 14265 (2017).
Minoo, P. et al. Prognostic significance of mammalian sterile20-like kinase 1 in colorectal cancer. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 20, 331–338 (2007).
Zlobec, I. et al. Role of RHAMM within the hierarchy of well-established prognostic factors in colorectal cancer. Gut 57, 1413–1419 (2008).
Ge, X. & Gong, L. MiR-590-3p suppresses hepatocellular carcinoma growth by targeting TEAD1. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 39, 1010428317695947 (2017).
Knight, J. F. et al. TEAD1 and c-Cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br. J. Cancer 99, 1849–1858 (2008).
Liu, Y. et al. Increased TEAD4 expression and nuclear localization in colorectal cancer promote epithelial-mesenchymal transition and metastasis in a YAP-independent manner. Oncogene 35, 2789–2800 (2016).
Xia, Y. et al. YAP promotes ovarian cancer cell tumorigenesis and is indicative of a poor prognosis for ovarian cancer patients. PloS One 9, e91770 (2014).
Takahashi, H. et al. Prognostic significance of Traf2- and Nck- interacting kinase (TNIK) in colorectal cancer. BMC Cancer 15, 794 (2015).
Jin, J. et al. Nuclear expression of phosphorylated TRAF2- and NCK-interacting kinase in hepatocellular carcinoma is associated with poor prognosis. Pathol. Res. Pract. 210, 621–627 (2014).
Zhang, Y. et al. TNIK serves as a novel biomarker associated with poor prognosis in patients with pancreatic cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 37, 1035–1040 (2016).
Jiao, S. et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166–180 (2014).
Yoshihama, Y. et al. High expression of KIBRA in low atypical protein kinase C-expressing gastric cancer correlates with lymphatic invasion and poor prognosis. Cancer Sci. 104, 259–265 (2013).
Wang, L. et al. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PloS One 8, e65539 (2013).
Sun, L. et al. Prognostic impact of TAZ and β-catenin expression in adenocarcinoma of the esophagogastric junction. Diagn. Pathol. 9, 125 (2014).
Guo, Y. et al. Functional and clinical evidence that TAZ is a candidate oncogene in hepatocellular carcinoma. J. Cell. Biochem. 116, 2465–2475 (2015).
Hayashi, H. et al. An Imbalance in TAZ and YAP Expression in Hepatocellular Carcinoma Confers Cancer Stem Cell-like Behaviors Contributing to Disease Progression. Cancer Res. 75, 4985–4997 (2015).
Xie, M. et al. Prognostic significance of TAZ expression in resected non-small cell lung cancer. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 7, 799–807 (2012).
Li, Z. et al. The Hippo transducer TAZ promotes epithelial to mesenchymal transition and cancer stem cell maintenance in oral cancer. Mol. Oncol. 9, 1091–1105 (2015).
Zhang, Y., Xue, C., Cui, H. & Huang, Z. High expression of TAZ indicates a poor prognosis in retinoblastoma. Diagn. Pathol. 10, 187 (2015).
Wei, Z. et al. Overexpression of Hippo pathway effector TAZ in tongue squamous cell carcinoma: correlation with clinicopathological features and patients’ prognosis. J. Oral Pathol. Med. Off. Publ. Int. Assoc. Oral Pathol. Am. Acad. Oral Pathol. 42, 747–754 (2013).
Zhan, M. et al. Prognostic significance of a component of the Hippo pathway, TAZ, in human uterine endometrioid adenocarcinoma. Oncol. Lett. 11, 3611–3616 (2016).
Abduch, R. H. et al. Unraveling the expression of the oncogene YAP1, a Wnt/beta-catenin target, in adrenocortical tumors and its association with poor outcome in pediatric patients. Oncotarget 7, 84634–84644 (2016).
Liu, J.-Y. et al. Overexpression of YAP 1 contributes to progressive features and poor prognosis of human urothelial carcinoma of the bladder. BMC Cancer 13, 349 (2013).
Cao, L., Sun, P.-L., Yao, M., Jia, M. & Gao, H. Expression of YES-associated protein (YAP) and its clinical significance in breast cancer tissues. Hum. Pathol. 68, 166–174 (2017).
Kim, H. M., Jung, W. H. & Koo, J. S. Expression of Yes-associated protein (YAP) in metastatic breast cancer. Int. J. Clin. Exp. Pathol. 8, 11248–11257 (2015).
Kim, S. K., Jung, W. H. & Koo, J. S. Yes-associated protein (YAP) is differentially expressed in tumor and stroma according to the molecular subtype of breast cancer. Int. J. Clin. Exp. Pathol. 7, 3224–3234 (2014).
Pei, T. et al. YAP is a critical oncogene in human cholangiocarcinoma. Oncotarget 6, 17206–17220 (2015).
Wang, Y., Xie, C., Li, Q., Xu, K. & Wang, E. Clinical and prognostic significance of Yes-associated protein in colorectal cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 34, 2169–2174 (2013).
