Abstract
To evolve rice varieties resistant to different groups of insect pests a fusion gene, comprising DI and DII domains of Bt Cry1Ac and carbohydrate binding domain of garlic lectin (ASAL), was constructed. Transgenic rice lines were generated and evaluated to assess the efficacy of Cry1Ac::ASAL fusion protein against three major pests, viz., yellow stem borer (YSB), leaf folder (LF) and brown planthopper (BPH). Molecular analyses of transgenic plants revealed stable integration and expression of the fusion gene. In planta insect bioassays on transgenics disclosed enhanced levels of resistance compared to the control plants. High insect mortality of YSB, LF and BPH was observed on transgenics compared to that of control plants. Furthermore, honeydew assays revealed significant decreases in the feeding ability of BPH on transgenic plants as compared to the controls. Ligand blot analysis, using BPH insects fed on cry1Ac::asal transgenic rice plants, revealed a modified receptor protein-binding pattern owing to its ability to bind to additional receptors in insects. The overall results authenticate that Cry1Ac::ASAL protein is endowed with remarkable entomotoxic effects against major lepidopteran and hemipteran insects. As such, the fusion gene appears promising and can be introduced into various other crops to control multiple insect pests.
Similar content being viewed by others
Introduction
Rice (Oryza sativa L.) is one of the major food grain crops which serves as a staple diet for more than two-thirds of the world’s population. Annually, about 24–41% of rice yield is lost due to diverse pests and diseases1, resulting from the widespread infestation of insect pests with rare feeding habits. Among the major insects of rice belonging to hemiptera (sap-sucking planthoppers) and lepidoptera (stem borers and leaf folders) are difficult to control and manage under field conditions. Amongst lepidopteran insects, yellow stem borer (YSB) (Scirphophaga incertulus Walker), striped stem borer (SSB) (Chilo supressalis Walker) and leaf folder (LF) (Cnaphalocrosis medinalis Guenee) proved to be major pests causing significant yield losses of up to 10–30% each year2. Brown planthopper (BPH) is one of the most notorious rice pest among hemipteran insects, which feeds mainly on stems and assimilates the phloem sap, causing wilted tillers and withered leaves. Further, BPH also serves as vector for transmitting viruses leading to severe decline in the rice productivity3.
Insecticidal Cry proteins from the bacterium Bacillus thuringiensis (Bt) are widely used as topical sprays besides their deployment in transgenic plants for insect control. These toxins have been expressed in various crops like maize, potato, tomato, cotton, brinjal, and rice for controlling major insect pests. However, they were found effective only against lepidopteran, coleopteran, and dipteran insect pests but were ineffective against hemipteran pests4,5,6. The current understanding of the mode of action of Cry toxins indicates that Bt inclusions get solubilized in the digestive tract of target insects, Cry protoxins are then activated and subsequently bind to the receptors on the epithelium of insect midgut and lyse the cells7. The toxic core structure of Cry1Ac (DI-II-III) is a soluble monomeric protein with Domain I being involved in toxin oligomerization, membrane insertion as well as pore formation8; while Domain II is associated with binding to specific larval midgut proteins and Domain III has a functional role in receptor recognition9. Strategies adopted for alteration of Bt toxin binding affinity and specificity can be divided into four classes, viz., domain or loop swapping between Cry toxins, site-directed mutagenesis, incorporation of binding peptides or fragments from non-Bt toxins, as well as generation and subsequent display of Cry toxin mutant libraries on phage10.
The proteins of plant origin such as lectins have been found effective against different sap-sucking insects. Lectins recognize and preferentially bind to carbohydrate complexes protruding from glycolipids and glycoproteins11. These carbohydrate-binding proteins contain two or more binding sites per subunit which can reversibly bind to specific sugar segments. In addition, they are involved in defense against phytopathogenic microorganisms, phytophagous insects and plant eating animals. The insecticidal activity of plant lectins against a large array of insect species belonging to several orders have been well documented1. Resistance to proteolytic degradation by the insect digestive enzymes and binding to insect gut membranes are the two important attributes that enable lectins to exert their deleterious effects on insects12. Garlic lectin ASAL binds to the carbohydrate part of the 55 kDa and 45 kDa brush border membrane vesicle receptor proteins in hemipteran insects. Binding of ASAL to these receptors decreases the permeability of the membrane and interfere with the digestive, protective or secretary functions of the intestine. Accordingly, lectins can adversely affect weight gain in the larvae, which in turn retard their development into pupae13,14.
In artificial diet assays, mannose-specific lectins revealed a significant anti-metabolic effect towards nymphs of the rice brown planthopper and a high mortality against the red cotton bug (Dysdercus cingulatus)15,16. The insecticidal activity of the ASAL expressed in transgenic cotton caused detrimental effects on larval development, besides growth and survival of the major lepidopteran pest Spodoptera littoralis17. Transgenic rice expressing ASAL exhibited explicit resistance against hemipteran insects BPH, green leafhopper (GLH) and white backed planthopper (WBPH)1,18. Similarly, transgenic cotton lines expressing ASAL displayed enhanced resistance against two major sap-sucking (jassid and whitefly) pests19.
The structural similarity between third domain of Cry1Ac and carbohydrate binding of ASAL lectin has been well studied20. In several cases, exchanging Domain III of the Cry1 toxins with other Cry1 toxins resulted in the enhanced toxicity of Cry proteins21,22,23,24. Previous studies indicated that Cry toxins, despite being activated in the gut of hemipteran insects, showed almost no toxicity25,26. In our earlier studies, based on the structural similarity between the receptor binding domain III of Cry and the carbohydrate binding domain of ASAL, the domain DIII was replaced with the carbohydrate binding domain of lectin and the resulting fusion-protein (Cry1Ac::ASAL) was evaluated for its toxic effects against major lepidoptern insects20,27. The overall results of in silico and in vitro studies on Cry1Ac::ASAL fusion protein revealed its higher binding energies and increased affinity towards insect receptors as compared to the parental proteins20,27.
In the present study, transgenic rice plants expressing Cry1Ac::ASAL fusion protein have been generated and tested against different insect pests using in planta bioassays. Transgenic rice plants exhibited significantly higher levels of resistance to three major insects of rice belonging to lepidopteran and hemipteran pests. Therefore, the engineered Bt toxin (fusion protein), which proved to be more effective against both the chewing (lepidopteran) and sucking (hemiptera) insects, holds great promise to meet the challenge of future pest management under the changed climatic conditions.
