Abstract
Shanxi aged vinegar (SAV), one of the famous Chinese vinegars, is produced by multispecies solid-state fermentation in which the acetic acid fermentation stage (AAF) is especially important. However, how bacterial succession and their metabolites change along with the different stages of AAF is still poorly understood. In this study, we investigated the dynamic bacterial succession and flavor formation in three batches of SAV using high-throughput sequencing and metabolomics approaches. It is interesting to find that AAF can be divided into three stages based on its bacterial community succession (early stage, days 0–4; medium stage, days 5–21; and later stage, days 22–26). Pantoea, Pediococcus, Lactococcus and Rhizobium played an important role in the early stage; Lactobacillus was dominant in the medium stage (67.72%); and Acetobacter, Komagataeibacter and Kroppenstedtia were the key bacteria in the later stage. A total of seven organic acids and 42 volatile constituents (esters, alcohol, ketones and aldehydes) were detected during the AAF. Spearman correlation analysis showed a significant correlation between the bacterial community and these flavor metabolites during the AAF of the SAV. This is the first report to explore the relationships between volatile flavor metabolites and bacterial community succession by a three-staged method and provide theoretical support for a flavor formation mechanism in traditional SAV.
Similar content being viewed by others
Introduction
Shanxi aged vinegar (SAV) is one of the traditional Chinese rice vinegars and is famous for its complex and pleasant aroma. The history of SAV can be traced back thousands of years1,2. SAV has attracted considerable interest because it is considered to be both a conventional and a functional food3. Recent evidence suggests that it has several types of therapeutic effects, such as antioxidant, hypotensive, hypoglycemic and cholesterol-lowering activities4,5,6. While the original production of these effects may have been unintentional, SAV is now indispensable due to its abundance of culinary and medicinal benefits.
SAV is traditionally produced from sorghum and other cereals by spontaneous solid-state fermentation via three major steps: starch saccharification, alcohol fermentation (AF) and acetic acid fermentation (AAF)7,8. Among these three steps, the AAF is thought to be the most complicated and critical in the formation of flavor metabolites and the accumulation of acetic acid9,10. During the AAF process, the alcoholic substances produced by AF are mixed with wheat bran, millet chaff and a small quantity of old Pei (acetic acid fermented product from the last batch used as vinegar seed) and followed by fermentation for approximately 20–30 days, stirring every morning to increase the oxygen content11,12. During the final step, NaCl is added to the mixture to inhibit the growth of Acetobacter. The characteristic aromatic components formed during AAF include organic acids, volatile compounds and amino acids13,14. It has been reported that 19 volatile compounds and 87 kinds of nonvolatile metabolites that are considered to be aroma-active compounds have been identified in SAV by using GC-MS15,16. Numerous studies suggest that the diverse volatile or nonvolatile compounds in SAV resulted from the bacterial activity during this period8,10. The rapid development in culture-independent techniques including denaturing gradient gel electrophoresis (DGGE)17,18,19 and high-throughput sequencing20 led to a deep understanding of the dynamics, diversity and function of microbial community succession in vinegar Pei (solid-state vinegar culture) during the AAF process. Four bacterial genera including Acetobacter, Lactobacillus, Escherichia and Klebsiella have been identified in the vinegar Pei of SAV during the AAF process. It was found that the bacterial diversity increased in the first three days of AAF and then decreased as the AAF proceeded19. Similar results have also been reported by Nie et al., who found that Acetobacter and Lactobacillus are the major bacterial genera in SAV during the AAF process. In addition, the results showed that the microbiota and nonvolatile flavor metabolites produced during AF and AAF were strongly correlated16. However, the relationship between the volatile metabolites and various bacteria, and which bacteria boosted the accumulation of the major flavor substance during the AAF process, is still unclear.
In this study, the bacterial community succession of vinegar Pei collected from three batches during AAF was analyzed by high-throughput sequencing. We identified organic acids and volatile flavor metabolites in each sample by HPLC and GC-MS, respectively. At this point, the acetic acid fermentation was divided into three stages. We aimed to determine the relationships between the dynamic succession of the bacterial community and the formation of flavor metabolites (including volatile substance and organic acids) in the different stages of AAF.
