Abstract
Gram-positive bacteria homeostasis and antibiotic resistance mechanisms are dependent on the intricate architecture of the cell wall, where amidated peptidoglycan plays an important role. The amidation reaction is carried out by the bi-enzymatic complex MurT-GatD, for which biochemical and structural information is very scarce. In this work, we report the first crystal structure of the glutamine amidotransferase member of this complex, GatD from Staphylococcus aureus, at 1.85 Å resolution. A glutamine molecule is found close to the active site funnel, hydrogen-bonded to the conserved R128. In vitro functional studies using 1H-NMR spectroscopy showed that S. aureus MurT-GatD complex has glutaminase activity even in the absence of lipid II, the MurT substrate. In addition, we produced R128A, C94A and H189A mutants, which were totally inactive for glutamine deamidation, revealing their essential role in substrate sequestration and catalytic reaction. GatD from S. aureus and other pathogenic bacteria share high identity to enzymes involved in cobalamin biosynthesis, which can be grouped in a new sub-family of glutamine amidotransferases. Given the ubiquitous presence of GatD, these results provide significant insights into the molecular basis of the so far undisclosed amidation mechanism, contributing to the development of alternative therapeutics to fight infections.
Similar content being viewed by others
Introduction
Staphylococcus aureus is considered one of the most important human Gram-positive bacterial pathogens due to its capacity to acquire and develop antibiotic resistance, causing high levels of mortality both in hospital and community settings1. Methicillin-resistant S. aureus (MRSA) are resistant to all ß-lactam antibiotics and show higher propensity to accumulate resistance to other classes of antibiotics. Fighting S. aureus is an urgent task that requires finding new drugs for new targets2. In this quest, peptidoglycan (PG) biosynthesis plays an important role since PG is essential for bacterial survival, cell shape maintenance and turgor pressure counterbalance3.
PG biosynthesis is a complex process that takes place in several sequential enzymatic steps2 (Supplementary Fig. S1). The first step of PG biosynthesis occurs in the cytoplasm, where the nucleotide carbohydrate UDP-MurNAc (UDP-N-acetyl muramic acid) is covalently linked to a small pentapeptide (L-alanine-D-iso-glutamate-L-lysine-D-alanine-D-alanine) forming UDP-MurNAc-pentapeptide. At the membrane level, an undecaprenyl-phosphate lipid carrier binds to the precursor molecule, generating lipid I (Step 1 in Supplementary Fig. S1). Subsequently, a GlcNAc unit is transferred from UDP-GlcNAc to the MurNAc carbohydrate of lipid I, leading to the formation of lipid II (Step 2 in Supplementary Fig. S1). This structure suffers several modifications, such as the addition of a pentaglycine bridge at the L-lysine residue of the pentapeptide, the O-acetylation of MurNAc carbohydrate and the amidation of the D-iso-glutamate residue into D-iso-glutamine (Step 3 in Supplementary Fig. S1). Finally, the PG monomer is translocated across the cytoplasmic membrane and assembled into the growing PG, through transglycosylation and transpeptidation reactions4 (Step 4 in Supplementary Fig. S1).
The PG of S. aureus is characterized by a high degree of crosslinking and almost complete lack of carboxyl groups, due to the amidation of the D-iso-glutamate residue of the stem peptide5. This amidation of PG structure has a major impact in β-lactam antibiotic resistance, as shown with the femC (glnRA) mutant of MRSA6. In vancomycin-resistant S. aureus strains, it was recently shown that the presence of D-iso-glutamine in lipid II stem peptide enhances the binding affinity to chloroeremomycin and oritavancin7.
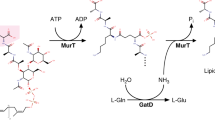
Recently, MurT and GatD enzymes were identified as responsible for catalyzing glutamate amidation in S. aureus PG stem peptides (Fig. 1)8,9. The murT and gatD gene arrangement found in S. aureus is conserved among Gram-positive bacteria8,9. Genetic studies showed that the impairment of the MurT-GatD complex reduces the bacterial growth rate, resistance to β-lactam antibiotics and to lysozyme9,10. Moreover, in vitro experiments showed that the MurT-GatD complex catalyzes glutamate amidation of lipid II using L-glutamine as a direct amine donor, when ATP and Mg2+ were present. Maximum lipid II amidation was achieved with a MurT:GatD molar ratio of 1:1, suggesting the formation of a heteromeric complex. Site-directed mutagenesis identified GatD C94 as an important residue for catalysis8. Similarly, Zapun and co-workers showed that lipid II amidation in Streptococcus pneumoniae strain R6 is catalyzed by MurT-GatD complex, which is essential for PG cross-linking and for cell viability11.
Schematic representation of lipid II amidation in S. aureus. The MurT-GatD-catalysed amidation reaction in the PG biosynthetic pathway is divided in two stages: first, in GatD, deamidation of glutamine releases ammonia, which travels to MurT, where it is used to amidate the stem peptide residue D-iso-Glu of lipid II to D-iso-Gln in the amidated lipid II.
MurT belongs to the family of Mur ligases, which are cytoplasmic enzymes responsible for the sequential addition of amino acid residues to the growing UDP-MurNAc precursor, containing the conserved motifs for ATP and Mg2+ binding. GatD shows high sequence homology to the glutaminase domain of glutamine amidotransferase (GATase) enzymes, in particular to cobyric acid synthase, which is involved in cobalamin biosynthesis9.