Results
Construction of a plant expression cassette containing cry1Ac::asal fusion gene
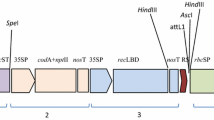
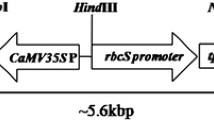
The cry1Ac::asal hybrid gene was constructed by fusion of DI and DII domains of Cry1Ac with carbohydrate binding domain of ASAL. The fusion gene (cry1Ac::asal) was amplified and was cloned into the pBSSK (+) vector, and restriction analysis of the recombinant plasmid showed the presence of a 1464 bp fragment (Supplementary Fig. S1a). The fusion gene was further cloned into pRT100 vector and restriction analysis of the recombinant vector with Nco I & Bam HI enzymes confirmed the presence of 1089 bp and 375 bp fragments corresponding to cry1Ac and asal (Supplementary Fig. S1b). The cry1Ac::asal gene driven by CaMV35S promoter along with its polyA terminator was excised and cloned at the Hind III site of pCAMBIA3300 binary vector, which when digested with Hind III released a 2.2 kb band corresponding to the pCAMBIA3300-cry1Ac::asal-bar expression unit (Supplementary Fig. S1c).
Generation of transgenic rice lines expressing the fusion gene (cry1Ac::asal) and identification of stable transformants
Embryogenic calli of Pusa Basmathi (PB1) were co-cultivated with the Agrobacterium strain EHA105 harbouring pCAMBIA3300-cry1Ac::asal-bar expression vector for the introduction of fusion gene into the rice genome (Fig. 1a). A total of twenty independent transgenic rice lines were generated from the selected calli grown on PPT (6–8 mg/l) containing medium. Among twenty transformants, six transgenic plants showed consistent high tolerance to the Basta (0.25%) herbicide in repeated tests (Supplementary Fig. S2a). PCR analyses of DNA extracts of Basta tolerant transgenic rice plants exhibited amplification of 1089 bp and 560 bp products corresponding to cry1Ac and bar transgenes, while no such bands were observed in the control plants (Supplementary Fig. S2b,c).
Restriction map of T-DNA region of pCAMBIA3300-cry1Ac::asal–bar and Southern blot analysis of transgenic plants. (a) Restriction map of T-DNA region containing CaMV 35S-cry1Ac::asal expression unit and bar selectable marker gene. (b) Genomic DNA digested with HindIII and probed with cry1Ac::asal coding sequence. (c) Genomic DNA digested with EcoRI and probed with bar coding sequence. Each lane was loaded with 15 µg of genomic DNA digested with respective enzymes. Lane P: Positive control (b) cry1Ac::asal expression unit and (c) bar expression unit. Lane UC: DNA from untransformed control plant. Lanes 1, 2, 3, 4, 5 & 6: DNA from PB-F1D2, PB-F3D4, PB-F4D2, PB-F8D4, PB-F17D1 and PB-F20A5 transgenic lines of Pusa Basmathi.
Southern blot, RT-PCR and Western blot analyses of transgenic plants
Southern blot analysis of six transformants, viz., PB-F1D, PB-F3D, PB-F4D, PB-F8D, PB-F17D, and PB-F20A when probed with the cry1Ac::asal coding sequence revealed a specific hybridizable band of ~2.5 kb (Fig. 1b). Further, when probed with the bar coding sequence hybridizable bands of varied sizes (>2.5 kb) were observed in different rice transformants (Fig. 1c). Whereas, untransformed control plants failed to show similar hybridization bands.
RT-PCR analysis of Southern-positive transgenic plants revealed the presence of cry1Ac::asal gene transcripts as evidenced by the amplification of 1089 bp and 375 bp bands corresponding to cry1Ac and asal regions (Fig. 2a,b). Furthermore, different transgenic plants showed various intensities of both cry and asal transcripts. Western blot analyses of transgenic plants demonstrated the presence of ~55 kDa Cry1Ac::ASAL protein when treated with ASAL antibodies (Fig. 2c). Moreover, the amount of fusion protein quantified by ELISA analysis using Cry1Ac anti-body which revealed the presence of Cry1Ac::ASAL in varied amounts ranging from 1.0% to 1.8% in different transgenic plants.
RT PCR and Western blot analyses of cry1Ac::asal transgenic rice plants along with control plants. (a) RT PCR analysis using RNA isolated form transgenic and control plants with primers corresponding to cry1Ac region. Amplified products were analyzed by agarose gel electrophoresis. Lane M: 1Kb DNA marker, Lane 1: Positive control, Lane 2: Untransformed control; Lanes 3–8: Different cry1Ac::asal transgenic rice plants. (b) RT PCR analysis using RNA isolated form transgenic and control plants with primers corresponding to asal region. Lane M: 1Kb DNA marker, Lane 1: Positive control, Lane 2: Untransformed control; Lanes 3–8: Different cry1Ac::asal transgenic rice plants. (c) Western blot analysis using the protein extracted from transgenic rice and control plants. Lane C: Protein extract (5 μg) from untransformed control plant, Lane P: Protein extract from purified protein; Lanes 1, 2, 3, 4, 5 & 6: Protein extracts (5 μg) from PB-F1D2, PB-F3D4, PB-F4D2, PB-F8D4, PB-F17D1, and PB-F20A5 transgenic lines.
Evaluation of the entomotoxic effects of Cry1Ac::ASAL fusion protein on the lepidopteran insects of rice
For evaluation of entomotoxic toxic effects of Cry1Ac::ASAL fusion-protein expressed in the transgenic rice plants, insect bioassays were carried out using two major lepidopteran insects, YSB and LF. Insect bioassays on six different transgenic lines confirmed the efficacy of Cry1Ac::ASAL fusion protein against two major pests of rice. Complete (100%) insect mortality was observed against YSB which were fed on cut stems of transgenic lines at the tillering stage (Fig. 3a). Similarly, mortality of LF fed on two-week-old transgenic rice lines was found to be 80–100% (Fig. 3b). Insect mortality was observed from third day in four transgenic rice lines, while in the remaining two lines it was delayed by two more days. The insect mortality was evident by the typical symptom of Cry toxin activity where larvae turned black. The percentage of leaf damage caused by LF larvae on PB-F4D2, PB-F3D4, PB-F17D1 and PB-F20A5 transgenic rice plants was 10 ± 1, 12 ± 2, 12 ± 1 and 12 ± 1, and on PB-F1D2 and PB-F8D4 lines it was 27 ± 3 and 24 ± 2, respectively, compared to 100% damage observed on untransformed control plants (Fig. 3b).
