Abstract
Carbapenem-resistant Enterobacteriaceae, including the increasingly reported OXA-48 Escherichia coli producers, are an emerging public health threat worldwide. Due to their alarming detection in our healthcare setting and their possible presence in the community, seven OXA-48-producing, extraintestinal pathogenic E. coli were analysed by whole genome sequencing as well as conventional tools, and tested for in vivo virulence. As a result, five E. coli OXA-48-producing subclones were detected (O25:H4-ST131/PST43-fimH30-virotype E; O25:H4-ST131/PST9-fimH22-virotype D5, O16:H5-ST131/PST506-fimH41; O25:H5-ST83/PST207 and O9:H25-ST58/PST24). Four ST131 and one ST83 isolates satisfied the ExPEC status, and all except the O16:H5 ST131 isolate were UPEC. All isolates exhibited local inflammatory response with extensive subcutaneous necrosis but low lethality when tested in a mouse sepsis model. The bla OXA-48 gene was located in MOBP131/IncL plasmids (four isolates) or within the chromosome (three ST131 H30-Rx isolates), carried by Tn1999-like elements. All, except the ST83 isolate, were multidrug-resistant, with additional plasmids acting as vehicles for the spread of various resistance genes. This is the first study to analyse the whole genome sequences of bla OXA-48-positive ST131, ST58 and ST83 E. coli isolates in conjunction with experimental data, and to evaluate the in vivo virulence of bla OXA-48 isolates, which pose an important challenge to patient management.
Similar content being viewed by others
Introduction
Escherichia coli is a common member of the intestine microbiota of warm-blooded vertebrates including humans. It can cause a range of conditions, from diarrheagenic diseases to extraintestinal infections1. Extraintestinal pathogenic E. coli (ExPEC) lineages are involved in the latter. Within them, the clonal group ST131 and its H30-R and H30-Rx subclones (clades C1 and C2), are associated with antimicrobial resistance and have successfully spread, creating a global epidemic of multidrug-resistant (MDR) E. coli infections1,2,3. Treatment of MDR E. coli infections has become a serious clinical issue, with carbapenems being one of the last therapeutic options4. Unfortunately, carbapenemase production is increasingly been reported in E. coli, as well as in other Enterobacteriaceae 4,5. The bla OXA-48 gene, which encodes the OXA-48 carbapenem-hydrolysing class D β-lactamase, has been detected in several E. coli clonal groups, including ST1316,7. The bla OXA-48 gene is usually located on self-transferable plasmids belonging to the IncL/M incompatibility group (recently re-classified into IncL and IncM8), largely responsible for its dissemination between members of the Entebacteriaceae 5,8. Occasionally, bla OXA-48 has also been found in the E. coli chromosome9,10. In both cases, plasmids and chromosomes, the gene is carried by Tn1999-like composite transposons, designated Tn1999.1 to Tn1999.4 9,11,12,13,14.
Due to the alarming emergence of OXA-48-producing E. coli in our healthcare area, this study aimed to (i) analyse the genomic diversity of the isolates, using a combination of whole genome sequencing and molecular typing; (ii) investigate the role of plasmids in the dissemination of resistance and virulence genes, and (iii) assess the lethality of the isolates using a mouse sepsis model. For this purpose, seven OXA-48-producing E. coli recovered between 2012 and 2015 from clinical samples in three different settings within a Spanish city (Oviedo), were selected (Table 1).
Results and Discussion
Resistance properties and molecular typing of the isolates
As shown previously for isolates Ec-HUCA 1 to 315, Ec-HUCA 4 to 7 showed increased MICs to ertapenem. Fortunately, the level of resistance to other carbapenems was low. In fact, all isolates except Ec-HUCA-4 were susceptible to meropenem, so this drug could have been included in the therapy regimen for the treatment of the affected patients (Table 1). In addition, all isolates were positive in the modified Hodge and the Carba NP tests, and contained the bla OXA-48 gene (Table 1). In Ec-HUCA 1 to 4 the bla OXA-48 gene was carried by conjugative plasmids of about 60 Kb, experimentally assigned to the IncL group. The bla OXA-48 gene was chromosomally located in Ec-HUCA 5 to 7. All isolates except Ec-HUCA 3 were also resistant to other antimicrobial agents, including fluoroquinolones, aminoglycosides, trimethoprim-sulfamethoxazole and broad spectrum cephalosporins (Table 1). The latter resistance was detected in four isolates (Ec-HUCA 2, 5, 6 and 7), which tested positive for bla CTX-M-15 by PCR amplification and sequencing.