Results
Composition and changes of bacterial succession in vinegar Pei during AAF
In prokaryotic microbes, high-quality 2,134,087 16S rRNA gene sequences of V3–V4 regions were obtained from 42 Pei vinegar samples. For all of the samples, rarefaction and Shannon diversity curves leveled off strongly, indicating that the majority of the diversity was captured in this analysis (Fig. 1). The sequences were classified into 351 operational taxonomic units (OTU) at a 97% similarity level and distributed among six major phyla (relative abundance >1%): Firmicutes, Proteobacteria, Tenericutes, Bacteroidetes, Actinobacteria and Cyanobacteria. Firmicutes and Proteobacteria were the predominant phyla, together contributing up to 90% of the sequences, while Cyanobacteria, Tenericutes, Bacteroidetes, Actinobacteria only accounted for 2%–40% of the sequences (Fig. 2). The abundance of Firmicutes decreased rapidly in the samples collected after the NaCl was added on the last day (<20%). In contrast, the proportion of Proteobacteria was significantly higher in 4_NaCl, 7_NaCl and 8_NaCl compared with the others. Cyanobacteria decreased gradually and finally disappeared on the 12th day.
The use of RDP classifier Bayesian algorithm analysis allowed bacterial identification at the genus level (Fig. 3). The results showed that the abundance of dominant bacteria differed substantially during the AAF process. The relative abundance of Lactobacillus and Acetobacter was greater than that of the other bacteria during the AAF. At the beginning of the AAF, Lactobacillus was the predominant bacterium, and then decreased during the fermentation process, while Acetobacter was highly enriched throughout the AAF and constituted the most dominant genus at the end of the AAF. In addition, Rhizobium, Methylobacterium, Pantoea, Pseudomonas, Streptomyces, Lactococcus, Xanthomonas and Saccharopolyspora were primarily observed in vinegar Pei collected during the first four days and then decreased as the fermentation (AAF) proceeded. Komagataeibacter was detected on the 6th day and dramatically increased as the fermentation (AAF) proceeded. Pedobacter and Aureimonas in the vinegar Pei gradually decreased during fermentation and finally disappeared in samples obtained on the 22th day, while Rhodococcus and Candidatus_Hepatoplasma increased slightly. The abundance of Bacillus and Ralstonia were relatively stable during the entire AAF process.
Comparison of bacterial community composition at the different stages of the AAF
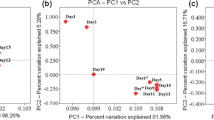
On the basis of the dynamic changes in the microbial community during the AAF (Figs 2 and 3), the fermentation process could be divided into three fermentation stages: group 1 (samples collected in the first 4 days of AAF) was the earlier fermentation stage; group 2 (samples collected from 6th day to 20th day of AAF) and group 3 (samples collected from the 22th day to the 26th day of AAF) were in the medium and later stages of the fermentation, respectively. The distance between sample points in the PCoA diagram was used to evaluate the similarity of the bacterial profile. It was observed that the samples from the same fermentation stage grouped tightly. A more clear discrimination was exhibited between the different fermentation stages in the first direction (PC1), which accounted for 50.56% of the variability (Fig. 4a).
Comparison of bacterial succession among the different stages of the AAF process. (a) Principal coordinates analysis to assess the differences of bacterial communities between the three stages of AAF. (b) One-way ANOVA showing the difference in bacterial richness between the three stages of AAF process. (c) LEfSe showing the dominant bacteria in each stage of AAF.
The Spearman correlation and linear discriminate analysis effect size (LEfSe) identified potential discriminating microorganisms between the three fermentation stages. It was observed that Lactobacillus, Acetobacter, Komagataeibacter, Pantoea, Rhizobium, Methylobacterium, Lactococcus and Candidatus_Hepatoplasma had statistically significant (p < 0.05) differences among the three stages that were consistent with the Alpha-diversity analysis (Fig. 4b). In the early stage, the abundance of Pantoea, Pediococcus, Lactococcus and Rhizobium was significantly higher than those in the other two stages. In the medium stages, Lactobacillus was the dominant type of bacterium that accounted for 67.62% of the total bacteria. In the later stage, Lactobacillus decreased remarkably, while Acetobacter, Komagataeibacter and Kroppenstedtia significantly increased. Acetobacter, Komagataeibacter and Kroppenstedtia prevailed during the later stage (Fig. 4c).
Analysis of the flavor metabolites and their correlation with the bacteria
In this study, a total of 49 kinds of flavor compounds were detected in vinegar Pei during the AAF of SAV (Table 1). These major compounds included seven organic acids and 42 volatile metabolites consisting of seven alcohols, 26 esters, five aldehydes and four ketones. Overall, the microbial diversity and metabolism changed during the AAF, and these metabolites participated in many chemical reactions that caused the varieties of flavor compounds in this period. The correlation Heatmap diagram was used to assess the correlation between microbial classification and environmental variables by the Spearman correlation test.