Bio-assay of transgenic rice plants against yellow stem borer and leaf folder. (a) Yellow stem borer insects fed on six weeks old untransformed control plant (UC) and PB-F3D4, PB-F4D2 & PB-F17D1 transgenic plants. (b) Leaf folder insects fed on six weeks old untransformed control plant (UC) and PB-F3D4, PB-F4D2 & PB-F17D1 transgenic plants.
Evaluation of the fusion protein against BPH insects
Brown planthopper bioassays using cry1Ac::asal transgenic plants revealed significant mortality of larval nymphs and reduction in the fecundity of insects. Transgenic plants expressing Cry1Ac::ASAL fusion protein exhibited varied levels (1–3 score on a 0–9 scale) of resistance to BPH on par with those of BPH-resistant check var. PTB33 (Fig. 4a). Honeydew assay revealed significant reduction of ~ 87% to 93% in the feeding ability of BPH on transgenic plants when compared to the insects fed on the untransformed control plants (Fig. 4b). A mean number of 4.3 ± 0.27, 5.0 ± 0.47 and 10.0 ± 0.48 honeydew units/plant were excreted by BPH, when fed on transgenic plants PB-F4D2, PB-F3D4 and PB-F17D1, respectively, compared with 124.0 ± 0.94 honeydew units/plant observed on the control plants (Fig. 5a). The survival of BPH on PB-F4D2, PB-F3D4 and PB-F17D1 transgenic rice plants was significantly reduced to 3.6 ± 1.4, 3.3 ± 1.6 and 4.3 ± 1.1 insects/plant, respectively, compared to 12.3 ± 2.9 insects/plant on untransformed control PB1 plants (Fig. 5b). Amongst survived BPH insects, only 20% to 30% could reach the adult stage on different transgenic lines as compared to insect survival on control plants (Fig. 5c).
Evaluation of transgenic rice lines expressing Cry1Ac::ASAL fusion protein against brown planthopper (BPH). (a) Six weeks old transgenic lines along with respective controls infested with BPH nymphs. TN-1 and untransformed PB1 rice plants represent controls, var. PTB33 a Resistant check for BPH and PB-F3D4, PB-F4D2 and PB-F17D1 represent transgenic rice lines of PB1. Photographs were taken after 14 days of infestation. (b) Honeydew excretion by female BPH insects after 24 hours of feeding on controls and PB-F3D4, PB-F4D2 & PB-F17D1 transgenic rice plants.
Survival, development, fecundity and feeding behaviour of BPH insects growing on transgenic rice lines expressing Cry1Ac::ASAL fusion protein. Twenty 1st instar nymphs of BPH were released on each plant on day 0. Bioassays were carried out on 20 plants sampled from each transgenic line PB-F3D4, PB-F4D2 & PB-F17D1 and three controls TN1, PTB33, and untransformed PB1 (a) Survival rate of insects, (b) Insect development was recorded after 24 days. (c) Total number of nymphs produced by a pair of adult BPH insects on controls and transgenic plants were counted and the data was used to plot the graph. Bars indicate mean ± SE. (d) Feeding behaviour of BPH was estimated by honeydew excretion method and the data was used to plot the graph. Bars indicate mean ± SE.
Ligand binding assays of BPH insects fed on transgenic rice
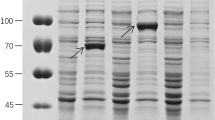
Ligand binding assays were carried out to detect the BPH receptor proteins’ specificity to Cry1Ac::ASAL fusion protein. Proteins isolated from BPH insects fed on both transgenics (asal, and cry1Ac::asal) and control plants were treated with ASAL antibodies (Fig. 6). BPH insects fed on cry1Ac-asal-transgenic (PB-F4D2) plants disclosed three receptor proteins of ~70 kDa, ~55 kDa and ~40 kDa compared to ~55 kDa and ~40 kDa of insects fed on asal-transgenic plants developed in our previous study1. The bands observed on ligand blots represent complexes formed between the fusion protein and receptor proteins of the insects (Fig. 6). Whereas, BPH insects fed on control plants failed to show any detectable receptor protein specific to fusion protein (Fig. 6).
Ligand blot analyses of proteins extracted from BPH insects fed on transgenic and untransformed control plants. Protein was isolated from BPH insects and analysis was carried out using the ASAL antibodies. Lane 1: Protein extract (5 µg) from BPH insects fed on untransformed control plant; Lane 2: Protein extract (5 µg) from BPH insects fed on asal-transgenic plant; Lanes 3 and 4: Protein extracts (5 µg) from BPH insects fed on cry1Ac::asal transgenic rice lines.
Discussion
Globally, insects cause about 15% of direct losses to different agricultural crops as well as indirect losses owing to impaired quality of the produce28. Insects also act as vectors of various plant pathogens such as bacterial, fungal, and viral29. The threat of development of resistance by insect pests to a broad spectrum chemical insecticides has prompted research for adoption of alternative strategies. Several crop plants have been engineered with different cry genes conferring resistance against various major insect pests30,31,32. Rapid adoption and commercial introduction of Bt crops led to the development of resistance by insects against Bt toxins. Major pests, such as the diamondback moth, tobacco budworm, Colorado potato beetle, Indian mealy moth, maize stalk borer, cotton bollworm and fall armyworm have shown resistance to Cry toxins33,34,35,36,37. Moreover, Cry toxins are effective against lepidopterans but are ineffective against sucking pests belonging to hemiptera.
Plants, in general, are known to synthesize a wide range of defense proteins against different pathogens. The insecticidal activity of plant lectins against various insects belonging to hemipteran have been well documented14. Transgenic rice expressing ASAL exhibited ample resistance against sucking insects BPH, GLH and WBPH1,18. Pyramided transgenic rice lines containing asal and gna lectin genes exhibited enhanced resistance to major sap-sucking insects38. A number of successful fusion proteins were developed using lectin as a carrier protein. The observed increases in the mortality of insects caused by fusion proteins have been ascribed to the lectin domain, which enhanced the binding process and facilitated the entry of toxin more efficiently into the insect39. GNA when fused as a carrier protein for different chimeric toxins such as, Manase-AC/GNA40, SFlI/GNA41, Chitinase/GNA42, ButalT/GNA43 and ω-ACTX-Hv1a/GNA44 resulted in higher toxicities against various insect pests.