Yeo, M.-K. et al. Correlation of expression of phosphorylated and non-phosphorylated Yes-associated protein with clinicopathological parameters in esophageal squamous cell carcinoma in a Korean population. Anticancer Res. 32, 3835–3840 (2012).
Li, M. et al. Yes-associated protein 1 (YAP1) promotes human gallbladder tumor growth via activation of the AXL/MAPK pathway. Cancer Lett. 355, 201–209 (2014).
Huang, S. et al. Significant association of YAP1 and HSPC111 proteins with poor prognosis in Chinese gastric cancer patients. Oncotarget 8, 80303–80314 (2017).
Sun, D. et al. YAP1 enhances cell proliferation, migration, and invasion of gastric cancer in vitro and in vivo. Oncotarget 7, 81062–81076 (2016).
Li, P. et al. Elevated expression of Nodal and YAP1 is associated with poor prognosis of gastric adenocarcinoma. J. Cancer Res. Clin. Oncol. 142, 1765–1773 (2016).
Song, M., Cheong, J.-H., Kim, H., Noh, S. H. & Kim, H. Nuclear expression of Yes-associated protein 1 correlates with poor prognosis in intestinal type gastric cancer. Anticancer Res. 32, 3827–3834 (2012).
Kang, W. et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 17, 2130–2139 (2011).
Liu, M. et al. Phosphorylated mTOR and YAP serve as prognostic markers and therapeutic targets in gliomas. Lab. Investig. J. Tech. Methods Pathol. 97, 1354–1363 (2017).
Lee, K. et al. The correlation between poor prognosis and increased yes-associated protein 1 expression in keratin 19 expressing hepatocellular carcinomas and cholangiocarcinomas. BMC Cancer 17, 441 (2017).
Wu, H. et al. Clinicopathological and prognostic significance of Yes-associated protein expression in hepatocellular carcinoma and hepatic cholangiocarcinoma. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 37, 13499–13508 (2016).
Xu, B. et al. Menin promotes hepatocellular carcinogenesis and epigenetically up-regulates Yap1 transcription. Proc. Natl. Acad. Sci. USA 110, 17480–17485 (2013).
Han, S. et al. Expression and clinical significance of YAP, TAZ, and AREG in hepatocellular carcinoma. J. Immunol. Res. 2014, 261365 (2014).
Sun, P.-L. et al. Cytoplasmic YAP expression is associated with prolonged survival in patients with lung adenocarcinomas and epidermal growth factor receptor tyrosine kinase inhibitor treatment. Ann. Surg. Oncol. 21(Suppl 4), S610–618 (2014).
Menzel, M. et al. In melanoma, Hippo signaling is affected by copy number alterations and YAP1 overexpression impairs patient survival. Pigment Cell Melanoma Res. 27, 671–673 (2014).
He, C. et al. YAP forms autocrine loops with the ERBB pathway to regulate ovarian cancer initiation and progression. Oncogene 34, 6040–6054 (2015).
Salcedo Allende, M. T. et al. Overexpression of Yes Associated Protein 1, an Independent Prognostic Marker in Patients With Pancreatic Ductal Adenocarcinoma, Correlated With Liver Metastasis and Poor Prognosis. Pancreas 46, 913–920 (2017).
Zhao, X. et al. A combinatorial strategy using YAP and pan-RAF inhibitors for treating KRAS-mutant pancreatic cancer. Cancer Lett. 402, 61–70 (2017).
Wei, H. et al. Hypoxia induces oncogene yes-associated protein 1 nuclear translocation to promote pancreatic ductal adenocarcinoma invasion via epithelial-mesenchymal transition. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 39, https://doi.org/10.1177/1010428317691684 (2017).
Acknowledgements
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC, grant number IG_10316_2010); and Progetti di Rilevante Interesse Nazionale (PRIN, grant number 2015HPMLFY).
Author information
Authors and Affiliations
Contributions
A.M.P., L.T. and F.B. designed the study, A.M.P. performed statistical analyses, F.B. and G.F. supervised the study, A.M.P. and R.B. write the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Poma, A.M., Torregrossa, L., Bruno, R. et al. Hippo pathway affects survival of cancer patients: extensive analysis of TCGA data and review of literature. Sci Rep 8, 10623 (2018). https://doi.org/10.1038/s41598-018-28928-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-28928-3
- Springer Nature Limited
This article is cited by
-
Activation of AMPK inhibits Galectin-3-induced pulmonary artery smooth muscle cells proliferation by upregulating hippo signaling effector YAP
Molecular and Cellular Biochemistry (2021)
-
The clinical relevance of the Hippo pathway in pancreatic ductal adenocarcinoma
Journal of Cancer Research and Clinical Oncology (2021)
-
Whole transcriptome targeted gene quantification provides new insights on pulmonary sarcomatoid carcinomas
Scientific Reports (2019)