Abstract
The SIX1 homeobox gene belongs to the six homeodomain family and is widely thought to play a principal role in mediating of skeletal muscle development. In the present study, we determined that the bovine SIX1 gene was highly expressed in the longissimus thoracis and physiologically immature individuals. DNA sequencing of 428 individual Qinchuan cattle identified nine single nucleotide polymorphisms (SNPs) in the promoter region of the SIX1 gene. Using a series of 5′ deletion promoter plasmid luciferase reporter assays and 5′-rapid amplification of cDNA end analysis (RACE), two of these SNPs were found to be located in the proximal minimal promoter region −216/−28 relative to the transcriptional start site (TSS). Correlation analysis showed the combined haplotypes H1-H2 (-GG-GA-) was significantly greater in the body measurement traits (BMTs) than the others, which was consistent with the results showing that the transcriptional activity of Hap2 was higher than the others in Qinchuan cattle myoblast cells. Furthermore, the electrophoretic mobility shift assays (EMSA) and chromatin immunoprecipitation assay (ChIP) demonstrated that NRF1 and ZSCAN10 binding occurred in the promoter region of diplotypes H1-H2 to regulate SIX1 transcriptional activity. This information may be useful for molecular marker-assisted selection (MAS) in cattle breeding.
Similar content being viewed by others
Introduction
Body measurement traits (BMTs) are relevant indicators in cattle selection and breeding. Qinchuan cattle are known to be good beef cattle in China, because of distinctive qualities including good adaptability and fine beef flavor among others. However, these cattle also exhibit certain drawbacks, such as slow growth and underdeveloped hind hip. Accordingly, it is necessary to select important functional genes to solve these problems and increase the BMTs of beef cattle through marker-assisted selection (MAS), which is recognized as a powerful and efficient strategy compared to traditional breeding methods1. However, quantitative traits such as BMTs are controlled by many genes with minor effects2. The identification of statistically significant associations between genetic variants within candidate genes provides a potentially powerful approach to accelerate breeding efforts related to these traits for Qinchuan cattle breed improvement3.
The SIX homeodomain family have been identified six members to date, and designated as SIX1–SIX64. The sine oculis homeobox homologue 1 (SIX1) gene is localized in both the cytoplasm and nucleus of mesenchymal stem cells during embryogenesis and plays important roles in the formation and development of various organs, such as the cranial ganglia5, inner ear6, kidney7, olfactory8 and skeletal muscle9, 10. During skeletal muscle development, SIX1 regulates the expression of the myogenic regulatory factors MyoD and myogenin to mediate skeletal muscle growth and regeneration10,11,12. Additionally, SIX1-null mice die at birth due to hypoplasia and an abnormal structure of primary myogenesis caused by a reduction in and delayed activation of the MyoD and myogenin genes in the limb buds6, 12. Moreover, SIX1 can be activated by the phosphorylation of its cofactor Eya and drives the transformation of the slow-twitch muscle phenotype towards the fast-twitch (glycolytic) phenotype5, 13. Additionally, the knockdown SIX1 expression causes a fibre-type shift towards a slower phenotype by affectings the MHC isoform, which is expressed in the myofibres14. Slow-twitch and fast-twitch muscle fibres are the two most important factors regulating meat tenderness15. Altogether, SIX1 is critical for embryonic development, particularly for skeletal muscle.
Despite its clear role in regulating the formation of muscle and other tissues, the mechanism by which the bovine SIX1 gene is associated with BMTs in cattle has not been reported to date. Promoter region variants can influence transcription activity by altering the transcription factor binding sites, thereby affecting individual development16. Herein, the objectives of this study were to identify the genetic polymorphisms at the 5′UTR of the bovine SIX1 gene and confirm potential cis-acting elements that are associated with single nucleotide polymorphisms (SNPs) and BMTs in Qinchuan cattle.
Results
Detection of SIX1 expression in bovine tissues and organs
The distribution of bovine SIX1 mRNA was determined through qPCR using cDNA from the following 15 bovine tissues and organs: heart, liver, spleen, kidney, rumen, reticulum, omasum, abomasa, small intestine, large intestine, subcutaneous fat, longissimus thoracis, soleus, psoas and testicular tissue. (Fig. 1a). SIX1 had a broad tissue distribution in the cattle tissues and organs. However, the bovine SIX1 gene was highly expressed in the longissimus thoracis, psoas and soleus, moderately expressed in the rumen, testicular and abomasa, and only slightly expressed in the large intestine, omasum, reticulum, subcutaneous fat, small intestine, kidney, spleen, liver and heart tissue.