The amidation reactions are catalyzed by different domains of a single polypeptide chain or by different subunits of a protein complex and it can be divided into two steps. The first step is the glutaminase reaction, in which glutamine is converted into ammonia and glutamate. The NH3 molecule is then transferred to the synthase domain/subunit through a solvent channel that prevents its protonation to ammonium. Finally, ammonia is added to a specific substrate, in a synthase reaction. Glutamine is used as an amide donor in several pathways that use amino acids, carbohydrates, nucleotides, co-enzymes and antibiotics as substrates12.
Based on amino acid sequence analysis, the glutaminase domain in GATases have been divided in two classes, with distinct protein folds and very different active sites. The enzymes that belong to class I have a conserved catalytic triad at the active site, formed by a cysteine, a histidine and a glutamate residue, and are usually called triad GATases. Structurally characterized triad GATases include: anthranilate synthase (AS), carbamoyl phosphate synthetase (CPS), cytidine triphosphate synthetase, formylglycinamidine ribonucleotide amidotransferase (FGAR-AT), guanosine monophosphate synthetase, imidazole glycerol phosphate synthase (IGPS) and pyridoxal 5′-phosphate synthase (PLPS)13. In class II enzymes, the main characteristics are the position of the essential catalytic cysteine at the N-terminal and no recognizable catalytic triad in the sequence12,14. Asparagine synthetase B, glucosamine-6-phosphate synthase, glutamate synthase and glutamine phosphoribosylpyrophosphate amidotransferase are examples of structurally characterized class II GATases13.
A reaction mechanism has been proposed for class I triad GATases, based on structural evidences13. Generally, the glutamate residue is hydrogen-bonded to the side chain of the catalytic histidine, restricting its rotation and inducing its polarization. The histidine imidazole ring can then deprotonate the nucleophilic cysteine, increasing its reactivity. The catalytic cysteine, in the form of a thiolate, attacks the carbonyl carbon atom of the GATase substrate, the glutamine carboxamide group, forming a tetrahedral intermediate15,16. The stabilization of this negatively charged intermediate is achieved through hydrogen bonds with main chain atoms at the conserved “oxyanion hole”. After the protonation of the amino group in the tetrahedral intermediate, an ammonia molecule is released via a channel to the synthase domain. Finally, the former glutamine/new glutamate is released from the glutamyl-thioester via an acid/base catalysis of a nucleophile attacking water12.
In this work, we combined X-ray crystallography and Nuclear Magnetic Resonance (NMR) spectroscopy to study the glutaminase activity of S. aureus strain COL GatD. The crystal structure was solved at 1.85 Å resolution, unraveling singular features of GATase architecture and important residues at the active site. Site-directed mutagenesis revealed the key amino acids involved in MurT-GatD glutaminase activity and substrate sequestration.
Results/Discussion
Overall structure
GatD crystallized in P212121 space group, with 2 molecules in the asymmetric unit, forming a non-physiological dimer via non-crystallographic symmetry operations (validated by visual inspection and using PISA software17). Molecule A (Fig. 2) corresponds to the full-length GatD protein with 243 amino acids, while molecule B lacks electron density for the last four residues of the polypeptide chain. A free glutamine molecule was found at the surface of GatD chain A. The GatD molecule has a globular shape with the approximate dimensions 50 × 54 × 33 Å3.
Ribbon representation of S. aureus GatD 3D structure. GatD chain A with secondary structural elements labelled. The active site residues (C94 and H189) are represented as sticks with carbon atoms colored green and free glutamine as ball and stick with carbon atoms colored black. The structure is shown in wall-eyed stereo view.
The enzyme adopts an open α/β structure fold, similar to the glutaminase domain from a wide range of GATases15. Indeed, using the GatD model as input in the protein structure comparison service PDBeFold at European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/ssm)18, several class I triad GATases were identified (Table 1). The most similar structure found is the Thermotoga maritima IGPS, with a RMSD of 2.58 Å (162 residues aligned) upon superposition, with an evident overall structure conservation (Fig. 3).
The S. aureus GatD crystal structure shows the characteristic β-sheet core composed by six strands (β1, β2, β3, β4, β10 and β11), all parallel except for β10 (Fig. 2). This β-sheet core is flanked on one side by two helices (α3 and α4) and a two-stranded antiparallel β-sheets (β5 and β6), similar to what has been described for other GATase structures19. The other side of the β-sheet core is covered by a three-stranded antiparallel β-sheets (β7, β8 and β9) and by four α-helices (α1, α2, α5 and α6) (Fig. 2). Interestingly, GatD holds an extra 17 residues long α-helix (α7) at the C-terminal that is absent in all GATases characterized so far19 (Fig. 3 and Supplementary Fig. S2). This α-helix is located at the surface of the protein, as depicted in Fig. 2.
The 3D models of bi-enzymatic complexes selected for structural comparison show a common region for the interaction between the GATase and the corresponding synthase domain. Upon protein-protein interaction, conformational changes occur in the GATase domain that allow the substrate to reach the active site and the catalytic reaction to occur with formation of the ammonia shuttle13. The S. aureus GatD structure here reported was crystallized without its synthase domain (MurT), consequently, in an unproductive enzymatic form.
Active site
As expected, the active site of S. aureus GatD shows high structural similarity to the triad GATases previously mentioned. For comparison, the model of AS from Serratia marcescens (PDB ID 1I7Q) was selected since the glutamyl-thioester intermediate was captured in this crystal structure, highlighting the important residues for the reaction. The superposition (RMSD of 3.07 Å, 148 residues aligned) shows only slight differences in the locations of the catalytic residues between both models (Fig. 4a), probably due to conformational changes that occur upon substrate binding, in the presence of the synthase domain.