To overcome resistance acquired by insects against Cry toxins different strategies were employed to modify Cry functional domains to improve their toxicity8,10. Different gene fusions, viz., Cry1Ca, Cry1Fb, Cry1Ba modified with Cry1Ac domain III, Cry1Ac/ricin-B, Cry1Ac/CpTI, Cry1Ac/HWTX-I, Cry1Ac/CDEP2, and Cry1Ab/ACTX-Ar1, employed for engineering of plants bestowed with enhanced insect resistance22,45,46,47,48,49.
In the present study, transgenic plants expressing Cry1Ac::ASAL fusion protein have been generated and tested against different insect pests adopting in planta bioassays. The binary vector pCAMBIA3300-CaMV35S-bar, containing cry1Ac::asal expression unit driven by CaMV35S promoter, was introduced into rice by employing the Agrobacterium-mediated genetic transformation method. PCR analyses of Basta tolerant plants showed amplification of 1089 bp and 560 bp products corresponding to cry1Ac and bar transgenes testifying the presence of these genes in the genomes of different transgenic plants (Supplementary Fig. S2). Southern blot analysis of transformants, when probed with the cry1Ac::asal coding sequence, revealed a specific hybridizable band of ~2.5 kb confirming stable integration of the fusion gene in different rice transformants (Fig. 1b). Appearance of different hybridizable bands of ≥2.5 kb, corresponding to the bar expression cassette with the EcoRI digested genomic DNA, attest the single copy and independent nature of the transformants (Fig. 1c). These observations further suggest that the T-DNA is integrated into the rice genome as a single copy without any rearrangement. Moreover, the presence of single copy integration of transgenes facilitate their stable expression and predictable pattern of inheritance50.
RT-PCR analysis revealed the presence of cry1Ac::asal gene transcripts in the transgenic plants as evidenced by the amplification of 1089 bp and 375 bp fragments corresponding to cry1Ac and asal (Fig. 2a,b). Western blot analyses of transgenic plants, when treated with ASAL antibodies, demonstrated the presence of ~55 kDa Cry1Ac::ASAL protein (Fig. 2c). Furthermore, ELISA analysis revealed varied amounts (1.0–1.8%) of Cry1Ac::ASAL protein in different transgenic rice plants owing to insertion of the transgene at different transcriptionally active sites in the rice genome.
For evaluation of entomotoxic effects of Cry1Ac::ASAL fusion-protein expressed in the transgenic rice plants, insect bioassays have been carried out employing YSB, LF and BPH insects. Insect bioassays have been done on six different transgenic lines for YSB and LF. For further evaluation against BPH, three best rice lines have been used. The results revealed 100% mortality of the stem borers when fed on cut stems of transgenics at the tillering stage. Also, the mortality of leaf folders grown on two-week-old transgenic rice plants ranged from 80–100% (Fig. 3a,b). The ability of Cry1Ac::ASAL fusion protein to induce high mortality of lepidopteran pests testify that the domains I and II of Cry1Ac in conjunction with ASAL domain can cause high level insect toxicity. Several hybrid Cry proteins Cry1Ab-Cry1B, Cry1Ac-Cry1Ab, and Polh–Cry1Ac developed by domain swapping exhibited increased toxicities and broader activities when compared to the parental proteins51,52,53. Transgenic rice plants expressing Cry1Ab and Cry1Ac fusion protein showed high resistance against leaf folder and yellow stem borer insect pests without any reduced yields54. Rice yellow stem borer and leaf folder fed on Cry1Ab- and Cry1Ac-expressing plants showed varied insect mortality (10–100%)55,56. A chimeric B. thuringiensis toxin Cry2AX1 expressed in rice proved to be effective against certain major lepidopteran insect pests57.
BPH bioassays on transgenic rice resulted in a significant mortality of larval nymphs as well as reduction in the fecundity of insects. Transgenic plants expressing Cry1Ac::ASAL fusion protein exhibited varied levels (1–3 score on a 0–9 scale) of resistance to BPH, on par with those of BPH-resistant check var. PTB33. The survival of BPH on transgenic rice plants was significantly reduced compared to untransformed control plants (Fig. 5b). Among BPH insects survived, only 20–30% could reach the adult stage on different transgenic lines as compared to the control plants, which is similar to the earlier reports of ASAL-expressing transgenic rice plants1. Honeydew assay revealed significant reduction in the feeding ability of BPH on transgenic plants as compared to the insects fed on control plants (Fig. 5a). The overall results amply indicate the enhanced efficacy of Cry1Ac::ASAL fusion protein against the three major insect pests of rice.
The ligand binding assays on BPH insects fed on the transgenic rice plants expressing ASAL and Cry1Ac::ASAL fusion proteins, separately, exhibited receptor protein binding patterns of ~55 kDa & ~40 kDa, and ~70 kDa, ~55 kDa & ~40 kDa, respectively, after treatment with ASAL antibodies (Fig. 6). The present results suggest the existence of an additional binding site for the Cry1Ac::ASAL in BPH gut cells (Fig. 6). Earlier it was shown that the replacement of loops in Cry1Ab domain II resulted in the identification of additional binding sites58. Modification of Cyt2Aa toxin by the addition of peptides caused higher toxicity against pea aphids as compared to the replacement of loops with the peptides59.
The major objective of the present study has been to custommake rice plants using a novel cry1Ac::asal fusion gene which encodes a potent insecticidal protein having the ability to bind to different receptors of the insect gut cells and bestow enhanced toxicity as well as durable resistance against an array of insect pests. The Cry1Ac::ASAL fusion protein containing domains of both bacterial (Cry) and plant (ASAL) origin would be helpful in delaying insect resistance besides minimizing pest populations. The increased entomotoxic effects of fusion protein against lepidopteran and hemipteran insects are attributable to its higher binding affinity towards more number of receptor proteins in the insect gut epithelial cells. An overview of the present results demonstrates that the presence of ASAL in combination with two domains of Cry1Ac culminates in accentuated toxicity of the fusion protein to yellow stem borer, leaf folder and brown planthopper insects of rice. Accordingly, the newly designed fusion protein holds promise and may be deployed as a potent toxin against major lepidopteran and sucking pests of various other crop plants.