Expression pattern analysis of the bovine SIX1. (a) Analysis of the bovine SIX1 expression pattern in tissues and organs. (b) Expression pattern of the bovine SIX1 mRNA at different developmental stages. (c) Bovine Six1 expression pattern at different developmental stages. The samples of the longissimus thoracis were obtained at 1, 3, 6, 12, 18, 24, 36 and 48 months after birth. SIX1 mRNA expression was normalized to the housekeeping gene GAPDH and the expression levels were calculated relative to the gene expression in the liver and 24 months, respectively. The value of each column represents the mean ± standard deviation of three independent experiments. The unpaired Student’s t-test was used to detect significant differences. “*”P < 0.05 and “**”P < 0.01.
Temporal expression of SIX1 in longissimus thoracis during different developmental stages
The expression of the bovine SIX1 gene in longissimus thoracis at 1, 3, 6 and 12 months of age was significantly higher than that during any other stages (Fig. 1b and c). However, the expression level was reduced dramatically after 18 months (Fig. 1b and c), but no significant difference was observed in the expression level after 24 months, and the expression level trend remained stable at both the mRNA and protein levels. Based on these results, there appears to be a close relationship between the growth rate traits and SIX1 gene expression in Qinchuan cattle.
Determination of the transcription start site of the SIX1 gene
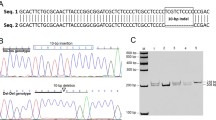
To analyze the structure and molecular mechanisms of the bovine SIX1 gene, we performed 5′-RACE to identify the transcriptional start site (TSS) of SIX1. Two successive rounds of PCR were performed using antisense primer R1 and nested primer R2 (Table S1). As shown in Fig. 2a, two bands of 555 and 475 bp were amplified. In total, 18 positive clones had three different 5′ ends in the first exons, i.e., at 316, 297 and 236 bp upstream of the TSS (Fig. 2b). Sequence alignment showed that the most upstream of the TSS was completely in accord with the published SIX1 mRNA sequence (XM_588692.7). Therefore, we verified the site and designated as +1.
Results of 5′RACE analysis of the SIX1 cDNA from skeletal muscle. (a) Products of the 5′ RACE of the SIX1 (arrow) from nested PCR were analysed by agarose gel electrophoresis. (b) Sequence of the SIX1 mRNA region. The primers (R1 and R2) were used for the 5′ RACE analysis are underlined. The positions of identified TSS are marked with arrows and red underline. The translational start site (ATG) is shown in red letters.
Seven SNPs were identified in the 1.8 kb upstream region of the bovine SIX1 gene
The bovine SIX1 gene located on chromosome 10, contains two exons and two introns and encodes a protein of 284 amino acids. In this study, nine SNPs were identified in the 5′ region of the bovine SIX1 gene by DNA pool sequencing [AC_000167.1: g. −1633 G > A (SNP 9); −1357G > T (SNP 8); −867 G > A (SNP 7); −742 T > C (SNP 6); −421 C > T (SNP 5); −409 T > C (SNP 4); −85 G > T (SNP1); g. −63 G > A (SNP1); g. −14 A > G (SNP 3)] (Fig. 3b).
Luciferase activities in the bovine SIX1 promoter constructs in QCMCs. (a) All plasmids containing 5′ unidirectional deletions of the promoter region of the SIX1 gene (pGL3 −1802, −1488, −1044, −708, −483, −216, −28 and pGL3-Basic) were transfected into QCMCs. After 5 h we replaced the transfection mixture with DMEM with 2% HS (myotubes). The cells were collected for the luciferase assay at 48 h. The results are expressed as the mean ± standard deviation in arbitrary units based on firefly luciferase activity normalized to the Renilla luciferase activity in triplicate transfections. The unpaired Student’s t-test was used to detect significant differences. “*”P < 0.05 and “**”P < 0.01. (b) A graphical representation of the bovine SIX1 gene proximal promoter region from +1 to −1802 base pairs, predicting the regions with a high GC content. Folded lines indicate the GC percentage, which is represented on the y-axis, and the x-axis denotes the bp position on the 5′ untranslated region; the bottom of the blue area indicates the relative positions of the CpG islands. Coordinates are given relative to the translational start site (shown as +1). Arrows indicate TSS-1, positions −216 and −28 bp and nine SNP loci in the promoter, of which SNP1 and SNP2 were located in the proximal minimal promoter of the SIX1 gene.