Representation of S. aureus GatD active site. (a) Structural comparison of the catalytic sites of S. aureus GatD (in green) and Anthranilate Synthase (AS) from S. marcescens (in grey). The AS crystal structure (PDB ID 1I7Q) includes the glutamyl-thioester intermediate with C85. In AS, the G58 and L86 form the oxyanion hole; the catalytic triad is formed by the C85, H172 and E174; and the G83, I84 and G87 belong to the nucleophile elbow. The superposition is shown in wall-eyed stereo view and the 2Fo-Fc map is contoured at 1.0 σ. (b) Partial sequence alignment (using the ClustalW algorithm) of GatD and PDBeFold hits presented in Table 2 showing the sequence conservation of class I GATases active site, apart from the catalytic glutamate. The secondary structural elements from the GatD 3D structure are shown above the aligned sequences, with α-helices represented as cylinders and β-sheets as arrows. (c) GatD active site architecture, with the catalytic residues (C94 and H189), the nucleophile elbow and the oxyanion hole residues represented as sticks with carbon atoms colored green. The free glutamine molecule is represented as ball and stick with carbon atoms colored black. The 2Fo-Fc electron density map is contoured at 1.0 σ.
An important difference between the S. aureus GatD and other GATases so far characterized, is the absence of the glutamate residue that usually completes the catalytic triad (E174 in S. marcescens AS protein, Fig. 4a). In the structure here reported, glutamate is replaced by a glycine (G190), thereby eliminating any possible polarization effect upon the catalytic histidine H189. In fact, the catalytic role of the conserved glutamate in other GATases is not well understood. In CPS from Escherichia coli, site directed mutagenesis analysis revealed that the catalytic cysteine (C269) and histidine (H353) residues are essential for catalysis, but not the conserved glutamate (E355)20. Additionally, Hart, E.J., et al. showed that the structurally conserved catalytic triad of CPS can act as a functional dyad, since substitution of the neighboring residues of the catalytic histidine (Q310, N311, D334 and Q351) by alanine had no effect on the reaction15. IGPS from T. maritima is another example of a triad GATase, where biochemical data supported the non-catalytic role of the conserved glutamate21. All these evidences point towards the hypothesis that glutamine deamidation can be carried out by cysteine/histidine dyads, which is in agreement with the GatD crystal structure here reported. However, we cannot exclude the hypothesis that GatD residues at the active site may rearrange upon MurT-GatD complex formation (in vitro and/or in vivo), bringing another acidic residue into play and generating a catalytic triad. The glutamate conservation in class I triad GATase and its absence in S. aureus GatD could be related to the specific function of MurT-GatD in the context of peptidoglycan synthesis pathway and should be further studied.
Additional features of GATases active sites, such as the oxyanion hole and the nucleophile elbow, affect glutamine deamidation, empowering the hydrolysis reaction. The oxyanion hole is formed by the amino acid residue next to the catalytic cysteine, commonly a leucine, and by a second residue adjacent to the so-called oxyanion strand13 (G58 in S. marcescens AS protein, Fig. 4a). The backbone NHs of these residues are oriented in such a way that enable two hydrogen bond interactions with a single oxygen atom from the substrate. In S. aureus GatD the residues G95 and G60 of strand β3 are structurally aligned with S. marcescens AS, forming the oxyanion hole (Fig. 4a).
Generally, in triad GATases, the catalytic cysteine is located at the edge of a central strand of the β-sheet core and adjacent to an α-helix, forming a “nucleophile elbow” structural motif. The sequence of this motif is conserved among triad GATases and corresponds to G-X-C-X-G22 (Supplementary Fig. S2). The nucleophile elbow is flanked by the loop containing the catalytic histidine and glutamate, and by the oxyanion strand, inducing the cysteine to adopt a disallowed backbone conformation23,24. In the S. aureus GatD crystal structure here reported, the catalytic C94 is located between β4 and α4 (Fig. 2) with a Ramachandran disallowed conformation (Φ = 49.5° and Ψ = −114.0°), similarly to other GATase triads such as CPS15 and AS25. Moreover, the GatD C94 shows very well-defined electron density. The sequence of the nucleophile elbow motif in S. aureus GatD, T92-I93-C94-G95-G96, is not fully conserved in comparison to other triad GATases since the first residue of the G-X-C-X-G motif (glycine) has been replaced by a threonine (Fig. 4a and b). However, the small size of the threonine residue will not hinder the required arrangement of the cysteine within the nucleophile elbow, for the catalytic reaction (Fig. 4c).
Glutamine sequestration
S. aureus GatD crystallized in the presence of its substrate, glutamine, which likely arose from the expression conditions of the protein, since it was not added during the purification/crystallization steps. The amino acid molecule is found at the surface of the protein, at 8.6 Å from the nucleophilic thiol group of C94 (Fig. 4c), only interacting with R128 from molecule A. The amide oxygen of the free glutamine is establishing hydrogen bonds with the guanidinium group of R128, with bond distances of 3.3 Å and 2.7 Å (Fig. 5). In this regard, R128 seems to be an important residue for sequestrating and accommodating the substrate at the surface of the protein before directing it to the catalytic site for hydrolysis. It is worth mentioning that, due to crystal packing effects, the glutamine binding site is accessible only in chain A. R128 of chain B is facing chain A, which prevents substrate binding.
S. aureus GatD-glutamine complex in wall-eyed stereo view. The glutamine substrate molecule is hydrogen-bonded to the protein surface residue R128, probably because the loop containing the Y17 amino acid is blocking its entrance to GatD active site. Y17 and R128 residues are represented as sticks with carbon atoms colored orange while glutamine is represented as ball and stick with carbon atoms in black. The catalytic dyad residues C94 and H189 are represented as sticks with carbon atoms colored green. The structure is shown in wall-eyed stereo view and the 2Fo-Fc map for R128 (in blue) contoured at 1.0 σ. Omit map mFo-DFc for glutamine is colored green and contoured at 3.0 σ.