Materials and Methods
Development of cry1Ac::asal fusion gene construct for rice transformation
Earlier, a fusion gene cry1Ac::asal was constructed in our laboratory containing DI and DII domains (1089 bp) of Cry1Ac and a 375 bp fragment encoding carbohydrate binding domain of ASAL20. The cry1Ac::asal fusion gene of 1464 bp was amplified by PCR using 5′-GGCCATGGAGTTCGCCAGGAACAAGG-3′ and 5′-CCCGGGTCAACCCACACTTCT TCTGTAGG-3′ as forward and reverse primers employing pET28 vector as a template. Primers used for amplification contained the restriction sites Nco I and Sma I for cloning into the plant expression vector, and amplified PCR product was cloned at Sma I site of pBSK(+) plasmid. The recombinant clone was confirmed by restriction analysis and DNA sequencing method. Later, the fusion gene was excised using Nco I and Sma I restriction enzymes and cloned into pRT100 vector between CAMV 35 S promoter and polyA terminator. Finally, the fusion gene cassette was excised and cloned at Hind III site in pCAMBIA3300 vector containing bar gene as a selectable marker. The resultant pCAMBIA3300-CaMV35S-bar-CAMV-cry1Ac::asal vector was mobilized into Agrobacterium (EHA105) by tri-parental mating using helper plasmid PRK2013. Plasmid DNA was isolated from EHA105 cells and digested with Hind III enzyme to confirm the presence of the gene cassette.
Agrobacterium-mediated genetic transformation and development of transgenic plants
Seeds of rice cultivar Pusa Basmathi (PB1), obtained from the Indian Institute of Rice Research (IIRR), Hyderabad, were used for rice transformation. Agrobacterium-mediated genetic transformation experiments were performed using EHA105 harbouring pCAMBIA3300-CaMV35S-bar-CAMV-cry1Ac::asal vector60. For selection of transformants, the co-cultivated calli were subjected to two rounds of selection on medium containing phosphinothricin (6 mg/l and 8 mg/L) for 2 weeks each. Later, the actively proliferating calli were selected and transferred onto regeneration medium60. Regenerated shoots were transferred to the rooting medium, and the rooted plants were transferred to pots. All the regenerated plants were grown to maturity in the glasshouse and putative transgenics were identified using herbicide (0.25%) Basta61.
Molecular analysis of transgenic plants
Genomic DNA was isolated from the putative transformants and untransformed control (UC) plants62. For PCR analysis, primers 5-′GAATTCGAGTTCGCCAGGAACCAG-3′ & 5′-GGATCCGATGATGCTCACGGAACTG-3′ for cry1Ac gene, and 5′-GGATCCGCTATTCTAACCATACTG-3′ & 5′-GAGCTCACCCACA CTTCTTCTGTAGG-3′ for asal gene, 5′-CTACCATGAGCCCAGAACG-3′ & 5′-TCAG ATCTCGGTGACGGG-3′ for bar gene, and 5′GCTCAACACATGAGCGAAAC-3′ polyA reverse primer were used. Later, the PCR products were separated on 1% agarose gel and analyzed.
For Southern blot analysis, 15 g of genomic DNA samples from transformants as well as UC plants were digested separately with Hind III and EcoRI enzymes. The digested DNAs were electrophoresed on a 0.8% agarose gel and subsequently transferred to N+ Nylon membranes (Amersham Biosciences), and were fixed by exposing to UV (1200 J for 60 s) in an UV cross-linker. The membranes were separately probed with cry1Ac::asal and bar coding sequences labeled with α-32P dCTP, employing ready-to-go random primer DNA labelling kit (Amersham Biosciences). Further, the processing of membranes was done according to the manufacturer’s instructions.
RT-PCR analysis
Total RNA was isolated from transgenic and UC plants using the TRIZOL method (Invitrogen, Carlsbad, CA, USA). The first strand cDNA generated was used as a template along with the primers 5′-GGCCATGGAGTTCGCCAGGAACAAGG-3′ & 5′-CCCGGGTCAACCC ACACTTCTTCTGTAGG-3′ for fusion gene (cry1Ac::asal), and 5′-GGATCCGCTA TTCTAACCATACTG-3′ & 5′GCTCAACACATGAGCGAAAC-3 for asal-polyA, to detect the presence of corresponding gene transcripts in the transgenic plants. Amplified products were analyzed by the gel electrophoresis on 1.0% agarose gel.
Western blot analysis of transgenic rice plants
Samples of transgenic and UC plants’ leaf tissue were homogenized in 50 mM Tris-HCl buffer (pH 9.0). The extracts were centrifuged at 5,000 g for 20 min at 4 °C, and supernatants were collected and the protein samples (5 μg) were subjected to 15% SDS-PAGE63. Subsequent to electrophoresis, the separated proteins were transferred onto nitrocellulose N-membrane (Amersham) by electroblotting64. After protein transferring, the membrane was blocked by incubating in PBS solution containing 10% non-fat dried milk and 0.1% Tween 20 for 2 h at room temperature. Later, the membrane was probed with polyclonal rabbit anti-ASAL serum (1:10,000 dilution) as primary and goat anti-rabbit IgG horse-radish peroxidase conjugate (GENEi) as secondary antibody (1:10,000 dilution). The membrane was washed and treated with saturated benzidine solution containing 20% ammonium chloride and 0.1% H2O2.
ELISA analysis
The microtitre plate wells were coated with 1 μg of crude protein extract of transgenic and UC rice plants and were kept overnight at 4 °C. The wells were washed thrice with 20 mM PBS containing 0.05% Tween 20 and were blocked with 10% non-fat dried milk for 2 h at 37 °C; later they were washed six times with PBS-T. The primary antibody of Cry1Ac (1:10,000) was added to the wells and incubated for 2 h at 4 °C. After incubation, the wells were washed thrice with PBS and incubated with secondary antibody, goat anti-rabbit IgG horse-radish peroxidase conjugate (GENEi) (1:10,000) for 1 h at room temperature. The plates were washed thrice with PBS, and 0.001% 3,3′,5,5′- tetramethylbenzidine (TMB) substrate in 0.05 M phosphate citrate buffer was added along with 0.1% H2O2 and kept in dark for 10 min. The reaction was stopped by adding 1 N H2SO4 and the absorbance was recorded on ELISA reader at 450 nm.
Insect bioassays
The brown planthopper, leaf folder, and yellow stem borer were maintained on 25 to 30-day-old Taichung Native 1 (TN1) plants under controlled conditions. The freshly hatched nymphs or the nymphs after attaining the desired age were utilized for various experiments. For insect bioassays, six-week-old transgenic rice plants of PB-F1D, PB-F3D, PB-F4D, PB-F8D, PB-F17D, and PB-F20A expressing the Cry1Ac::ASAL toxin were used and each experiment was repeated three times.