Five out of the seven isolates were identified as ST131 by the Achtman scheme16 and further differentiated into PST43 (Ec-HUCA 5 to 7), PST9 (Ec-HUCA 4) and PST506 (Ec-HUCA 1) according to the Pasteur Institute scheme17 (Table 1). All ST131 isolates belonged to serotype O25:H4, except Ec-HUCA 1 which was O16:H5. The three O25:H4-ST131/PST43 isolates carried fimH30, were resistant to fluoroquinolones and positive for bla CTX-M-15, thus belonged to subclone H30-Rx. The remaining two isolates (Ec-HUCA 2 and 3) showed different serotypes (O9:H25 and O25:H5), STs (ST58 and ST83), PSTs (PST24 and PST207) and fimH alleles (H27 and H21) (Table 1). ST58 has been identified in isolates of different origins (human, animal and environment), and was frequently associated with CTX-M production, particularly CTX-M-118,19. However, besides Ec-HUCA-2, only one other ST58 OXA-48-producer has been reported20. In contrast to ST131 and ST58, little information exists on ST83, which was previously found in uropathogenic E. coli from cats21. In addition, a query of the Enterobase database (version 6th May 2017) identified 22 ST83 entries from different sources (human, companion animals, wildlife and environment) and geographical regions. The presence of bla OXA-48 in the otherwise susceptible Ec-HUCA 3, demands further surveillance of the ST83 lineage. By PCR, the ST131 and ST83 isolates were assigned to phylogroup B2, while the ST58 isolate belonged to phylogroup B1.
The isolates were also analysed by XbaI-PFGE electrophoresis. The dendrogram generated from comparison of the macrorestriction profiles was consistent with the genetic diversity of the isolates. Thus, only the three ST131/PST43 H30-Rx isolates of virotype E clustered with similarity ≥85%. The remaining two ST131 isolates (Ec-HUCA 1 and 4) presented 76.7% and 65% similarity with that cluster (Supplementary Figure S1). Although the high diversity of the ST131 clonal group has been widely reported3,22, it was unexpected for this specific group of five ST131 OXA-48-producing isolates, since they were obtained in the same health area over a relatively short period of time.
Sequencing of OXA-48-producing isolates and plasmid reconstruction
The draft genomes of the seven E. coli isolates yielded 22 to 135 contigs larger than 1 Kb, with assembly sizes ranging from 4.9 Mb (Ec-HUCA 3) to 5.4 Mb (Ec-HUCA 4); (average of 5.2 ± 0.137 Mb). This is the first time that the genomes of OXA-48-producing ST131 E. coli isolates of subclones H30-Rx, H22 and H41, ST58 and ST83 were sequenced. In silico determinations of serotypes, STs/PSTs and fimH alleles fully agreed with experimental data.
In order to establish the plasmid content of the seven E. coli genomes, the PLACNET protocol23 was used for plasmid reconstruction (Supplementary Figure S2). As shown in Table 2, plasmids were found in all isolates in numbers ranging from one (Ec-HUCA 5) to seven (Ec-HUCA 4). In the genomes of Ec-HUCA 2 and 4, PLACNET identified two plasmids that could not be separated and accounted for a total of 348 Kb [IncF (F2/51/40:A-:B1)] and 253 Kb [IncF (F2:A-:B1) plus IncI1 (ST48)], respectively. Plasmid extraction followed by visualization on agarose gels resolved two plasmids of ca. 140 and 110 Kb in Ec-HUCA 4, a single plasmid band of 150 Kb in Ec-HUCA 2 (maybe two plasmids of similar sizes), and fully confirmed the existence of all other plasmids reconstructed by PLACNET (Supplementary Figure S3).
Overall, the seven E. coli genomes included 23 plasmids, which were classified as MOBP51/ColE1-like, MOBP131/IncL, MOBF12/IncF, MOBF11/IncN, MOBP3/IncX and MOBP12/IncI (Table 2). For one MOBQ12 plasmid the incompatibility group was not determined and two other plasmids could not be affiliated with any of the MOB categories. Finally, a MOBH2 relaxase was identified in the chromosome of Ec-HUCA 1, consistent with the presence of an integrative and conjugative element.
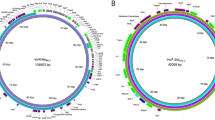
In four out of the seven E. coli isolates (Ec-HUCA 1 to 4; Table 2), the MOBP131/IncL plasmids were experimentally and in silico linked with the bla OXA-48 gene. Their genetic relationship was assessed by building a phylogenetic tree, which also included other IncL and IncM plasmids present in the GenBank-NCBI database (Fig. 1). Assignment to one or the other Inc group was corroborated in silico, or newly determined using the primers reported by Carattoli et al.8. The tree separated IncL and IncM plasmids in two clusters, the latter with two subclusters, IncM1 and IncM2, as previously observed8. Interestingly, bla OXA-48 was not only carried by IncL plasmids but also by IncM1 plasmids, formerly classified as IncL/M24,25. Besides, it was found also on IncA/C plasmids26. The backbones of the IncL plasmids from the HUCA isolates were identical to each other and to nine other IncL plasmids, originating in Klebsiella pneumoniae, Citrobacter freundi, Raoultella planticola and E. coli. These results underline the prevalence of an IncL plasmid lineage which plays a major role in the horizontal spread of bla OXA-48 between members of the Enterobacteriaceae 5. BRIG comparison of the IncL plasmids is shown in Fig. 2. The pEc-HUCA and pE71T27 plasmids are highly similar to pOXA-4811. pCF29 (Accession Number LN864820) showed a large gap at the mobilization-transfer region (24–39 Kb); whereas pCTX-M328 and pNDM-HK29 lack the bla OXA-48 region (2.5–8 Kb), as expected.