Thoden, J.B., et al.26 reported a crystal structure of the CPS enzyme from E. coli (PDB ID 1JDB), in which the glutamine substrate also did not enter the GATase active site, but was located at the surface, near the interface with the synthase domain. In this case, the carboxylic group of the glutamine main chain is hydrogen-bonded to two arginine residues of the glutaminase domain, R120 and R123. Additionally, in the structure of cytidine triphosphate synthetase from Thermus thermophilus (PDB ID 1VCO), the enzyme was crystallized in the presence of glutamine, which was found at the active site19. The authors claim that a hydrogen bond between the amine group of R472 main chain and the α-carboxylate group of glutamine is potentially important for substrate stabilization at the active site. Together, these results suggest that the presence of arginine residues can play an important role in glutamine sequestration for subsequent hydrolysis.
According to the related structures described in the literature, acceptor binding or complex assembly promote the reorganization of a loop at the surface of the glutaminase domain that allows glutamine entrance13. This loop may shield the glutaminase active site, preventing the deamidation reaction in the absence of the synthase domain. In S. aureus GatD, this corresponds to residues 14LNLYSD19, which are flanked by helices α1 and α2. The GatD-glutamine complex here reported shows the Y17 side chain nearly co-planar to the H189 imidazole ring and establishing a hydrophobic interaction with C94 (Fig. 5). In this conformation, Y17 is obstructing the access of the free glutamine to the active site.
Structural insights into the S. aureus GatD sequence homologs
GatD has been assigned as belonging to the group of prokaryotic enzymes involved in cobalamin biosynthesis, as cobyrinic acid a,c-diamide synthase (CobB) or cobyric acid synthase (CobQ)9. CobB27 and CobQ28 have been mostly studied in Pseudomonas denitrificans. CobB has glutaminase activity, catalyzing the conversion of cobyrinic acid to cobyrinic acid a,c-diamide27, while CobQ is responsible for the amidation reaction of 5′-deoxy-5′-adenosyl-cobyrinic acid a,c-diamide to cobyric acid28. Both proteins are organized in two domains, the N-terminal synthase domain with an ATP-binding motif, and a glutaminase domain at the C-terminal region.
Using S. aureus GatD sequence as query in a BLASTP search retrieves a set of targets, all belonging to the CobQ family of proteins, with higher homology towards a probable cobyric acid synthase protein from Methanocella arvoryzae (Table 2). Differentiation between CobQ and CobB proteins is not clear since their functional and structural information is scarce and different views about domain organization have been discussed, namely including them in the class I triad GATase24 or in a, so far, non-classified group of GATases12. Sequence alignment of CobB and CobQ proteins with triad GATases shows that only the catalytic cysteine and histidine residues are conserved29. The lack of the catalytic glutamate suggests that these enzymes can perform the glutaminase activity as functional dyads, as S. aureus GatD, or might have other residue to perform the role of glutamate in a potential triad29.
As expected, S. aureus GatD only aligns with the C-terminal domain of Cob proteins, which corresponds to the glutaminase domain of these enzymes (Supplementary Fig. S3). Based on this sequence alignment, the important features in the GatD crystal structure here described seem to be conserved in CobB/CobQ proteins, mainly in CobQ. In addition to the catalytic dyad, the residues next to C94 and H189 (using GatD numbering) are conserved (Supplementary Fig. S3), suggesting that the architecture of the active site of both CobQ and GatD are probably very similar. High sequence homology is also observed for the oxyanion hole and the nucleophile elbow (although, for CobQ proteins, the first residue of the motif is a glycine while in GatD it is a threonine). Interestingly, R128 of GatD aligns in most cases with positively charged lysine residues that might be responsible for substrate interaction prior to binding at the glutaminase active site. Both CobB and CobQ proteins show the presence of extra C-terminal amino acids that match the extra α-helix of GatD (Supplementary Fig. S3) but are absent in all other GATases characterized so far (Supplementary Fig. S2). According to secondary structure prediction software30, these residues can also form an α-helix that will likely superimpose with the C-terminal region of GatD. Grounded on all these structural evidences, we suggest that the S. aureus GatD crystal structure might be the first representative of a third class of GATases, where CobQ and CobB enzymes are included.
Comparison between the S. aureus GatD and pathogenic Gram-positive homologs
Given the importance of PG amidation in cell viability, the S. aureus GatD structure can be used to derive valuable information for the MurT-GatD homologous systems in other pathogenic Gram-positive bacteria. Protein sequence alignment of S. aureus GatD and its homologs from Staphylococcus epidermidis, Mycobacterium tuberculosis, S. pneumoniae and Streptococcus pyogenes reveals, as expected, high homology with sequence identity of 86%, 36%, 40% and 43%, respectively (Fig. 6). The catalytic cysteine and histidine of S. aureus GatD are conserved in all the aligned proteins, for which the putative triad glutamate residue is also missing. The oxyanion hole and the nucleophile elbow are also highly conserved, suggesting that the architecture of the S. aureus GatD active site is preserved. Interestingly, S. aureus GatD R128 and extra C-terminal residues are conserved in all sequences. These results indicate that Gram-positive GatD proteins diverge from the well-known triad GATases. The conservation of the loop capping the active site in these systems, especially Y17, makes this region a good target for inhibition strategies, imprisoning the enzyme in a nonproductive conformation.