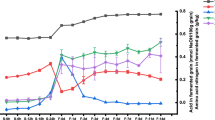
The compositions of organic acids (acetic acid, lactic acid, succinic acid, citric acid, oxalic acid, tartaric acid and α-ketoglutaric acid) were detected by HPLC. Acetic acid, lactic acid, succinic acid and citric acid were the primary organic acids in vinegar Pei accounting for more than 98% of the total contents of the seven organic acids detected. The amount of lactic acid was augmented slightly in the medium stage (74.1 g/kg dry Pei) and then reduced to 53.1 g/kg dry Pei, while the content of acetic acid increased rapidly up to 175 g/kg dry Pei by the end of the AAF. The data showed the sustained growth tendency of succinic acid and citric acid during the AAF. As seen from Fig. 5a, Komagataeibacter and Acetobacter had a significant positive correlation with organic acids, especially acetic acid, suggesting that these microorganisms could contribute to the yield of acids. Further analysis (Fig. 5b) showed that Lactobacillus was positively correlated with the lactic acid that probably explained why the lactic acid increased during the early and medium stages. The accumulation of acetic acid, succinic acid and citric acid might contributeto the increasingabundance of Acetobacter, Komagataeibacter and Kroppenstedtia.
Analyses of flavor metabolites and their correlation to the bacteria. The red color represents the positive correlation and the green color represents the negative correlation. (a) Correlations between bacterial community and flavor metabolites. *Represents the significance of relationships, *0.01 < p ≤ 0.05, **0.001 < p ≤ 0.01, ***p < 0.001 (b) Correlations between the dominant bacteria and the major organic acids. (c) Correlations between the dominant bacteria and the major esters. (d) Correlations between the dominant bacteria and the major alcohols. (e) Correlations between the dominant bacteria and the major aldehydes. (f) Correlations between the dominant bacteria and the major ketones.
As important flavor metabolites, esters could improve the aroma of vinegar and were previously identified21. Among the 26 esters identified in the vinegar Pei, ethyl acetate was the most abundant ester and decreased gradually during stage three as a majority of the esters in the samples. However, the concentrations of acetic acid-2-phenylethyl ester increased during the AAF process and showed a higher positive correlation with Acetobacter and Komagataeibcter. The concentration of 1-butanol-3-methyl-acetate increased in the medium stage and then decreased in the later stage (Table 1). The results of the correlation analysis (Fig. 5a,c) revealed a close relationship of Pantoea, Pediococcus, Rhizobium and Lactococcus with esters in which propanoic acid-2-hydroxy-ethyl ester, hexanoic acid ethyl ester and ethyl acetate were the most significant. The result also indicated that Lactobacillus only contributed to the content of the propanoic acid-2-hydroxy-ethyl ester.
Alcohols mainly originated from the alcohol fermentation stage that provided the precursors for the synthesis of organic acids. In this study, seven alcohols were identified, and ethanol accounts for the highest proportion of all the types of alcohol. Interestingly, 2,3-butanediol was not detected in the early stage and was formed during the medium and late stages, while others decreased during this fermentation process. In addition, Fig. 5d indicated that Pantoea, Pediococcus, Rhizobium and Lactococcus had a positive correlation with the three major alcohols including ethanol and 3-methyl-1-butanol. In contrast, Acetobacter, Kroppenstedtia and Komagataeibacter showed a negative correlation with ethanol and 3-methyl-1-butanol.
This study identified four ketones and five aldehydes in the vinegar Pei of SAV. These metabolites are usually derived from lipid and amino acid degradation22 and contribute honey and fruity odors23. Among the substances, acetoin was an important contributor to synthesize pyrazines that could be added to the food to develop an aroma24. Figure 5a shows Acetobacter and Komagataeibacter bound with these compounds at the p = 0.01 level. In addition, further statistical tests (Fig. 5e,f) revealed that they were positively correlated with the increasing furfural, benzaldehyde, acetoin and 3-acetoxy-2-butanone, while Pantoea, Pediococcus, Rhizobium and Lactococcus showed the opposite result. In addition, the correlations between Lactobacillus and benzaldehyde, as well as between Rhizobium and Lactococcus and 2-octanone, suggested that they were the predominant contributors to form these constituents.