Phylogenetic tree of IncL plasmids from OXA-48-producing isolates of Escherichia coli. The tree is based on SNPs found in the core genome (19,995 bp +/− 45 bp; 26 CDS with ≥80% identity, ≥60% pairwise alignment coverage), common to 31 IncL (18), IncM1 (seven) and IncM2 (six) plasmids. Bootstrap support values of 1,000 replicates are shown at the nodes. Clusters corresponding to each group/subgroup are enclosed in yellow, pink and blue boxes. IncL plasmids are more closely related to IncM2 than to IncM1 plasmids.
BRIG (Blast Ring Image generator) comparison of the IncL plasmids from OXA-48-producing Escherichia coli isolates and representative IncL, IncM1 and IncM2 plasmids. Each ring corresponds to a plasmid (indicated at the right of the figure together with the color code). Plasmid pOXA-48 (inner black ring) was used as a reference. Their genes are shown in the outer ring, represented by black arrows that also indicate the direction of transcription.
Genetic environment of the blaOXA-48 gene
As already indicated, bla OXA-48 was carried by MOBP131/IncL plasmids in four E. coli isolates, but chromosomally located in another three (Table 2). Although the DNA surrounding bla OXA-48 could not be assembled from the short Illumina reads, the location of the gene within Tn1999-like transposons and the genetic environment of the transposons was established by PCR mapping. Specifically, bla OXA-48 was carried by Tn1999.2 in Ec-HUCA 4; an inverted Tn1999.2 in Ec-HUCA 1 to 3; and an inverted and deleted version of Tn1999.2 in Ec-HUCA 5 to 7 (Fig. 3). Tn1999.2 differs from Tn1999.1 by insertion of IS1R into the IS1999 copy upstream of bla OXA-48, while in the deleted version, the IS1R-containing copy of IS1999 is truncated at the 5′ end9. IS1R enhances transcription of the downstream bla OXA-48 gene by providing an efficient promoter −35 box with an optimal 17-bp spacing with regard to the −10 box supplied by IS1999 12. This fact could explain the more frequent detection of Tn1999.2 and variants in comparison with Tn1999.1 6. Recently, the complete sequences of the first two ST131 bla OXA-48 plasmids have been reported25; in both, the gene was carried by Tn1999.2, like in Ec-HUCA 4.
Genetic environment of the bla OXA-48 gene. (A) Structures of the Tn1999-like transposons and adjacent DNA from the IncL bla OXA-48 plasmids of Ec-HUCA 1, 2, 3 and 4. (B) Structure of the inverted and deleted Tn1999.2 transposon and the adjacent DNA including the insertion sites in the chromosomes of Ec-HUCA 5, 6 and 7. Open reading frames are represented by arrows indicating the direction of transcription and having different fillings: orange, IS1999; black dots, IS1; red, bla OXA-48; purple, lysR; blue, IncL plasmid genes; green, E. coli chromosomal genes. The Tn1999-like structures are highlighted by yellow boxes.
To identify the insertion sites of bla OXA-48 within the chromosomes of Ec-HUCA 5 to 7, assembled contigs were analysed. Three contigs containing sequences homologous to pOXA-48a (NC_01915411) and pRA35 (LN8648219) were identified, and two of them were flanked by E. coli chromosomal genes. The three isolates shared an identical 21.9 Kb fragment in which the inverted and deleted version of Tn1999.2 was found, followed by genes previously detected in pOXA-48a and pRA35. This fragment, which is flanked by IS1R, constitutes an IS1R composite transposon, termed Tn6237 9. With the single exception of an isolate from Lebanon9, this is the first report of a Tn6237 insertion into the chromosome of ST131. The precise insertion site of the transposon was established by PCR mapping (Supplementary Figure S4). In Ec-HUCA 6 and 7, Tn6237 was placed between the tdcA (DNA-binding transcriptional activator) and tdcB (threonine dehydratase) genes, which are contiguous in other E. coli genomes. In the case of Ec-HUCA 5, Tn6237 was followed by tdcB but morA (putative dehydrogenase) was placed upstream. This organization may have resulted from inversion of the chromosomal segment spanning tdcA to morA. Consistent with its expected lack of specificity, IS1-flanked Tn6237, was also found in three other insertion sites within the E. coli chromosome, in isolates from the United Kingdom, Czech Republic and Lebanon9,30,31.