Multiple sequence alignment of GatD proteins from pathogenic Gram-positive bacteria. S. aureus GatD (represented in the alignment as SaGatD) was aligned using the ClustalW algorithm with the amino acid sequences of GatD proteins from S. epidermidis, S. pneumonia, S. pyogenes and M. tuberculosis (with UNIPROT codes A0A0H2VH11, Q8DNZ8, Q1JH53 and I6XI14, respectively). Important GatD residues are highlighted such as the catalytic C94 and H189 dyad in red, oxyanion hole G60 and G95 in gray, nucleophile elbow T92, I93 and G96 in cyan, glutamine sequestration R128 in blue and the extra C-terminal α-helix in orange. The secondary structural elements are shown above the aligned sequences with α-helices represented as cylinders and β-sheets as arrows.
In vitro glutaminase activity of S. aureus MurT-GatD
The analysis of GatD crystallographic data provides strong evidence for the involvement of R128 in the glutamine sequestration, together with some important residues in the catalytic dyad, such as C94 and H189. To further validate this hypothesis, the cloning construct containing murT and gatD genes was modified to introduce point-mutations, replacing the highlighted amino acids by alanines in GatD protein. The resulting protein complexes MurT-GatD wt and mutants C94A, H189A, R128A were produced and their glutaminase activity evaluated using 1H-NMR spectroscopy. Expression and purification of S. aureus MurT-GatD wt and the selected mutants led to similar yields of pure and homogeneous MurT-GatD protein complexes. The final samples showed a molar ratio of MurT-GatD of 1:1 (Fig. 7), confirming the heteromeric complex formation in solution, suggested by Münch, D., et al.8. The stability of the produced MurT-GatD variants show that the mutations imposed in GatD protein did not affect complex formation or solubility.
Glutaminase activity of S. aureus MurT-GatD wt and variants was followed by 1H-NMR spectroscopy (Supplementary Fig. S4). The NMR based activity assay is possible because glutamine and glutamate can be easily distinguished in the 1H-NMR spectrum, allowing to follow and quantify the glutamine conversion into glutamate simply by recording and analyzing 1H-NMR spectra over time (spectra of MurT-GatD wt and H189A mutant in Fig. 8 and C94A and R128A spectra in Supplementary Figure S5). The clearly resolved resonances of glutamine and glutamate γ-CH2 protons at 2.458 ppm and 2.348 ppm, respectively, were integrated to quantify the relative concentration of both amino acids in solution for each time point, as depicted in Fig. 9. These results show that MurT-GatD wt can convert glutamine into glutamate with a first order rate constant of k = 0.0080 ± 0.0001 min−1. This kinetic constant is in agreement with T. maritima IGPS glutaminase kinetics in the absence of the synthase substrate21.
Glutaminase activity kinetics of MurT-GatD protein complexes. Kinetics of glutamine conversion into glutamate was determined from the integration of their γ-CH2 peaks for MurT-GatD wt and mutants C94A, H189A, R128A. Peak intensities were normalized to the initial concentration of glutamine at t = 0. The error estimated from signal to noise ratio was 4.3% for all experiments. The curves for MurT-GatD wt were obtained by fitting the data to a first order rate law but the curves for the mutants are just guidelines for better visualization and comparison.
The glutaminase activity of the MurT-GatD wt complex show that GatD is active in the absence of lipid II, which is the ammonia acceptor and MurT substrate. Since the 1H-NMR signals of glutamate can be detected, we conclude that the product of the glutaminase activity is released from the MurT-GatD complex, allowing a new glutamine molecule to be deamidated. During the catalytic cycle, the Y17 loop is likely displaced, promoting glutamine entrance to the GatD active site.
In the presence of lipid II, it is likely that the GatD active site will rearrange to a fully active state. The new position of the catalytic residues may affect the conversion rate of glutamine into glutamate. This hypothesis is supported by the 1000-fold increase in glutaminase kinetics of T. maritima IGPS when the synthase substrate is present21.
The conversion of glutamine into glutamate could be followed in MurT-GatD wt glutaminase reaction, while the concentration of glutamine maintained constant along the reaction time for the mutant forms and no glutamate was detected in solution (Fig. 9). This confirms that GatD mutations in C94, H189 and R128 residues impair completely the glutaminase activity of the MurT-GatD complex. This establishes the selected residues as essential for the glutaminase activity. Similar studies performed on class I triad GATase CPS protein also showed that point mutation in the catalytic cysteine and histidine produced completely inactive protein forms31,32. These results reinforce the crystallographic data confirming the catalytic architecture of GatD active site, composed by these two residues. Furthermore, our results show univocally that R128 is determinant for glutaminase activity. The amine-based side chain must be detrimental to capture the substrate and drive it into the active site for catalysis.
Conclusions
Multidrug resistant Gram-positive bacteria are a major threat to human health, Staphylococcus aureus being one of the leading causes of infection worldwide. Many efforts have been made to find new antibiotics and develop new strategies to fight antibiotic resistant bacteria. The enzymatic machinery involved in PG biosynthesis is one of the most common targets for drug development, since it is vital for shape maintenance and turgor pressure counterbalance. The amidation reaction is one of the crucial steps of this biosynthetic process and is achieved by the bi-enzymatic complex MurT-GatD. GatD is responsible for metabolizing glutamine and providing ammonia to MurT, the ligase that converts D-glutamate into D-iso-glutamine. At the sequence level, S. aureus GatD is similar to other pathogenic Gram-positive bacteria and to CobB and CobQ-related enzymes, suggesting that these enzymes can be grouped in a new sub-family of GATase domains. The high-resolution crystal structure of S. aureus GatD showed unique features when compared to other GATase domain architectures. Complementary MurT-GatD glutaminase activity results revealed that GatD residues C94 and H189 are the catalytic amino acids of the active site. This arrangement suggests that GatD may rely on a catalytic dyad although, we cannot exclude putative conformational changes upon MurT/lipid II binding that may bring a third residue to participate in the enzymatic reaction. Glutamine sequestration by GatD is mediated by R128, while Y17 seems to block glutamine entrance, in the absence of MurT and lipid II. MurT binding and full complex formation seem to induce a GatD conformational change, adopting its active structural configuration. Even in the absence of lipid II, the MurT-GatD complex has glutaminase activity.