Bioassays against leaf folder (LF)
The length of each individual second instar larvae of leaf folder was measured and allowed to starve for 5 h. Later, they were placed onto leaves of transgenic and UC plants (kept in Petri plates containing moist filter papers) and were incubated at 25 ± 2 °C in dark at 70% relative humidity for three days. After three days, insect mortality and leaf area damage were recorded. Percentage of leaf area damage was calculated using the formula: % leaf area damage = consumed leaf area/total leaf area before bioassay × 100.
Cut-stem bioassays against yellow stem-borer (YSB)
Egg masses of yellow stem borer were collected from the rice field and allowed to hatch in the laboratory. Fresh stems of transgenic and control plants were harvested at the tillering stage and were cut into 5–6 cm long pieces. Twelve first-instar larvae of yellow stem borers and five pieces of freshly cut stems from each plant were placed in a sealed glass bottle and were incubated at 25 ± 2 °C in dark at 70% relative humidity for five days. After 5 days, the mortality rate of the insects was recorded.
Bioassay with brown planthopper (BPH)
Thirty day old homozygous transgenic rice plants and UC plants were used to assess insect mortality/survival using no choice method. Early first instar nymphs of BPH (20 each) were released on each plant that was confined in an insect proof mylar cage. Nymphal survival was monitored and observations were taken at 3 day intervals up to 24 days under controlled environmental conditions (25 ± 2 °C and 70% relative humidity). The delay in the development of insects was also observed daily by scoring the number of adults and insects still in nymphal stage.
The effect of fusion protein on the fecundity of insects was estimated by scoring the nymphs emerged from the hatched eggs. For this study, surviving male and the female insects were pooled and confined again in a 1: 1 ratio, so that there is no difference in the nymph production based on the sex ratio. The nymphs emerging from a pair of adults were counted daily up to 7 days, after which surviving adults were removed and the plants were observed for unhatched eggs by adopting the staining technique1. A sum of the emerged nymphs and unhatched eggs were used for estimating the fecundity. Plant damage was assessed visually and compared with susceptible TN1 rice plants (100% damage).
Semi-quantitative assay of honey dew production (feeding behavior) of BPH
The extent of insect feeding was estimated by semi-quantitative honeydew assay (liquid excreta) produced by the insects. Whatman No.1 filter paper, dipped in bromocresol green solution (2 mg/ml in ethanol) was placed at the base of each plant and covered with a plastic cup. On each plant, five adult female BPH insects, pre-starved for two hours, were released separately and allowed to feed for 24 h. Care was taken not to release gravid adult females. Honeydew, excreted by the insects, reacts with bromocresol green on the filter paper resulting in blue colour. The area of blue spots developed on filter papers were measured using millimeter graph paper and expressed in units (1 unit = 1mm2)1.
Western blot analysis to detect insect receptor proteins binding to the fusion protein
Insects fed on transgenic rice lines and untransformed control plant were collected and homogenized in 50 mM Tris-HCl buffer (pH 9.0) to isolate total proteins of the insects. The extract was centrifuged at 5,000 g for 20 min at 4 °C, and the supernatant was collected and the protein samples (5 μg) were subjected to 15% SDS-PAGE63. The separated proteins were transferred onto nitrocellulose N- membrane (Amersham) by electroblotting64. After protein transfer, the membrane was blocked by incubating in PBS solution containing 10% non-fat dried milk and 0.1% Tween 20 for 2 h at room temperature. The membrane strips were probed with polyclonal rabbit anti-ASAL serum (1:10,000 dilution), followed by goat anti-rabbit IgG horse-radish peroxidase conjugate (GENEi) as a secondary antibody (1:10,000 dilution). Membrane strips were washed and treated with saturated benzidine solution containing 20% ammonium chloride and 0.1% H2O2.
References
Yarasi, B., Vijaya, K. S., Pasalu, I. C., Reddy, V. D. & Rao, K. V. Transgenic rice expressing Allium sativum leaf agglutinin (ASAL) exhibits high-level resistance against major sap sucking pests. BMC Plant Biol. 8, 102, https://doi.org/10.1186/1471-2229-8-102 (2008).
Li, H. et al. Isolation of five rice nonendosperm tissue-expressed promoters and evaluation of their activities in transgenic rice. Plant Biotech. J. https://doi.org/10.1111/pbi.12858 (2017).
Lu, G., Zhang, T., He, Y. & Zhou, G. Virus altered rice attractiveness to planthoppers is mediated by volatiles and related to virus titre and expression of defense and volatile-biosynthesis genes. Sci Rep. 6, 38581, https://doi.org/10.1038/srep38581 (2016).
Arenas, I., Bravo, A., Soberón, M. & Gómez, I. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J Biol Chem. 285, 12497 (2010).
Bravo, A., Gill, S. S. & Soberon, M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 49, 423–435 (2007).
Baum, J. A. et al. Cotton plants expressing a hemipteran-active Bacillus thuringiensis crystal protein impact the development and survival of Lygus hesperus (Hemiptera: Miridae) nymphs. J Econ Entomol. 105, 616–624 (2012).
Tabashnik, B. E. et al. Dual mode of action of Bt proteins: protoxin efficacy against resistant insects. Sci Rep. 5, 15107, https://doi.org/10.1038/srep15107 (2015).
Bravo, A. et al. Evolution of Bacillus thuringiensis Cry toxins insecticidal activity. Microbial Biotech. 6, 17–26 (2013).
Bravo, A., Likitvivatanavong, S., Gill, S. S. & Soberon, M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem Mol Biol. 41, 423–431 (2011).
Deist, B. R., Rausch, M. A., Fernandez-Luna, M. T., Adang, M. J. & Bonning, B. C. Bt toxin modification for enhanced efficacy. Toxins. 6, 3005–3027 (2014).
Ghazarian, H., Idoni, B. & Oppenheimer, S. B. A glycobiology review: carbohydrates, lectins and implications in cancer therapeutics. Acta Histochem 113, 236–247 (2011).
Lagarda-Diaz, I., Guzman-Partida, A. M. & Vazquez- Moreno, L. Legume lectins: proteins with diverse applications. Int J Mol Sci. 18, 1242, https://doi.org/10.3390/ijms18061242 (2017).