Resistance genes for other β-lactam and non-β-lactam antimicrobials
According to their phenotypes (Table 1), additional resistance genes were found in all OXA-48 isolates, contained in MOBF12/IncF, MOBF12/IncF + IncI1 and MOBF11/IncN plasmids or in the chromosome (Table 2). Thus, diverse plasmids act as vehicles for the spread of resistance genes between E. coli clones/subclones circulating in the same health area. Remarkably, bla CTX-M-15 appeared in different locations within the four ST131 isolates carrying it. Specifically, bla CTX-M-15 was chromosomally located in two out of the three isolates belonging to the H30-Rx subclone (Ec-HUCA 5 and 6), whereas in Ec-HUCA 7 it was carried by a MOBF11/IncN (ST7) plasmid of 48.7 Kb, together with qnrS1 and dfrA14, for plasmid-mediated quinolone resistance (PMQR) and trimethoprim resistance, respectively. Interestingly, a slightly smaller MOBF11/IncN (ST7) plasmid of 45.8 Kb contained qnrS1 and dfrA14, but not bla CTX-M-15, in Ec-HUCA 6. The bla CTX-M-15 gene of the non-ST131 isolate (Ec-HUCA 2), was carried by a conjugative MOBF12/IncF plasmid of ca. 150 Kb15.
In the three ciprofloxacin resistant ST131 H30-Rx-OXA-48 isolates (Ec-HUCA 5 to 7; MIC > 32 mg/L), in silico analysis of the genomes revealed the distinct gyrA/parC allele combination (gyrA-S83L/D87N; parC-S80I/E84V) previously reported for the H30 subclone32. The S83L mutation was also found in the gyrA gene of Ec-HUCA 1 and 4, which were resistant to nalidixic acid but susceptible to ciprofloxacin. Apart from qnrS1 found in Ec-HUCA 6 and 7, other PMQR genes, including additional qnr genes and qep or oqx genes, tested negative by bioinformatic methods. The genetic bases of most other resistances could be also established by in silico analysis (Tables 1 and 2), being of note the presence of a W30R mutation in the folA gene (dihydrofolate reductase) of Ec-HUCA 5, which was resistant to trimethoprim-sulfamethoxazole but lacked sul and dfr genes.
Virulence gene content and studies of virulence “in vivo”
Based on PCR-screening of 50 genes/alleles characteristic of ExPEC, Ec-HUCA 2 and 4 showed the lowest and highest virulence scores, with 16 and 33 virulence factors (VF), respectively. Of the five ST131 isolates, Ec-HUCA 4 was virotype D5 while Ec-HUCA 5 to 7 were virotype E (Supplementary Figure S1; Supplementary Table S1). The virulence profile of the O16:H5-ST131 isolate did not match any of the 12 virotypes included in the scheme33. Four out of the five ST131 (Ec-HUCA 4 to 7) and the ST83 (Ec-HUCA 3) isolates showed the ExPEC status, and all isolates except Ec-HUCA 1 were classified as UPEC.
PCR-screening results were complemented by in silico analysis of the genome sequences, which allowed the identification of additional VFs, mainly encoded by genes located in the chromosome but also on plasmids (Table 2). Like before, the highest VF score was obtained for Ec-HUCA 4, followed by the ST131 H30-Rx-OXA-48 isolates (Ec-HUCA 5 to 7), which shared the same profile consisting of four operons and 15 individual genes. In contrast, a score of 16 was obtained for Ec-HUCA 1, the ST131 isolate with undefined virotype.
With regard to non-ST131 isolates, 27 VFs were detected in Ec-HUCA 2 and only 13 in Ec-HUCA 3. Interestingly, the plasmid virulence genes of Ec-HUCA 2 and 4 were nearly identical, and some virulence genes have chromosomal and plasmid copies (iss in Ec-HUCA 2, and mchF and iss in Ec-HUCA 4), providing regions of homology for interaction between the two replicons.
Finally, the intrinsic extraintestinal virulence of the OXA-48-producing E. coli was assessed in a mouse sepsis model. Within a seven day experiment, all mice challenged with E. coli CFT703 (positive control) died, compared with none of the mice challenged with E. coli MG1655 (negative control), which remained healthy. All OXA-48 isolates showed lethality as low as ≤10% and only three (Ec-HUCA 2, 6 and 7) caused the death of one out of the ten inoculated mice. In contrast, all isolates caused local inflammatory response, with extensive subcutaneous necrosis, in the surviving mice (Supplementary Table S2). Previously, we observed different virulence patterns in the final lethality, the rapidity in causing death and the inflammation-causing ability of ST131 isolates in correlation with the virotype, with the highest lethality (≥80% of mice challenged killed) shown by virotypes A, B, C and D1. By contrast, isolates within virotypes D2, D3 and D4 led to different outcomes, and isolates of virotype E showed the lowest final lethality, varying from 10 to 40% of the challenged mice. We also observed that certain ST131 isolates of virotypes C, D, and E induced an acute inflammatory response in the inoculation region22, like those in this study. There are few comparable in vivo studies and this is the first one assaying OXA-48-producing isolates. Results derived from the sepsis model might be consistent with the clinical nature of the isolates, not involved in severe disease but recovered from surgical wounds and UTIs. Future studies would be necessary to investigate the mechanisms responsible for the differences in lethality within ST131 virotypes.