Isolation of the functional proximal minimal promoter of the SIX1 gene
The proximal minimal promoter of the SIX1 gene was isolated to determine the SNPs that are responsible for influencing the function of promoter. Seven reporter constructs with progressive deletions from the 5′ end of the promoter were generated. The luciferase reporter constructs, named pGL3-1802, pGL3-1488, pGL3-1044, pGL3-708, pGL3-483, pGL3-216 and pGL3-28, were transfected into Qinchuan cattle myoblast cells (QCMCs). The luciferase assays revealed a 5-7-fold increase in the promoter activity of pGL-1802/+142 compared to that in the empty vector, indicating a functional promoter in the −1802/+142 region of the SIX1 gene. When the promoter was deleted to position −1044, the promoter activity of pGL-1044/+142 was decreased by 32% compared with the pGL-1488/+142 (Fig. 3a). After inducing the QCMCs via HS after the transfection, the results of the luciferase assays were similar to those during the undifferentiated stage (Fig. 3a). This result demonstrated that positive regulatory elements were located in the −1488/−1044 region. However, when the promoter was further deleted to −28 bp, the promoter activity of pGL-28/+142 was almost abolished in both the undifferentiated and differentiated QCMCs compared to that of pGL-216/+142. These results indicate that the proximal minimal promoter of the SIX1 gene is located within the region −216/−28 relative to TSS-1 and that the undifferentiated cell model is better for determining transcriptional activity.
Genetic polymorphism of in the Qinchuan Cattle SIX1 proximal minimal promoter region
In the present study, the genetic parameters of the bovine SIX1 gene, including the genotype and allele frequencies, were directly calculated for all 428 animals. SNP1 (g. −85 G > T) and SNP2 (g. −63 G > A) were located in the proximal minimal promoter region −216/−28 of the SIX1 gene. We also determined the genotypic and allelic frequencies, genetic diversity parameters of heterozygosity (He), effective allele numbers (Ne), polymorphism information content (PIC) and Hardy-Weinberg equilibrium (Table 1). The results indicated that for the g. −85 G > T mutation, the TT genotype (14.96%) was less frequent than the wild allele GG (44.16%). The allele frequencies, He, Ne and PIC at the current locus were 0.6460 (G), 0.3540 (T), 0.4574, 1.8428 and 0.3528, respectively. For the g. −63 G > A mutation, the GG genotype was the most prevalent (62.15%) followed by GG (30.84%) and AA (7.01%). The values of the allele frequencies, He, Ne, and PIC at this locus were 0.7757 (G), 0.2243 (A), 0.3480, 1.5537 and 0.2874, respectively. In our study, g. −85 G > T and g. −65 G > A had an intermediate level of polymorphism (high level, PIC > 0.5; moderate level, 0.5 < PIC > 0.25; low level, PIC < 0.25)17. The genotypic distributions of SNP1 and SNP2 were in Hardy-Weinberg disequilibrium (chi-square test, χ2 < χ20.05).
Effects of single marker on BMTs
The association between the two SNPs (g. −85 G > T and g. −63 G > A) and the seven economic traits of body length (BL), withers height (WH), chest depth (CD), chest circumference (CC), back fat thickness (BF), ultrasound loin muscle area (ULA) and intramuscular fat content (IFC) are presented in Table 2. Individuals with genotype the GG had a significantly greater CC and IFC than those with the TT genotype (P < 0.05) for g. −85 G > T. For g. −63 G > A, individuals with the AA genotype had a significantly greater BL, CD, CC, BF and ULA than those with the GA and GG genotypes (P < 0.05) (Table 2). These results indicated that the two loci g. −85 G > T and g. −63 G > A were associated with the BMTs in Qinchuan cattle.