Discussion
AAF is responsible for the flavor metabolite formation and organic acid production attributed to microbial succession, in which the bacteria played key roles9,10. The structure and composition of the microbial community of SAV had been investigated previously in daqu, jiulao and cupei samples using cultural-dependent and cultural-independent methods12,16,19,25,26. However, it was difficult to explore the AAF using traditional pure cultural technique due to the complexity of the bacterial community. For instance, only four bacterial genera including Acetobacter, Lactobacillus, Escherichia and Klebsiella were detected in the AAF using the DGGE method27. With the rapid development of sequencing techniques, it is easier to comprehensively characterize the bacterial community of SAV16,17,19,20. In this study, more than 15 genera of bacteria (relative abundance > 1%) were identified in vinegar Pei during the AAF fermentation using high-throughput sequencing that was more diverse than those detected by DGGE. In addition, Lactococcus and Kroppenstedtia are detected as key microorganisms during the AAF of the SAV for the first time in this study. Among the predominant bacteria, Acetobacter, Pantoea and Pseudomonas probably arose from the starter cultures,28 and Lactobacillus, Saccharopolyspora, Streptomyces and Lactococcus primarily originated from the alcohol mash16. Bacillus and the other genera might have come from the raw materials. Acetobacter, Lactobacillus and Komagataeibacter were dominant as the key bacteria of the SAV that was confirmed by the previous study8,16,19,27. In this study, we identified Acetobacter and Lactobacillus as the most abundant genera, and the variety of the bacterial community decreased during the AAF of SAV. This result is similar to that of Tianjin Duliu aged vinegar29. This result confirms that they have a lot in common with different kinds of vinegar.
In this study, we first used a three-stage classification method (early stage, days 0–4; medium stage, days 5–21; and later stage, days 22–26) based on the microbial assembly to analyze the bacterial community succession during the AAF of SAV. Interestingly, a distinct difference of the bacterial community exhibited among the three stages during AAF of SAV advanced our understanding of the detailed dynamic bacterial succession. This division provided a succession profile of the microbiota during the AAF, which could be used to search for microbial markers characterizing the AAF process and to develop a microbiota-based strategy to monitor the AAF process. In the previous study, Nie et al. reported that the bacterial composition was slightly different during the AAF, and the period of AAF only lasted 7 days16. However, Lactobacillus increased in the medium stage and then decreased gradually in the later stage during this study that differed from the previous report. Nie et al. reported that Lactobacillus decreased gradually during the whole process of the AAF of SAV, and a similar trend was also observed in Zhenjiang Aroma vinegar (ZAV)10. Although SAV and ZAV are both made with a cycle-inoculation technique8,10,27, diverse crude material and geographical locations contribute to the diverse compositions and changes of the bacteria. In addition to Acetobacter, Kroppenstedtia and Komagataeibacter were found to be dominant acid-producing bacteria in the later stages of AAF. The main reason could be attributed to the acid tolerance of these microorganisms30. The result confirms that the microbial diversity relies not only on the fermentation process but also the fermentation environment. The strategy of grouping helps us to monitor the composition and dynamic succession of microbes during the AAF, and the succession profile of the bacteria in the three stages of AAF provide a theoretical guidance for the production of SAV.
Microbial behavior contributes to the production of flavor metabolites9,11,16. More recently, high-throughput sequencing technology was used to investigate the behavior of bacterial in a natural environment31,32, but it was difficult to correlate the bacterial behavior with volatile substances during the AAF due to the complexity and diversity of microorganisms and flavors. The three-stage method used in this paper can reduce the amount of post-production data and can be adopted to easily analyze the relationships. The result suggested a high correlation between the dynamics of the bacterial community and the metabolites, which was confirmed by the previous study on SAV and ZAV9,16,27. It is notable that several dominant acid-producing microbes exhibited a high correlation not only with the acid but also with the volatile flavors. Generally, Acetobacter and Komagataeibacter could oxidize ethanol to acetic acid during the AAF33,34,35. Additives (bran and rice husk) and daily manual stirring could provide glucose and oxygen for the growth of aerobic acid-producing microbes25,36. Interestingly, we found that Acetobacter and Komagataeibacter also had vital impacts on the formation of acetic acid-2-phenylethyl ester, acetoin, 3-acetoxy-2-Butanone butanone and furfural during the late stage of AAF. Lactobacillus was another dominant bacterial group which played an important role in the AAF16,21. In this study, the concentration of lactic acid increased in the medium stage, which corresponded to the dynamic changes of Lactobacillus, suggesting that Lactobacillus contributed to the lactic acid production. Lactobacillus resisted the high concentration of ethanol and decreased its amino acid transport and metabolism under ethanol stress37. To our knowledge, most of the published papers focused on the acid-producing function of Lactobacillus in vinegar. In contrast, we found that the content of benzaldehyde was gradually augmented with the increasing abundance of Lactobacillus. The statistical analysis further suggested that Lactobacillus played an important role in the formation of benzaldehyde. The dominant status of Acetobacter and Lactobacillus and their correlation with organic acids indicated that acetic acid and lactic acid are the main organic acids as described in the previous report38. In addition, we also found that Lactococcus, Pantoea, Pediococcus and Rhizobium dominance in the early stage represented positive relevant esters including ethyl acetate, hexanoic acid ethyl ester, propanoic acid 2-hydroxy-ethyl ester, ethanol and 3-methyl-1-butanol which suggests that these bacteria participate in esterification. These bacteria were first identified as the key microbes in the early stage of AAF. SAV possesses antioxidant properties primarily due to its total polyphenols and flavonoids4, and ketones and aldehydes primarily accumulated in the later stage. Esters that provide an important factor for the aroma of SAV are mainly produced in the early stage. Therefore, the early stage and late stage might be vital periods during the brewing process of SAV.