Phylogenomics of the OXA-48-producing isolates
The phylogenetic context of the OXA-48-producing E. coli was assessed by comparison with 28 E. coli genomes with different STs, PSTs, virotypes and fimH alleles (Fig. 4, Supplementary Table S3). All ST131 isolates grouped in a single cluster which was further divided into subclusters according to clade, virotype and serotype. In agreement with their assignment to phylogroup B2, the ST131 cluster was genetically closer to ST83 (also B2) than to ST58 (phylogroup B1). It is of note that only two SNPs differences were found between the COG-based core genomes of Ec-HUCA 6 and 7, and that both differed from Ec-HUCA 5 by 14 SNPs. Comparing these three highly similar isolates using SNP analysis and conventional PFGE, we found that both approaches discriminated between Ec-HUCA 5 vs 6 and 7, the latter displaying 100% identity between them and 88.4% with Ec-HUCA 5 by PFGE (Supplementary Figure S1). As shown in Table 1, these clonal isolates were detected in a primary-care centre, in the emergency unit of the HUCA and in the geriatric unit of a long-term facility. Thus, the H30-Rx-OXA-48 subclone, with the chromosomally located carbapenemase gene, is circulating both in the community and health-care institutions, which indicates transmission between the two settings. The number of SNPs between these and other isolates progressively increased according to the ST/PST and phylogroup affiliation, with averages of 3,755, 8334, 19,787 and 79,640, with regard to Ec-HUCA 4, 1, 3 and 2, respectively.
Phylogenetic tree of the OXA-48-producing Escherichia coli genomes. The tree is based on the core genome SNPs (3,154,218 bp ± 1716 bp; 3185 CDS with ≥80% identity, ≥60% pairwise alignment coverage). Bootstrap support values of 1,000 replicates are shown at the nodes. The ST131 clades are indicated with blue (Clade A, PST506/fimH41), green (Clade B, PST9/fimH22-234) and red (Clade C, PST43/fimH30) circles.
Concerning Ec-HUCA 5, 6 and 7, they are classified as virotype E and belong to clade C (PST43/H30), which comprises previously reported ST131 genomes with virotypes A, B and C23. None of these ST131 references carried the bla OXA-48 gene, although BIDMC20B, BWH24 and MNCRE-44 (virotype C) harbour the bla KPC-3 carbapenemase gene, always located on large conjugative plasmids23,34,35. It is important to note that this is the first study where ST131 virotype E genomes have been sequenced. In the clades A (PST506/H41) and B (PST9/H22-234), containing the Ec-HUCA 1 and 4 genomes, respectively, only Ecol_743 harbours the bla OXA-48 gene (located on a 69 Kb plasmid named pEC743_OXA48), while Ecol_448 contains the closely related class D beta-lactamase bla OXA-163 gene (pEC448_OXA163, 71 Kb plasmid)25.
According to our results, diverse OXA-48-producing E. coli clones are circulating in Oviedo, a situation justified in part by the conjugative potential of the IncL plasmid carrying the bla OXA-48 gene. Additional plasmids also play a role as vehicles of resistance and/or virulence genes. Treatment of the affected patients represents a serious challenge since all except one isolate were MDR. So, antimicrobial stewardship policies, new antimicrobial therapy approaches and control measures are necessary to combat the infections caused by these bacteria, and to control further dispersal.
Material and Methods
Epidemiological background of the OXA-48-producing isolates
The isolates were recovered from wound infections or UTIs between 2012 and 2015 (Table 1). Like Ec-HUCA 1 to 315, Ec-HUCA 4 caused a hospital-acquired infection and affected a critical patient exposed to long-term hospitalization and prolonged antimicrobial treatment in the HUCA. The remaining isolates were recovered from patients attended at a primary care centre (Ec-HUCA 5), the emergency unit of the HUCA (Ec-HUCA 6), and the geriatric unit of a long-term care facility associated with the hospital (Ec-HUCA 7).
Antimicrobial susceptibility testing and plasmid analysis
For the new isolates, antimicrobial susceptibility was determined by disk (Oxoid, Madrid, Spain or Becton Dickinson, Sparks, MD, USA) diffusion assays and the Microscan system (MicroScan, Beckman Coulter, CA, USA), which also allows bacterial identification. MICs for erythromycin were determined by broth microdilution following CLSI guidelines36. MICs for carbapenems (ertapenem, imipenem and meropenem) were obtained with Etest strips (bioMérieux, Marcy-l’Étoile, France). Results were interpreted according to CLSI breakpoints36. Carbapenemase production was confirmed by the modified Hodge and Carba NP tests37. Identification of genes encoding resistance to carbapenems and broad-spectrum cephalosporins, and plasmid analysis were performed as reported8,15,38. The genetic context of the bla OXA-48 gene was determined by PCR mapping (see Supplementary Table S4 and Supplementary Figure S4).