The structural features of S. aureus GatD identified in this work are promising hints for the development of small molecules capable to impair lipid II amidation, placing MurT-GatD as a potential new drug target to fight multidrug resistant bacteria.
Methods
GatD expression, purification and crystallization
S. aureus GatD was cloned, expressed, purified and crystallized as previously described33. Briefly, the PCR amplified sequence of GatD was cloned into the pOPINF vector (Supplementary Table 1) using the In-Fusion method34. The protein was expressed by the auto-induction method35 in E.coli strain Lemo21(DE3) and purified in two chromatographic steps: IMAC and SEC. The crystallization screens were performed at a high-throughput facility (OPPF-UK) using a Cartesian instrument33. Native and Se-Met labeled protein crystals were obtained by vapor diffusion using polyethylene glycol (PEG) as precipitating agent. The procedure from protein expression to crystallization is described in33.
Data collection ans structure determination
X-ray diffraction data of native GatD crystals were collected at temperature 100 K at beamline I02 (λ = 1.0000 Å), Diamond Light Source, UK. GatD crystallized in the space group P212121 with unit cell dimensions of a = 48.61 Å, b = 93.92 Å, c = 110.08 Å, containing 2 molecules per asymmetric unit and a solvent content of 47%. The data was processed using Xia236,37 through the automatic software pipeline available at Diamond Light Source. The phase problem was solved by SAD using a Se-Met derivative, as described in33. The experimental phases were later used to solve the structure of GatD in the native dataset (at 1.85 Å resolution). The structure was manually built using Coot38 and refined using Phenix, considering restraint refinement with 15 TLS groups and isotropic B-factors39. After several rounds of manual rebuilding and refinement the R and Rfree of the final model converged to 0.149 and 0.186. To avoid bulk solvent mask effect on the modeled glutamine molecule, an omit mFo-DFc difference map was generated for the glutamine using phenix. polder program40, where the glutamine region within a radius of 10 Å was excluded (Supplementary Figure S6).
Automatic water picking followed by manual examination allowed identifying 625 water molecules, one PEG molecule, and one glutamine molecule. The structure quality was validated using MolProbity41. Data collection and refinement statistics are summarized in Table 3. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with the accession code 5N9M.
Sequence homology studies
The BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was achieved using GatD sequence as query against UniProtKB/Swiss-Prot database in the standard protein blast (blastp). All multiple sequence alignments were performed using the ClustalW algorithm42.
Cloning and production of S. aureus MurT-GatD wt and mutants
The coding regions for S. aureus murT-gatD genes were amplified by PCR from total DNA obtained from S. aureus strain COL (Supplementary Table 1) and the flanking restriction sites Nco I and Xho I were introduced in the resulting DNA fragments by PCR primers. The amplified murT-gatD operon was cloned into the Nco I – Xho I sites of the expression plasmid pET28a (Supplementary Table 1). The resulting plasmid pET28a-murT-gatD-His6 was introduced into E. coli (DE3) CodonPlus RIPL (Stratagene).
Site-directed mutagenesis was used to produce three variants of MurT-GatD wt, where C94, H189 and R128 from GatD protein were altered to alanine. The base pair exchanges were introduced in gatD gene on plasmid pET28a-murT-gatD-His6 by PCR mutagenesis using Phusion DNA Polymerase (Thermo Fisher Scientific) resulting in the plasmids pET28a-murT-gatD-His6C94A, pET28a-murT-gatD-His6H189A and pET28a-murT-gatD-His6R128A (Supplementary Table 1). All primers are shown in Supplementary Table 2. Presence of the mutations was confirmed by sequencing.
S. aureus MurT-GatD wt and mutants expression and purification
Escherichia coli BL21-CodonPlus (DE3)-RIPL cells (Agilent) were used for heterologous expression of S. aureus MurT-GatD wt and mutants C94A, H189A and R128A, after transformation with plasmids pET28a-murT-gatD-His6C94A, pET28a-murT-gatD-His6H189A and pET28a-murT-gatD-His6R128A, respectively. Cells were grown aerobically in LB medium at 310 K and then the media was supplemented with 0.5 mM isopropyl β-D-1-thiogalactopyranoside to induce protein expression at 293 K and 150 rpm for 16 hours. The cells were harvested by centrifugation at 7500 × g for 15 minutes. The cell pellet was resuspended in buffer A (100 mM Tris-HCl, pH 8.2, 500 mM NaCl, 10 mM MgCl2, 5 mM 2-mercaptoethanol) supplemented with 10 mM imidazole, 10 μg/mL DNaseI and ½ tablet of cOmplete™ ULTRA Tablets, Mini, EDTA-free, EASYpack Protease Inhibitor Cocktail (Roche). Cells lysis was completed by sonication and the resulting supernatant was applied onto a 5 mL HisTrap column (GE Healthcare). Bound protein was eluted using a linear gradient of imidazole. The fractions, where the presence of the complex MurT-GatD was confirmed by SDS-PAGE, were pooled and concentrated. The resulting sample was applied onto a Superdex 200 10/300 GL (GE Healthcare) pre-equilibrated with buffer A. The peak fractions were pooled and concentrated to a final concentration of 70 µM. Buffer exchange to 50 mM Tris-d11, 500 mM NaCl, 10 mM MgCl2, 5 mM 2-mercaptoethanol in D2O was performed using a PD MiniTrap G-25 (GE Healthcare).