In conclusion, the AAF process was systematically elaborated in this study. We investigated the correlation between bacterial succession and flavor metabolites including organic acids and volatile substances in each stage of the AAF. This is the first report to explore the relationships between volatile flavor metabolites and bacterial community succession using a three-staged-method and provides the theoretical support for the flavor formation mechanism of traditional SAV.
Methods
Sample collection
The vinegar Pei of SAV in the process of fermentation was sampled from the branch of the Shanxi Aged Vinegar Group (Dingzhou, China). A sterilized cylinder-shaped sampler was used to collect vinegar Pei every other day in process of the AAF from the top to bottom in the middle of three parallel pools (#4, #7, #8, 11.7 m × 1.41 m × 0.9 m) and four corners. Forty-two samples of vinegar Pei (4_0, 4_2, 4_4, 4_6, 4_8, 4_10, 4_12, 4_14, 4_16, 4_18, 4_20, 4_22, 4_24, 4_NaCl, 7_0, 7_2, 7_4, 7_6, 7_8, 7_10, 7_12, 7_14, 7_16, 7_18, 7_20, 7_22, 7_24, 7_NaCl, 8_0, 8_2, 8_4, 8_6, 8_8, 8_10, 8_12, 8_14, 8_16, 8_18, 8_20, 8_22, 8_24, 8_NaCl,) were collected for metagenomic sequencing and flavor analyses. The collected vinegar Pei samples were mixed thoroughly and divided into two parts stored at 4 °C and −80 °C, respectively, for further analysis.
Organic acids analyses
The contents of seven organic acids (oxalic acid, lactic acid, acetic acid, succinic acid, tartaric acid, α-ketoglutaric acid and citric acid) of vinegar Pei were analyzed by using high performance liquid chromatography (HPLC; 1260 series, Agilent Technologies, Santa Clara, Calif, USA). Each vinegar Pei (10 g) was placed in a 250 mL flask and added to 30 mL deionized water. The flask sealed with plastic wrap was shaken at 100 rpm for 2 h on a rotary shaker at room temperature and then filtered through a double layer of filter paper. The water extract of vinegar Pei (5 mL) was mixed with 2 mL zinc sulfate solution (300 g L−1) and potassium ferrocyanide solution (106 g L−1) to precipitate the protein and fixed capacity in a 50 mL volumetric flask standing for 20 min. After filtration by double layer filter paper and a 0.22 μm filter membrane, the extract was purified using Sep-Pak C18 Cartridges (500 mg 3 mL−1, Waters, Milford, Massachusetts,USA) for HPLC analysis. The reversed-phase separation of organic acid substances was performed on a ZORBAX Eclipse Plus C-18 (250 mm × 4.6 mm i.d, 5 μm particle size) at 30 °C, and ultraviolet detection (UVD) was performed at 210 nm. The mobile phase of HPLC was NaH2PO4 (20 mmol L−1, pH 7.2) and the flow rate was 0.9 mL/min. The injection column was 10 μL, and it was parsed for 15 min for each sample21.
Volatile flavor metabolite analyses
The compositions of volatile metabolites in vinegar Pei were determined using headspace solid-phase microextraction/gas chromatography-mass spectrometry (HS-SPME/GC-MS)(Trace MS/GC; Thermo Quest Finnigan Co. (Silicon Valley, CA, USA). Each vinegar Pei (2 g) was mixed with 5 mL saturated sodium chloride solution and 5 μL 2-octanol solution at 2.1 mg mL−1 in methanol as an internal standard in a 15 mL gas chromatography vial. Then, the vial was shaken slightly and equilibrated for 10 min at 40 °C in an incubation furnace, and the volatile metabolites were adsorbed by SPME fiber (50/30 µm divinylbenzene/carboxen on polydimethylsiloxane, DVB/CAR on PDMS (SupelCo Co., Bellefonte, PA, USA) for 40 min at 40 °C. The fiber was inserted into the injection port for 5 min at 250 °C to desorb the adsorbate for mass spectral analysis.