Bandyopadhyay, S., Roy, A. & Das, S. Binding of garlic (Allium sativum) leaf lectin to the gut receptors of homopteran pests is correlated to its insecticidal activity. Plant Sci. 161, 1025–1033 (2001).
Macedo, M. L. R., Oliveira, C. F. R. & Oliveira, C. T. Insecticidal activity of plant lectins and potential application in crop protection. Molecules. 20, 2014–2033 (2015).
Roy, A., Banerjee, S., Majumder, P. & Das, S. Efficiency of mannose binding plant lectins in controlling a homopteran insect, the red cotton bug. J Agric Food Chem. 50, 6775–6779 (2002).
Powell, K. S. et al. Antimetabolic effects of related plant lectins towards nymphal stages of Nilaparvata lugens. Entomol Exp Appl. 75, 61–65 (1995).
Sadeghi, A. et al. Expression of garlic leaf lectin under the control of the phloem-specific promoter Asus1 from Arabidopsis thaliana protects tobacco plants against the tobacco aphid (Myzus nicotianae). Pest Manage Sci. 63, 1215–1223 (2007).
Saha, P. et al. Transgenic rice expressing Allium sativum leaf lectin with enhanced resistance against sap-sucking insect pests. Planta. 223, 1329–1343 (2006).
Vajhala, C. S. K. et al. Development of transgenic cotton lines expressing Allium sativum agglutinin (ASAL) for enhanced resistance against major sap-sucking pests. PLoS ONE. 8, e72542 (2013).
Tajne, S. et al. Molecular modeling of Bt Cry1Ac (DI-DII)-ASAL (Allium sativum lectin)-fusion protein and its interaction with aminopeptidase N (APN) receptor of Manduca sexta. J Mol Graph Model. 33, 61–76 (2012).
De Maagd, R. A. et al. Domain III substitution in Bacillus thuringiensis delta-endotoxin CryIA(b) results in superior toxicity for Spodoptera exigua and altered membrane protein recognition. Appl Environ Microbiol. 62, 1537–1543 (1996).
De Maagd, R. A., Weemen-Hendriks, M., Stiekema, W. & Bosch, D. Bacillus thuringiensis delta-endotoxin Cry1C domain III can function as a specificity determinant for Spodoptera exigua in different, but not all, Cry1-Cry1C hybrids. Appl Environ Microbiol. 66, 1559–1563 (2000).
Karlova, R. et al. Bacillus thuringiensis delta-endotoxin cry1ac domain III enhances activity against Heliothis virescens in some, but not all Cry1-Cry1Ac hybrids. J Invertebr Pathol. 88, 169–172 (2005).
Naimov, S., Weemen-Hendriks, M., Dukiandjiev, S. & de Maagd, R. A. Bacillus thuringiensis delta-endotoxin Cry1 hybrid proteins with increased activity against the colorado potato beetle. Appl Environ Microbiol. 67, 5328–5330 (2001).
Li, H., Chougule, N. P. & Bonning, B. C. Interaction of the Bacillus thuringiensis delta endotoxins Cry1Ac and Cry3Aa with the gut of the pea aphid, Acyrthosiphon pisum (Harris). J Invertebr Pathol. 107, 69–78 (2011).
Shao, E., Liu, S., Lin, L. & Guan, X. Proteolytic processing of Bacillus thuringiensis toxin Cry1Ab in rice brown planthopper, Nilaparvata lugens (Stål). J Invertebr Pathol. 114, 255–257 (2013).
Tajne, S., Boddupally, D., Sadumpati, V., Vudem, D. R. & Khareedu, V. R. Synthetic fusion-protein containing domains of Bt Cry1Ac and Allium sativum lectin (ASAL) conferred enhanced insecticidal activity against major lepidopteran pests. J Biotechnol. 171, 71–75 (2014).
Maxmen, A. Crop pests: under attack. Nature. 501, 15–17 (2013).
Whitfield, A. E., Falk, B. W. & Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology. 479–480, 278–289, https://doi.org/10.1016/j.virol.2015.03.026 (2015).
Tabashnik, B. E. et al. Efficacy of genetically modified Bt toxins alone and in combinations against pink bollworm resistant to Cry1Ac and Cry2Ab. PLoS ONE. 8, e80496, https://doi.org/10.1371/journal.pone.0080496 (2013).
Nicolia, A., Manzo, A., Veronesi, F. & Rosellini, D. An overview of the last 10 years of genetically engineered crop safety research. Crit Rev Biotechnol. 34, 77–88 (2013).
Pardo-Lopez, L., Soberon, M. & Bravo, A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 37, 3–22 (2013).
McGaughey, W. H., Gould, F. & Gelernter, W. Bt resistance management. Nat Biotechnol. 16, 144–146 (1998).
Tabashnik, B. E. et al. Insect resistance to transgenic Bt crops: lessons from the laboratory and field. J Econ Entomol. 96, 1031–1038 (2003).
Van Rensburg, J. B. J. First report of field resistance by stem borer Busseola fusca (Fuller) to Bt-transgenic maize. S Afr J Plant Soil. 24, 147–151 (2007).
Tabashnik, B. E., Gassmann, A. J., Crowder, D. W. & Carriere, Y. Insect resistance to Bt crops: evidence versus theory. Nat Biotechnol. 26, 199–202 (2008).
Blanco, C. A. et al. Susceptibility of isofamilies of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Cry1Ac and Cry1Fa proteins of Bacillus thuringiensis. Southwestern Entomologist. 35, 409–415 (2010).
Bharathi, Y. et al. Pyramided rice lines harbouring Allium sativum (ASAL) and Galanthus nivalis (GNA) lectin genes impart enhanced resistance against major sap-sucking pests. J Biotechnol. 152, 63–71 (2011).
Yang, Y. Y., Mei, F., Zhang, W., Shen, Z. & Fang, J. Creation of Bt rice expressing a fusion protein of Cry1Ac and Cry1I-like using a green tissue-specific promoter. J Econ Entomol 107, 1674–1679 (2014).
Fitches, E., Audsley, N., Gatehouse, J. A. & Edwards, J. P. Fusion proteins containing neuropeptides as novel insect control agents: Snowdrop lectin delivers fused allatostatin to insect haemolymph following oral ingestion. Insect Biochem Mol Biol. 32, 1653–1661 (2002).