NMR experiments
All NMR experiments were performed at 293 K using a Bruker Avance III 400 operating at 400.15 MHz for protons, equipped with a 5 mm BBO probe head. Enzyme kinetics was followed by 1H-NMR spectroscopy measuring the conversion of glutamine into glutamate by integration of their γ-CH2 resonance over time. In a typical experiment, the MurT-GatD protein sample (wt, H189A, R128A or C94A) was added in a 5 mm NMR tube containing a solution of glutamine and 1H-NMR spectra were recorded approximately every 12 minutes for 320 minutes. The initial glutamine concentration was 875 µM in a 99.9% D2O buffered solution (pH meter reading pH 7.9, uncorrected for deuterium isotope effect, 50 mM Tris-d11, 500 mM NaCl, 10 mM MgCl2, 5 mM 2-mercaptoethanol, and 50 µM TSP) and the sample protein concentration was 35 µM (i.e. protein:glutamine ratio of 1:25). 1H-NMR spectra were acquired with 16 K data points and 256 scans in a spectral window of 6009.6 Hz centered at the water resonance (1880.5 Hz) with a T1ρ relaxation filter of 35 ms to suppress protein signals. Chemical shifts were calibrated relative to TSP as internal reference. NMR data was processed in Bruker TopSpin™ 3.5 and analysed with Bruker Dynamics Center™ 2.5. Errors were determined based on the signal to noise ratio.
References
Boucher, H. W. & Corey, G. R. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46, S344–S349 (2008).
Roemer, T., Schneider, T. & Pinho, M. G. Auxiliary factors: A chink in the armor of MRSA resistance to β-lactam antibiotics. Curr. Opin. Microbiol. 16, 538–548 (2013).
Holtje, J. V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62, 181–203 (1998).
van Heijenoort, J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 18, 503–519 (2001).
Schleifer, K. H. & Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477 (1972).
Gustafson, J., Strassle, A., Hachler, H., Kayser, F. H. & Berger-Bachi, B. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J. Bacteriol. 176, 1460–1467 (1994).
Münch, D. et al. Structural variations of the cell wall precursor lipid II and their influence on binding and activity of the lipoglycopeptide antibiotic oritavancin. Antimicrob. Agents Chemother. 59, 772–781 (2015).
Münch, D. et al. Identification and in vitro analysis of the GatD/MurT enzyme-complex catalyzing lipid II amidation in Staphylococcus aureus. PLoS Pathog. 8, 1–11 (2012).
Figueiredo, T. A. et al. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog. 8 (2012).
Figueiredo, T. A., Ludovice, A. M. & Sobral, R. G. Contribution of peptidoglycan amidation to beta-lactam and lysozyme resistance in different genetic lineages of Staphylococcus aureus. Microb. Drug Resist. 20, 1–12 (2014).
Zapun, A. et al. In vitro reconstitution of peptidoglycan assembly from the gram-positive pathogen Streptococcus pneumoniae. ACS Chem. Biol. 8, 2688–2696 (2013).
Massière, F. & Badet-Denisot, M. A. The mechanism of glutamine-dependent amidotransferases. Cell. Mol. Life Sci. 54, 205–222 (1998).
Mouilleron, S. & Golinelli-Pimpaneau, B. Conformational changes in ammonia-channeling glutamine amidotransferases. Curr. Opin. Struct. Biol. 17, 653–664 (2007).
Raushel, F. M., Thoden, J. B. & Holden, H. M. The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia. Biochemistry 38, 7891–7899 (1999).
Hart, E. J. & Powers-lee, S. G. Mutation analysis of carbamoyl phosphate synthetase: does the structurally conserved glutamine amidotransferase triad act as a functional dyad? Protein Sci. 17, 1120–1128 (2008).
Thoden, J. B., Huang, X., Raushel, F. M. & Holden, H. M. The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway. Biochemistry 38, 16158–16166 (1999).
Henrick, E. K. K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D 60, 2256–2268 (2004).
Goto, M., Omi, R., Nakagawa, N., Miyahara, I. & Hirotsu, K. Crystal structures of CTP synthetase reveal ATP, UTP, and glutamine binding sites. Structure 12, 1413–1423 (2004).
Huang, X. & Raushel, F. M. Deconstruction of the catalytic array within the amidotransferase subunit of carbamoyl phosphate synthetase. Biochemistry 38, 15909–15914 (1999).
List, F. et al. Catalysis uncoupling in a glutamine amidotransferase bienzyme by unblocking the glutaminase active site. Chem. Biol. 19, 1589–1599 (2012).
Chaudhuri, B. N., Lange, S. C., Myers, R. S., Davisson, V. J. & Smith, J. L. Toward understanding the mechanism of the complex cyclization reaction catalyzed by imidazole glycerolphosphate synthase: Crystal structures of a ternary complex and the free enzyme. Biochemistry 42, 7003–7012 (2003).
Ollis, D. L. et al. The α/β hydrolase fold. 5, 197–211 (1990).
Zalkin, H. & Smith J. L. Advances in enzymology and related areas of molecular biology: Amino acid metabolism, part A Vol. 72: (eds Purich, D. L.) Ch. Enzymes utilizing glutamine as an amide donor, 87–144 (John Wiley & Sons, Inc., 1998).
Knöchel, T. et al. The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications. Proc. Natl. Acad. Sci. USA 96, 9479–9484 (1999).
Thoden, J. B., Frank, M. & Benning, M. M. The structure of carbamoyl phosphate synthetase determined to 2.1 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. D-55, 8–24 (1999).