The volatile metabolites of vinegar Pei were analyzed using GC-MS as described by Yu et al.39 Separations were performed using a DB-WAX capillary column (30 m × 0.320 mm i.d, 0.25 μm film thickness). GC-MS conditions were performed as follows: the carrier gas was helium at a constant rate of 1.0 mL L−1, and the mode of injection was split-less. The initial temperature of the oven was 32 °C, and it was then heated to 120 °C at a rate of 3 °C/min, and finally 10 °C/min up to 250 °C that was held for 10 min. The temperature of the nozzle and ion source were set to 280 °C and 230 °C, respectively. The electronic impact energy was 70 eV, and the mass spectral scanning range was 18 m/z to 500 m/z. NIST 05a library (Finnigan Co., USA) was used to identify the volatile metabolites by comparing the mass spectral data. Quantification analysis of each compound was conducted by comparing its peak areas with that of 2-octanol in the spectrogram.
Genomic DNA extraction, PCR amplification and pyrosequencing
Microbial DNA was extracted from vinegar Pei using an E.Z.A.N.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s instructions. Pei (500 mg) was mixed with 978 μL sodium phosphate buffer and 122 μL MT buffer in a lysing Matrix E tube, and then centrifuged at 14,000 × g for 5–10 min to pellet the debris. The supernatants were mixed with 250 μL Protein Precipitation Solution (PPS) by shaking the tube by hand 10 times. After shaking, the supernatants from an additional centrifugation were transferred to a clean 15 mL tube; 1.0 mL Binding Matrix suspension was added to bind the DNA, stabilized for 3 min to allow settling of silica matrix, resuspended in Binding Matrix after discarding 500 μL supernatants, and the remaining mixture was transferred to a SPIN Filter. The supernatants collected after centrifugation at 14,000 × g for 1 min two times were mixed with 500 μL prepared SEWS-M and gently resuspended using the force of the liquid from the pipet tip and centrifuged as before. After centrifugation a second time at 14,000 × g for 2 min, the SPIN Filter was placed for 5 min at room temperature to dry out the matrix of the residual wash solution. DNA was collected by resuspending the Binding Matrix in 50 μL DES and centrifuging at 14,000 × g for 1 min. The final DNA purity was determined at A260/A280, and the DNA quality was evaluated by 1% agarose gel electrophoresis under ultraviolet light. All of the extracted DNA was ready for PCR and further experiments.
The V3–V4 hypervariable regions of the bacterial 16S rRNA genes were amplified with primers 338 F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′) using a thermocycler PCR system (GeneAmp 9700, ABI, USA). The PCR reactions were conducted using the following program: 3 min denaturation at 95 °C, 30 cycles 30 s at 95 °C, 30 s annealing at 55 °C, and 45 s elongation at 72 °C, and a final extension at 72 °C for 10 min. PCR reactions were performed in triplicate in 20 μL mixtures containing 4 μL 5 × FastPfu Buffer, 2 μL 2.5 mM dNTPs, 0.8 μL each primer (5 μM), 0.4 μL FastPfu Polymerase and 10 ng template DNA. The resulted PCR products were extracted from a 2% agarose gel.
Processing of sequencing data
Raw FASTQ files were de-multiplexed, quality-filtered using Trimmomatic and merged using FLASH. Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version7.1 http://drive5.com/uparse/), and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed using an RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (Release128 http://www.arb-silva.de) 16S rRNA database using a confidence threshold of 70%.
Alpha-diversity was used to reflect the community diversity and richness using the Chao1 richness estimator and Simpson diversity index. Differences and similarities between the samples were reflected by Beta-diversity that was calculated using the Hellinger distance between samples for bacterial 16S rRNA reads. The relationships between samples were determined by unconstrained dimensionality reduction analysis and mapping through distance matrices known as principal coordinates analysis (PCoA). A one-way analysis of variance (ANOVA) was used to determine whether there were any significant differences between the sample groups of the different periods of vinegar fermentation, and the bacteria with significant differences in each group were determined using LDA EffectSize (LEfSe analysis). The correlation between the microbial classification and environmental variables was explored using the Spearman correlation coefficient, and the thermography could intuitively show the distance matrix.
References
Liu, D. et al. Chinese vinegar and its solid-state fermentation process. Food Rev. Int. 20, 407–424 (2004).
Zhang, Q. et al. Aroma-enhancing role of pichia manshurica isolated from daqu in the brewing of shanxi aged vinegar. Int. J. Food Prop. 9, 2169–2179 (2017).
Haruta, S. et al. Succession of bacterial and fungal communities during a traditional pot fermentation of rice vinegar assessed by PCR-mediated denaturing gradient gel electrophoresis. J. Int. J. Food Microbiol. 109, 79–87 (2006).
Xie, X. et al. Antioxidant Activity of Chinese Shanxi Aged Vinegar and Its Correlation with Polyphenols and Flavonoids During the Brewing Process. J. Food Sci. 82, 2479–2486 (2017).
Xia, T. et al. Protective effects of shanxi aged vinegar against hydrogen peroxide-induced oxidative damage in LO2 cells through Nrf2-mediated antioxidant responses. Rsc Adv. 7, 17377–17386 (2017).