Effects of diplotypes on BMTs
To further analyse the associations between the diplotypes of the two the SNPs and the BMTs, six diplotypes were combined by four haplotypes (Hap1, Hap2, Hap3 and Hap4) (Table 3) in this sample of Qinchuan cattle. Compared with the other diplotype results, the H1-H2 diplotypes had a significantly greater BL, CD, CC, BF, ULA and IFC (P < 0.05) than the H1-H1, H1-H3 and H3-H3 diplotypes (Table 4). Additionally, the H3-H4 diplotypes had a significantly higher BL, CD, CC and BF (P < 0.05) than the H1-H1, H1-H3 and H3-H3 diplotypes (Table 4). In contrast, the H3-H4 diplotypes displayed a reduced IFC (P < 0.05).
Potential transcription-factors in the SIX1 haplotypes possessed different transcriptional activities
To detect the transcriptional activities of the haplotypes, the various haplotypes were cloned, and then luciferase reporter, named pGL3-Hap1, pGL3-Hap2, pGL3-Hap3 and pGL3-Hap4, were constructed. The transcriptional activities of these haplotypes were determined using a dual-luciferase reporter assay system in the QCMCs. The results showed that Hap1 had a 0.83-fold (P < 0.01), 0.04-fold (P > 0.05) and 0.91-fold (P < 0.01) lower activity than Hap2, Hap3 and Hap4, respectively (Fig. 4). A pairwise comparison of the transcriptional activities of Hap1 and Hap2 (different at the SNP2 locus, PHap1/2 < 0.01), Hap3 and Hap4 (different at the SNP2 locus, PHap3/4 < 0.01), Hap1 and Hap3 (different at the SNP1 locus, PHap1/3 > 0.05), Hap2 and Hap4 (different at the SNP1 locus, PHap2/4 > 0.05) was performed. We hypothesized that SNP2 in the g. −63 G > A mutation potentially resulted in the binding of the transcription-factor Nuclear respiratory factor 1 (NRF1) and Zinc finger and SCAN domain containing 10 (ZSCAN10) proteins, which may play a prominent role in the regulation of transcription activities (Table S2, Fig. 5a).
Constitutive activities of the bovine SIX1 haplotypes in QCMCs. The relative lucferase of the transcriptional activities in the candidate haplotypes was used to normalize the promoter activity. Haplotypes of the proximal minimal promoter region of the SIX1 gene were transfected into the QCMCs, and pGL3-Basic served as a control. The cells were collected for the luciferase assay at 48 h. The results are expressed as the mean ± standard deviation in arbitrary units based on the firefly luciferase activity normalized to the renilla luciferase activity in triplicate transfections. The unpaired Student’s t-test was used to detect significant differences. “*”P < 0.05 and “**”P < 0.01.
EMSA assays showing direct binding of NRF1 and ZSCAN10 to the SIX1 promoter in vitro. (a) Sequence and putative transcription factor-binding sites in the wild type (WT) and mutation type (MT) in the proximal minimal promoter of SIX1 gene. The putative transcription factor binding sites are boxed. The primers for the unidirectional deletions are underlined. (b) Nuclear protein extracts were incubated with a free probe containing the NRF1 binding site in the presence or absence of any competition (lane 2), 50× mutation probe (lane 3) and 50× unlabelled probe (lane 4). The super-shift assay was conducted using 10 μg anti-NRF1 antibodies (lane 5). (c) Nuclear protein extracts were incubated with a free probe containing the ZSCAN10 binding site in the presence or absence of any competition (lane 2), 25× mutation probe (lane 3) and 25× unlabelled probe (lane 4). The super-shift assay was conducted using 10 μg anti-ZSCAN10 antibodies (lane 5). The arrows mark the main complexes.
NRF1 and ZSCAN10 bind to the promoter region of the bovine SIX1 gene in vitro and in vivo
Electrophoretic mobility shift assays (EMSAs) and a chromatin immunoprecipitation assay (ChIP) were used to determine whether NRF1 and ZSCAN10 bind to the promoter region of the bovine SIX1 gene (Fig. 5a), resulting in increased transcriptional activities of Hap2 and Hap4. As shown in Fig. 5b, the nuclear protein from the QCMCs bound to the 5′-biotin labelled NRF1 probes and formed two main complexes (lane 2, Fig. 5b). The competition assays verified that the mutant probe had a slight effect on the main complexes (lane 3 Fig. 5b). However, specificity of the NRF1/DNA interaction was prevented by competition from excess non-labelled DNA (lane 4, Fig. 5b). The final lane shows that the complex was super-shifted when it was incubated with the NRF1-antibody (lane 5, Fig. 5b). ZSCAN10 yielded similar results as NRF1 (Fig. 5c). Although the EMSA in the ZSCAN10 experiments did not reveal a super-shifted product at the ZSCAN10 binding sites, however, the brand of the main complex was clearly decreased. The super-shifted may have formed a high molecular weight polymer, which caused a reduced gel mobility shift (lane 5, Fig. 5c). The ChIP results revealed that NRF1 and ZSCAN10 interacted with the binding sites; the relative enrichment levels were ~4.9 and ~6.6-folds over the IgG control respectively (Fig. 6a and b), respectively, based on three independent experiments.