Fitches, E. et al. Fusion proteins containing insect-specific toxins as pest control agents: snowdrop lectin delivers fused insecticidal spider venom toxin to insect haemolymph following oral ingestion. J Insect Physiol. 50, 61–71 (2004a).
Fitches, E. et al. Cloning, expression and functional characterisation of chitinase from larvae of tomato moth (Lacanobia oleracea): A demonstration of the insecticidal activity of insect chitinase. Insect Biochem Mol Biol. 34, 1037–1050 (2004b).
Fitches, E. C. et al. Insecticidal activity of scorpion toxin (ButalT) and snowdrop lectin (GNA) containing fusion proteins towards pest species of different orders. Pest Manag Sci. 66, 74–83 (2010).
Nakasu, E. Y., Edwards, M. G., Fitches, E., Gatehouse, J. A. & Gatehouse, A. M. Transgenic plants expressing ω-ACTX-Hv1a and snowdrop lectin (GNA) fusion protein show enhanced resistance to aphids. Front Plant Sci. 5, 673 (2014).
Mehlo, L. et al. An alternative strategy for sustainable pest resistance in genetically enhanced crops. Proc Natl Acad Sci. 102, 7812–7816 (2005).
Wu, H. et al. Effects of transgenic Cry1Ac + CpTI cotton on non-target mealybug pest Ferrisia virgata and its predator Cryptolaemus montrouzieri. PLoS ONE. 9, e95537, https://doi.org/10.1371/journal.pone.0095537 (2014).
Xia, L., Long, X., Ding, X. & Zhang, Y. Increase in insecticidal toxicity by fusion of the cry1Ac gene from Bacillus thuringiensis with the neurotoxin gene hwtx-I. Curr Microbiol. 58, 52–57 (2009a).
Xia, L., Zheng, Z., Ding, X. & Huang, F. The expression of a recombinant cry1Ac gene with subtilisin-like protease CDEP2 gene in acrystalliferous Bacillus thuringiensis by Red/ET homologous recombination. Curr Microbiol. 59, 386–392 (2009b).
Cao, C. W. et al. Response of the gypsy moth, Lymantria dispar to transgenic poplar, Populus simonii × P. nigra, expressing fusion protein gene of the spider insecticidal peptide and Bt-toxin C-peptide. J Insect Sci. 10, 1–13 (2010).
Finnegan, J. & McElroy, D. Transgene inactivation: plants fight back. Nat Biotechnol. 12, 883–888 (1994).
Ho, N. H. et al. Translational fusion hybrid Bt genes confer resistance against yellow stem borer in transgenic elite Vietnamese rice (Oryza sativa L.) cultivars. Crop Sci. 46, 781–789 (2006).
Honee, G., Vrienzen, W. & Visser, B. A. Translation fusion product of two different insecticidal crystal protein genes of Bacillus thuringiensis exhibits an enlarged insecticidal spectrum. Appl Environ Microbiol. 56, 823–825 (1990).
Seo, J. H., Yeo, J. S. & Cha, H. J. Baculoviral polyhedrin-Bacillus thuringiensis toxin fusion protein: a protein-based bio-insecticide expressed in Escherichia coli. Biotechnol Bioeng. 92, 166–172 (2005).
Tu, J. M. et al. Field performance of transgenic elite commercial hybrid rice expressing Bacillus thuringiensis delta endotoxin. Nat Biotechnol. 18, 1101–1104 (2000).
Fujimoto, H., Itoh, K., Yamamoto, M., Kyozuka, J. & Shimamoto, K. Insect resistant rice generated by introduction of a modified δ-endotoxin gene of Bacillus thuringiensis. Biotechnol. 11, 1151–1155 (1993).
Nayak, P. et al. Transgenic elite indica rice plants expressing Cry1Ac δ-endotoxin of Bacillus thuringiensis are resistant against yellow stem borer (Scirpophaga incertulas). Proc Natl Acad Sci USA 94, 2111–2116 (1997).
Chakraborty, M. et al. Transgenic rice expressing thecry2AX1 gene confers resistance to multiple lepidopteran pests. Transgenic Res. 25, 665–678 (2016).
Shao, E. et al. Loop replacements with gut-binding peptides in Cry1Ab domain II enhanced toxicity against the brown planthopper, Nilaparvata lugens (Stal). Sci Rep. 6, https://doi.org/10.1038/srep20106 (2016).
Chougule, N. P. et al. Retargeting of the Bacillus thuringiensis toxin Cyt2Aa against hemipteran insect pests. Proc Natl Acad Sci USA 110, 8465–8470 (2013).
Ramesh, S., Nagadhara, D., Reddy, V. D. & Rao, K. V. Production of transgenic indica rice resistant to yellow stemborer and sap-sucking insects, using super-binary vectors of Agrobacterium tumefaciens. Plant Sci. 166, 1077–1085 (2004).
Nagadhara, D. et al. Transgenic indica rice resistant to sap-sucking insects. Plant Biotechnol J. 1, 231–240 (2003).
McCouch, S. R. et al. Molecular mapping of rice chromosomes. Theor Appl Genet. 76, 815–829 (1988).
Laemmli, U. K. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature. 227, 680–685 (1970).
Towbin, H., Steahelin, T. & Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76, 4350–4354 (1979).
Acknowledgements
We are grateful to the Osmania University for the support to carry out the present work. BD and TS are thankful to the University Grants Commission, New Delhi and Department of Biotechnology, New Delhi for the award of Research Fellowships. We thank Dr. Madan Mohan for his help in BPH insect bioassays. Authors are grateful to Prof. T. Papi Reddy for his suggestions and critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: V.R.K., D.R.V. Performed the experiments: D.B., S.T. S.R.G. Analyzed the data: V.R.K., D.B., S.T. Wrote the paper: D.B., S.T., V.R.K.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Boddupally, D., Tamirisa, S., Gundra, S.R. et al. Expression of hybrid fusion protein (Cry1Ac::ASAL) in transgenic rice plants imparts resistance against multiple insect pests. Sci Rep 8, 8458 (2018). https://doi.org/10.1038/s41598-018-26881-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26881-9
- Springer Nature Limited
This article is cited by
-
Diversity of transgenes in sustainable management of insect pests
Transgenic Research (2023)
-
35 years in plant lectin research: a journey from basic science to applications in agriculture and medicine
Glycoconjugate Journal (2022)
-
Transgenic plants expressing immunosuppressive dsRNA improve entomopathogen efficacy against Spodoptera littoralis larvae
Journal of Pest Science (2022)