Typing, subtyping and phylogenetic grouping of the isolates
E. coli isolates were characterized with regard to O:H serotype and fimH alleles (for type 1 fimbrial adhesion)39,40,41. The STs were established following the MLST schemes of Achtman (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) and the Pasteur Institute (http://bigsdb.pasteur.fr/ecoli/ecoli.html). The XbaI-PFGE profiles were determined according to PulseNet protocol (http://www.pulsenetinternational.org/), imported into BioNumerics (Applied Maths, St-Martens-Latern Belgium) and clustered by Dice/UPGMA. Fifty genes/alleles encoding virulence factors (VF) were screened by PCR (Supplementary Table S5). Isolates were presumptively designated as ExPEC if positive for two or more of five markers and as uropathogenic (UPEC) if positive for three or more of four markers (Supplementary Table S1)42,43. The virotype of the ST131 isolates was established according to the scheme described by Dahbi et al.33. Assignment to the main phylogroups (A, B1, B2 and D) was based on the protocol of Clermont et al.17.
Genome sequencing, assembly and analysis
Total DNA from E. coli isolates was extracted with the QIAmp DNA Mini Kit (Qiagen). Libraries were prepared using the TruSeq PCR-free DNA Sample Preparation Kit (Illumina) at the sequencing facility of the University of Cantabria. Paired-end 100 bp reads (550 bp insert size) were sequenced in a HiSeq. 2500 (Health in Code Facility). Reads were assembled with the VelvetOptimiser.pl script of Velvet software44. Serotype, MLST, fimH alleles and virulence gene profiles were in silico determined with SerotypeFinder v1.145,46, and homemade MLST (Achtman and Pasteur schemes), fimH and virotype databases47. Antimicrobial resistance genes were detected using the ARG-ANNOT48 and ResFinder49 databases; virulence gene content was established with VirulenceFinder and homemade databases50.
For phylogenetic analysis, core genome was defined as described by Lanza et al.23, using the genomes of the E. coli isolates from the HUCA plus reference full-genomes retrieved from GenBank-NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/) and Enterobase (http://enterobase.warwick.ac.uk/species/index/ecoli).
Plasmid reconstruction from WGS data and analysis
Plasmid reconstructions were based on the PLACNET method23. Contig analysis was performed against complete bacterial genomes and plasmids from GenBank-NCBI. Relaxase proteins (REL) and Replication Initiation Proteins (RIP) were identified using in-house databases23,51. Incompatibility groups and pMLST subtypes were experimentally8,52 and/or in silico determined (http://pubmlst.org/plasmid/). Reconstructed bla OXA-48 IncL plasmids and references belonging to the IncL and IncM groups were compared using BRIG53, and a phylogenetic tree was built from variable positions (SNPs) in genes encoding core proteins.
Mouse lethality assay
A mouse sepsis model was used to assess extraintestinal virulence of the isolates22. For each one, 10 outbred female RjOrl:Swiss mice (3–4 weeks old; Janvier Labs, France) received a subcutaneous injection into the nape of the neck of approximately 2 × 108 CFU of log-phase bacteria. After inoculation, mice were clinically inspected along one week. Time of death and local presence of lesions (acute inflammation in the region of inoculation) were recorded for each mouse. Surviving mice were euthanatized on day seven by cervical dislocation. In each assay, two control isolates were included: E. coli K-12 MG1655 which does not kill mice by seven days post-challenge, and E. coli CFT073 which shows a lethality of ≥80% by seven days post-challenge. Results of lethality were indicated as the number of mice killed within 24 h and within seven days post-injection.
Ethics statement
All experimental protocols dealing with bacteria from human samples were approved by the ethics committee of the HUCA.
All animal experimentation was conducted following European (Directive 2010/63/EU on the protection of animals used for scientific purposes) and National (RD 53/2013) regulations for transport, housing and care of laboratory animals. The protocol used was approved by the Animal Welfare Committee of the Veterinary Faculty in Lugo, University of Santiago de Compostela (AE-LU-002/14-1).
Data availability
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under accession numbers NBSZ00000000, NBSY00000000, NBSX00000000, NBSW00000000, NBSV00000000, NBSU00000000, NBST00000000 for Ec-HUCA 1 to 7, respectively. All the samples are part of BioProject PRJNA381431 and correspond to BioSample IDs SAMN06676454 to SAMN06676460.
References
Riley, L. W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 20, 380–390 (2014).
Petty, N. K. et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. 111, 5694–5699 (2014).
Nicolas-Chanoine, M.-H. et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61, 273–281 (2008).
Patel, G. & Bonomo, R. A. ‘Stormy waters ahead’: Global emergence of carbapenemases. Frontiers in Microbiology 4 (2013).
Poirel, L., Potron, A. & Nordmann, P. OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67, 1597–1606 (2012).