Budak, N. H., Aykin, E., Seydim, A. C., Greene, A. K. & Guzel-Seydim, Z. B. Functional properties of vinegar. J. Food Sci. 79, 757–764 (2014).
Solieri, L. & Giudici, P. Vinegars of the World. 243–259. (Springer Milan, 2009).
Lu, Z. M. et al. Microbial ecology of cereal vinegar fermentation: insights for driving the ecosystem function. Curr. Opin. Biotech. 49, 88–93 (2017).
Wang, Z. M., Lu, Z. M., Shi, J. S. & Xu, Z. H. Exploring flavour-producing core microbiota in multispecies solid-state fermentation of traditional Chinese vinegar. Sci. Rep. 6, 26818 (2016).
Wang, Z. M. et al. Batch-to-batch uniformity of bacterial community succession and flavour formation in the fermentation of Zhenjiang aromatic vinegar. Food Microbiol. 50, 64–69 (2015).
Nie, Z. et al. Exploring microbial succession and diversity during solid-state fermentation of Tianjin duliu mature vinegar. Bioresource Technol. 148, 325–333 (2013).
Wu, J. J., Ma, Y. K., Zhang, F. F. & Chen, F. S. Biodiversity of yeasts, lactic acid bacteria and acetic acid bacteria in the fermentation of “shanxi aged vinegar”, a traditional chinese vinegar. Food Microbiol. 30, 289–297 (2012).
Lu, Z. M. et al. Recovery of aroma substances from Zhenjiang aromatic vinegar by supercritical fluid extraction. Int. J. Food Sci. Technol. 46, 1508–1514 (2011).
Chen, J. C. et al. Simultaneous determination of acetoin and tetramethylpyrazine in traditional vinegars by HPLC method. Food Chem. 122, 1247–1252 (2010).
Hong, Z., Jie, Z., Wang, L. & Li, Z. Development of a spme-gc-ms method for the determination of volatile compounds in shanxi aged vinegar and its analytical characterization by aroma wheel. J. Food Sci. Technol. 53, 171–183 (2015).
Nie, Z. et al. Unraveling the correlation between microbiota succession and metabolite changes in traditional Shanxi aged vinegar. Sci. Rep. 7, 9240 (2017).
Mamlouk, D., Hidalgo, C., Torija, M. J. & Gullo, M. Evaluation and optimisation of bacterial genomic dna extraction for no-culture techniques applied to vinegars. Food Microbiol. 28, 1374–1379 (2011).
Xu, W. et al. Monitoring the microbial community during solid-state acetic acid fermentation of Zhenjiang aromatic vinegar. Food Microbiol. 28, 1175–1181 (2011).
Nie, Z., Zheng, Y., Du, H., Xie, S. & Wang, M. Dynamics and diversity of microbial community succession in traditional fermentation of shanxi aged vinegar. Food Microbiol. 47, 62–68 (2015).
Valera, M. J., Torija, M. J., Mas, A. & Mateo, E. Acetic acid bacteria from biofilm of strawberry vinegar visualized by microscopy and detected by complementing culture-dependent and culture-independent techniques. Food Microbiol. 46, 452–462 (2015).
Wu, L. H. et al. Metagenomics reveals flavour metabolic network of cereal vinegar microbiota. Food Microbiol. 62, 23–31 (2016).
Luo, T., Fan, W. & Xu, Y. Characterization of volatile and semi-volatile compounds in Chinese rice wines by headspace solid phase microextraction followed by gas chromatography-mass spectrometry. J. Inst. Brewing. 114, 172–179 (2012).
Niu, Y. et al. Use of Stir Bar Sorptive Extraction and Thermal Desorption for Gas Chromatography-Mass Spectrometry Characterization of Selected Volatile Compounds in Chinese Liquors. J. Food Anal. Method. 8, 1771–1784 (2014).
Sabik, H., Fortin, J. & Martin, N. Identification of pyrazine derivatives in a typical maple syrup using headspace solid-phase microextraction with gas chromatography– mass spectrometry. Food Chem. 133, 1006–1010 (2012).
Wu, J. J., Gullo, M., Chen, F. S. & Giudici, P. Diversity of acetobacter pasteurianus strains isolated from solid-state fermentation of cereal vinegars. Curr. Microbiol. 60, 280–286 (2010).
Wu, J. J., Ma, Y. K., Zhang, F. F. & Chen, F. S. Culture-dependent and culture-independent analysis of lactic acid bacteria from shanxi aged vinegar. Ann. Microbiol. 62, 1825–1830 (2012).
Li, S., Li, P., Liu, X., Luo, L. & Lin, W. Bacterial dynamics and metabolite changes in solid-state acetic acid fermentation of shanxi aged vinegar. J. Appl. Microbiol. Biotechnol. 100, 4395–4411 (2016).