Debuscche, L., Thibaut, D., Cameron, B., Crouzet, J. & Blanche, F. Purification and characterization of cobyrinic acid a,c-diamide synthase from Pseudomonas denitrificans. J. Bacteriol. 172, 6239–6244 (1990).
Blanche, F. et al. Biosynthesis of vitamin B12: stepwise amidation of carboxyl groups b, d, e, and g of cobyrinic acid a,c-diamide is catalyzed by one enzyme in Pseudomonas denitrificans. J. Bacteriol. 173, 6046–6051 (1991).
Galperin, M. Y. & Grishin, N. V. The synthetase domains of cobalamin biosynthesis amidotransferases CobB and CobQ belong to a new family of ATP-dependent amidoligases, related to dethiobiotin synthetase. Proteins 41, 238–247 (2000).
Drozdetskiy, A., Cole, C., Procter, J. & Barton, G. J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015).
Miran, S. G., Chang, S. H. & Raushel, F. M. Role of the four conserved histidine residues in the amidotransferase domain of carbamoyl phosphate synthetase. Biochemistry 30, 7901–7907 (1991).
Rubino, S. D., Nyunoya, H. & Lusty, C. J. Catalytic domains of carbamyl phosphate synthetase. J Biol Chem 261, 11320–11327 (1986).
Vieira, D. et al. Purification, crystallization and preliminary X-ray diffraction analysis of GatD, a glutamine amidotransferase-like protein from Staphylococcus aureus peptidoglycan. Acta Crystallogr. Sect. F Struct. Biol. Commun. 70, 1–4 (2014).
Bird, L. E. High throughput construction and small scale expression screening of multi-tag vectors in Escherichia coli. Methods 55, 29–37 (2011).
Studier, F. W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Winter, G. Xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 133–144 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 (2010).
Liebschner, D. et al. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. Sect. D Struct. Biol. 73, 148–157 (2017).
Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. MolProbity: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, 615–619 (2004).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Douangamath, A. et al. Structural evidence for ammonia tunneling across the (βα)8 barrel of the imidazole glycerol phosphate synthase bienzyme complex. Structure 10, 185–193 (2002).
Zein, F. et al. Structural insights into the mechanism of the PLP synthase holoenzyme from Thermotoga maritima. Biochemistry 45, 14609–14620 (2006).
Morar, M., Hoskins, A. A., Stubbe, J. & Ealick, S. E. Formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima: structural insights into complex formation. Biochemistry 47, 7816–7830 (2008).
Smith, A. M., Brown, W. C., Harms, E. & Smith, J. L. Crystal structures capture three states in the catalytic cycle of a pyridoxal phosphate (PLP) synthase. J. Biol. Chem. 290, 5226–5239 (2015).
Strohmeier, M. et al. Structure of a bacterial pyridoxal 5′-phosphate synthase complex. Proc. Natl. Acad. Sci. USA 103, 19284–19289 (2006).
Guédez, G. et al. Assembly of the eukaryotic PLP-Synthase Complex from Plasmodium and activation of the Pdx1 Enzyme. Structure 20, 172–184 (2012).
Gengenbacher, M. et al. Vitamin B6 biosynthesis by the malaria parasite Plasmodium falciparum: biochemical and structural insights. J. Biol. Chem. 281, 3633–3641 (2006).
Spraggon, G. et al. The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan. Proc. Natl. Acad. Sci. USA 98, 6021–6026 (2001).
Acknowledgements
The authors thank the Oxford Protein Production Facility, Research Complex (Harwell, Didcot, UK) for access to high-throughput facilities and assistance during the experiments; beamline staff at I02 and I04 for assistance during data collection at Diamond Light Source (Didcot, UK). DVV, TAF, FL and MS were supported by fellowships SFRH/BD/62415/2009, SFRH/BD/36843/2007, PD/BD/105737/2014 and PD/BD/128202/2016 from Fundação para a Ciência e Tecnologia (FCT-MCTES), Portugal. This project was supported by Project PTDC/BIA-MIC/3195/2012 from FCT-MCTES, Portugal, Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER - COMPETE2020 - Programa Operacional Competitividade e Internacionalização (POCI), Pest-OE/BIA/UI0457/2001 (CREM) and Unidade de Ciências Biomoleculares Aplicadas-UCIBIO which is financed by national funds from FCT/MEC (UID/Multi/04378/2013) and co-financed by the ERDF under the PT2020 Partnership Agreement (POCI-01-0145-FEDER-007728). The NMR spectrometers are part of the National NMR Network (PTNMR) and are supported by Infrastructure Project N° 022161 (co‐financed by FEDER through COMPETE 2020, POCI, and PORL and FCT through PIDDAC).
Author information
Authors and Affiliations
Contributions
The cloning assays of the project were carried out by F.L., T.A.F., R.G.S., A.M.L. and H.L. D.V.V., T.A.F. and F.L. performed protein production and D.V.V. and J.T. carried out protein crystallization and structure determination. M.J.R., T.S.-S. and F.L. carried out model refinement and structural analysis. NMR experiments were performed by M.S. under the supervision of E.J.C. All authors contributed to the manuscript writing. H.L. and T.S.-S. directed and supervised the team members.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leisico, F., V. Vieira, D., Figueiredo, T.A. et al. First insights of peptidoglycan amidation in Gram-positive bacteria - the high-resolution crystal structure of Staphylococcus aureus glutamine amidotransferase GatD. Sci Rep 8, 5313 (2018). https://doi.org/10.1038/s41598-018-22986-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-22986-3
- Springer Nature Limited
This article is cited by
-
Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae
Nature Communications (2018)
-
Structural basis of cell wall peptidoglycan amidation by the GatD/MurT complex of Staphylococcus aureus
Scientific Reports (2018)