Potron, A., Poirel, L., Rondinaud, E. & Nordmann, P. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill. 18 (2013).
Peirano, G. et al. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg. Infect. Dis. 20, 1928–31 (2014).
Carattoli, A., Seiffert, S. N., Schwendener, S., Perreten, V. & Endimiani, A. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10, 1–14 (2015).
Beyrouthy, R. et al. IS1R-mediated plasticity of IncL/M plasmids leads to the insertion of bla OXA-48 into the Escherichia coli Chromosome. Antimicrob. Agents Chemother. 58, 3785–90 (2014).
Beyrouthy, R. et al. Chromosome-mediated OXA-48 carbapenemase in highly virulent Escherichia coli. J. Antimicrob. Chemother. 68, 1558–61 (2013).
Poirel, L., Bonnin, R. A. & Nordmann, P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56, 559–62 (2012).
Carrër, A. et al. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 52, 2950–4 (2008).
Giani, T. et al. Escherichia coli from Italy producing OXA-48 carbapenemase encoded by a novel Tn1999 transposon derivative. Antimicrob. Agents Chemother. 56, 2211–3 (2012).
Potron, A., Nordmann, P., Rondinaud, E., Jaureguy, F. & Poirel, L. A mosaic transposon encoding OXA-48 and CTX-M-15: towards pan-resistance. J. Antimicrob. Chemother. 68, 476–7 (2013).
Fernández, J., Montero, I., Fleites, A. & Rodicio, M. R. Cluster of Escherichia coli isolates producing a plasmid-mediated OXA-48 β-lactamase in a Spanish hospital in 2012. J. Clin. Microbiol. 52, 3414–7 (2014).
Wirth, T. et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–51 (2006).
Clermont, O. et al. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11, 654–62 (2011).
van Hoek, A. H. A. M. et al. Molecular Characteristics of Extended-Spectrum Cephalosporin-Resistant Enterobacteriaceae from Humans in the Community. PLoS One 10, e0129085 (2015).
Gerhold, G., Schulze, M. H., Gross, U. & Bohne, W. Multilocus sequence typing and CTX-M characterization of ESBL-producing E. coli: a prospective single-centre study in Lower Saxony, Germany. Epidemiol. Infect. 144, 3300–3304 (2016).
Ben Tanfous, F. et al. First Description of KPC-2-Producing Escherichia coli and ST15 OXA-48-Positive Klebsiella pneumoniae in Tunisia. Microb. Drug Resist. 23, 365–375 (2017).
Liu, X., Thungrat, K. & Boothe, D. M. Multilocus Sequence Typing and Virulence Profiles in Uropathogenic Escherichia coli Isolated from Cats in the United States. PLoS One 10, e0143335 (2015).
Mora, A. et al. Virulence patterns in a murine sepsis model of ST131 Escherichia coli clinical isolates belonging to serotypes O25b:H4 and O16:H5 are associated to specific virotypes. PLoS One 9, e87025 (2014).
Lanza, V. F. et al. Plasmid Flux in Escherichia coli ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences. PLoS genetics 10, e1004766 (2014).
Chen, L. et al. First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza, Palestine. Antimicrob. Agents Chemother. 59, 4305–7 (2015).
Stoesser, N. et al. Complete Sequencing of Plasmids Containing blaOXA-163 and blaOXA-48 in Escherichia coli Sequence Type 131. Antimicrob. Agents Chemother. 60, 6948–6951 (2016).
Ktari, S. et al. Spread of Klebsiella pneumoniae isolates producing OXA-48 β-lactamase in a Tunisian university hospital. J. Antimicrob. Chemother. 66, 1644–6 (2011).
Power, K. et al. Molecular Analysis of OXA-48-Carrying Conjugative IncL/M-Like Plasmids in Clinical Isolates of Klebsiella pneumoniae in Ireland. Microb. Drug Resist. 20, 270–274 (2014).
Golebiewski, M. et al. Complete Nucleotide Sequence of the pCTX-M3 Plasmid and Its Involvement in Spread of the Extended-Spectrum -Lactamase Gene blaCTX-M-3. Antimicrob. Agents Chemother. 51, 3789–3795 (2007).
Ho, P. L. et al. Complete Sequencing of pNDM-HK Encoding NDM-1 Carbapenemase from a Multidrug-Resistant Escherichia coli Strain Isolated in Hong Kong. PLoS One 6, e17989 (2011).
Turton, J. F. et al. Clonal expansion of Escherichia coli ST38 carrying a chromosomally integrated OXA-48 carbapenemase gene. J. Med. Microbiol. 65, 538–46 (2016).
Skálová, A. et al. Molecular characterization of OXA-48-like-producing Enterobacteriaceae in the Czech Republic: evidence for horizontal transfer of pOXA-48-like plasmids. Antimicrob. Agents Chemother. AAC, 01889–16, doi:https://doi.org/10.1128/AAC.01889-16 (2016).