Wang, X., Ban, S., Hu, B., Qiu, S. & Zhou, H. Bacterial diversity of moutai-flavour daqu based on high-throughput sequencing method. J. Inst. Brewing. 123, 138–143 (2017).
Peng, Q., Yang, Y., Guo, Y. & Han, Y. Analysis of Bacterial Diversity During Acetic Acid Fermentation of Tianjin Duliu Aged Vinegar by 454 Pyrosequencing. Curr. Microbiol. 71, 195–203 (2015).
Yang, X. et al. Transcriptome responses of Lactobacillus acetotolerans F28 to a short and long term ethanol stress. Sci. Rep. 7, 2650 (2017).
Gandolfi, I., Bertolini, V., Ambrosini, R., Bestetti, G. & Franzetti, A. Unravelling the bacterial diversity in the atmosphere. Appl. Microbiol. Biotechnol. 97, 4727–4736 (2013).
De, R. K., Marzorati, M., Van, dA. P., Van, dW. T. & Boon, N. Synthetic microbial ecosystems: an exciting tool to understand and apply microbial communities. Environ. Microbiol. 16, 1472–1481 (2014).
Trček, J., Toyama, H., Czuba, J., Misiewicz, A. & Matsushita, K. Correlation between acetic acid resistance and characteristics of PQQ-dependent ADH in acetic acid bacteria. Appl. Microbiol. Biotechnol. 70, 366–373 (2006).
Matsushita, K., Takaki, Y., Shinagawa, E., Ameyama, M. & Adachi, O. Ethanol Oxidase Respiratory Chain of Acetic Acid Bacteria. Reactivity with Ubiquinone of Pyrroloquinoline Quinone-dependent Alcohol Dehydrogenases Purified from Acetobacter aceti and Gluconohacter suhoxydans. Biosci. Biotechnol. Biochem. 56, 304–310 (1992).
Giudici, P., Gullo, M. & Solieri, L. Vinegars of the World. 157–177. (Springer Milan 2009).
Andrés-Barrao, C. et al. Metaproteomics and Ultrastructure Characterization of Komagataeibacter, spp. Involved in High-Acid Spirit Vinegar Production. Food Microbiol. 55, 112–122 (2016).
Ishii, Y., Akasaka, N., Sakoda, H., Hidese, R. & Fujiwara, S. Leucine responsive regulatory protein is involved in methionine metabolism and polyamine homeostasis in acetic acid bacterium komagataeibacter europaeus. J. Biosci. Bioeng. https://doi.org/10.1016/j.jbiosc.2017.07.017 (2017).
Chen, T., Gui, Q., Shi, J. J., Zhang, X. Y. & Chen, F. S. Analysis of variation of main components during aging process of Shanxi Aged Vinegar. Acetic Acid Bacteria. 2, 6 (2013).
Yu, Y. J. et al. HS-SPME/GC-MS and chemometrics for volatile composition of Chinese traditional aromatic vinegar in the Zhenjiang region. J. Inst. Brewing. 118, 133–141 (2012).
Acknowledgements
This research was supported financially by the National Key R&D Program of China (2016YFD0400500), the National Natural Science Foundation of China (No. 31571872, No. 31771960) and the Construction of technological innovation service capability - basic research business fee (No. PXM2018_014213_000033).
Author information
Authors and Affiliations
Contributions
Y.P.Zhu designed and performed the experiment, analyzed the data and wrote the main manuscript. F.F.Zhang performed the experiments and analyzed the data. C.N.Zhang and L.Yang analyzed the data. G.S.Fan and Y.Q.Xu commented on the manuscript. B.G.Sun provided essential materials and commented on the manuscript. X.T.Li design the research and provided essential materials. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, Y., Zhang, F., Zhang, C. et al. Dynamic microbial succession of Shanxi aged vinegar and its correlation with flavor metabolites during different stages of acetic acid fermentation. Sci Rep 8, 8612 (2018). https://doi.org/10.1038/s41598-018-26787-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26787-6
- Springer Nature Limited
This article is cited by
-
Estamaran date vinegar: chemical and microbial dynamics during fermentation
Brazilian Journal of Microbiology (2024)
-
Exploring the correlation of metabolites changes and microbial succession in solid-state fermentation of Sichuan Sun-dried vinegar
BMC Microbiology (2023)
-
Processing Technologies and Flavor Analysis of Chinese Cereal Vinegar: a Comprehensive Review
Food Analytical Methods (2023)
-
Determination of the aroma changes of Zhengrong vinegar during different processing steps by SPME–GC–MS and GC-O
Journal of Food Measurement and Characterization (2020)