Johnson, J. R. et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 207, 919–28 (2013).
Dahbi, G. et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int. J. Med. Microbiol. 304, 1247–57 (2014).
Johnson, T. J. et al. Complete Genome Sequence of a Carbapenem-Resistant Extraintestinal Pathogenic Escherichia coli Strain Belonging to the Sequence Type 131 H 30R Subclade. Genome Announc. 3, e00272–15 (2015).
Cerqueira, G. C. et al. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc. Natl. Acad. Sci. USA. 114, 1135–1140 (2017).
Franklin R. Cockerill III, M. & Jean B. Patel, D. M100-S25 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Clin. Lab. Stand. Inst. 44–49, doi:https://doi.org/10.1186/1476-0711-9-23 (2015).
Nordmann, P., Poirel, L. & Dortet, L. Rapid Detection of Carbapenemase-producing. Enterobacteriaceae. Emerg. Infect. Dis. 18, 1503–1507 (2012).
Fernández, J. et al. Dissemination of multiresistant Enterobacter cloacae isolates producing OXA-48 and CTX-M-15 in a Spanish hospital. Int. J. Antimicrob. Agents 46, 469–74 (2015).
Guinée, P. A. M., Jansen, W. H., Wadström, T. & Sellwood, R. In Laboratory diagnosis in neonatal calf and pig diarrhoea: Current Topics in veterinary and animal Science (eds Leeww, P. W. & Guinée, P. A. M.) 126–162 (Martinus Nijhoff Publishers, 1981).
Clermont, O. et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64, 274–7 (2009).
Weissman, S. J. et al. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl. Environ. Microbiol. 78, 1353–60 (2012).
Johnson, J. R. et al. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47, 2161–8 (2003).
Spurbeck, R. R. et al. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 80, 4115–22 (2012).
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–9 (2008).
Joensen, K. G., Tetzschner, A. M. M., Iguchi, A., Aarestrup, F. M. & Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 53, 2410–26 (2015).
Larsen, M. V. et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 50, 1355–1361 (2012).
Blanco, J. et al. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J. Clin. Microbiol. 51, 3358–67 (2013).
Gupta, S. K. et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–20 (2014).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644 (2012).
Joensen, K. G. et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–10 (2014).
Garcillán-Barcia, M. P., Francia, M. V. & de la Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33, 657–87 (2009).
Carattoli, A. et al. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228 (2005).
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L. & Beatson, S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402 (2011).
Acknowledgements
Work at USC-LREC was supported by projects AGL2013–47852-R from the Ministerio de Economía y Competitividad (MINECO, Spain) and Fondo Europeo de Desarrollo Regional (FEDER); AGL2016–79343-R from the Agencia Estatal de Investigación (AEI, Spain) and FEDER; PI16/01477 from Plan Estatal de I + D + I 2013–2016, Instituto de Salud Carlos III (ISCIII), Subdirección General de Evaluación y Fomento de la Investigación, and FEDER; CN2012/303 from the Consellería de Cultura, Educación e Ordenación Universitaria, (Xunta de Galicia) and FEDER. Work at the FdlC laboratory was financed by the Spanish Ministry of Economy and Competitiveness (projects BFU2014–55534-C2-1-P and RTC-2015-3184-1). Work at the UO was supported by projects FIS PI11-00808 (Fondo de Investigación Sanitaria, ISCIII, Ministerio de Economía y Competitividad, Spain) co-funded by FEDER and UO-15-INVES-09 (Consejería de Educación, Cultura y Deporte del Principado de Asturias, Spain). A. Mora acknowledges the Ministerio de Educación, Cultura y Deporte (Spain) for the mobility grant for teachers and researchers from the Programa Estatal de Promoción del Talento y su Empleabilidad, Plan Estatal de Investigación Científica y Técnica y de Innovación 2013–2016.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: J.B., F.C. and M.R.R. Performed the experiments: M.T., J.F., V.G. and A.M. Analysed and interpreted the data: M.T., J.F., V.G., A.M., J.B., F.C. and M.R.R. Drafted the manuscript: M.T., J.F., V.G., A.M., J.B., F.C. and M.R.R. Provided critical input and approved the final manuscript: M.T., J.F., V.G., A.M., J.B., F.C. and M.R.R.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Toro, M., Fernández, J., García, V. et al. Whole genome sequencing, molecular typing and in vivo virulence of OXA-48-producing Escherichia coli isolates including ST131 H30-Rx, H22 and H41 subclones. Sci Rep 7, 12103 (2017). https://doi.org/10.1038/s41598-017-12015-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-12015-0
- Springer Nature Limited
This article is cited by
-
Uropathogenic Escherichia coli endeavors: an insight into the characteristic features, resistance mechanism, and treatment choice
Archives of Microbiology (2023)
-
Production of OXA-48 carbapenemase acts as an independent risk factor for poor outcome in Klebsiella pneumoniae infection
European Journal of Clinical Microbiology & Infectious Diseases (2023)