ChIP assay of NRF1 and ZSCAN10 binding to the SIX1 promoter in vivo. We analysed the immunoprecipitated products of the NRF1 (a) and ZSCAN10 (b) antibodies via RT-PCR and ChIP-QPCR. We used total chromatin from muscles as the input, and normal rabbit IgG served as the negative control antibody. “**”P < 0.01. Error bars represent the SD (n = 3).
Discussion
Identifying QTLs and candidate genes that can be utilized in marker-assisted breeding through the manifestation of economically important traits will facilitate Qinchuan cattle breeding programmes. Variants of candidate genes can be associated with economically important traits, such as growth and carcass traits18, 19. SIX1 has emerged as a candidate gene that is an important regulator of vertebrate development and the maintenance of differentiated tissues states20, 21. Our work supports that SIX1 is one of the genes that controls myogenesis and may influence BMTs.
In the present study, the tissue distribution of SIX1 mRNA showed the highest expression in the longissimus thoracis (Fig. 1a), which is consistent with previous findings observed in other species, such as in human22, porcine23 and duck24. However, the SIX1 expression level is very low in the kidney in this study, which differs from the results of previous studies in which SIX1 was shown to be a crucial factor for early kidney development6, 7. The main reason for this discrepancy is that SIX1 expression is regulated temporally in the kidney during early embryonic development6, 7. Previous studies have mainly studied the regulatory mechanism of the SIX1 gene in early embryogenesis, particularly in the successive steps of myogenesis, but less is known about its expression pattern and regulatory mechanism during later development. In the present study, we detected an up-regulation of SIX1 expression from 1 to 12 months after birth; however, the expression of SIX1 decreased dramatically after 18 months. Increases in skeletal muscle mass are largely determined by hypertrophy of muscle fibers during postnatal growth because the number and size of muscle fibres do not change after birth25. An increase in muscle mass solely through muscle fibres hypertrophy could influence the body measurement and meat quality26. Qinchuan cattle require on average of 12 to 24 months attaining physiological and skeletal maturity, which appears closely related to the decreasing SIX1 expression during this time (Fig. 1b and c). Overall, the mechanism of the bovine SIX1 gene may be associated not only with the early events of myogenesis but also with muscle growth and meat quality.
Previous studies have shown that genetic polymorphisms in SIX1 are associated with BMTs. In pigs, Wu et al.23 reported that a C/T and A/G polymorphism in the SIX1 promoter and intron were significantly associated with the meat color value (MCV1) and meat marbling (MM1) of longissimus dorsi and dressing percentage (DP). In the present study, the correlation analysis showed that cattle with the G allele at g. −85 G > T and the A allele at g. −63 G > A loci had better BMTs. A pairwise comparison revealed that a SNP2 mutation at g. −63 G > A associated with the prominent role of transcription-factors NRF1 and ZSCAN10 in the proximal minimal promoter region, which play a major role in transcriptional regulation27, 28, thus contributing to regulate the transcriptional activity of the SIX1 gene.
NRF1, which encodes a protein that homodimerizes and functions as a transcription factor, is associated with neurite outgrowth through the regulation of globin gene expression29. Previous studies have shown that the activation of NRF1 in fibroblasts induces increases in cytochrome c expression and mitochondrial respiratory capacity. Overexpressing NRF1 in skeletal muscle resulted in an increased expression of myocyte enhancer factor (MEF) 2 A and GLUT430, which were associated with a proportional increase in insulin-stimulated glucose transport in muscles31. Moreover, NRF1 can interact with DYNLL132, PPARGC1A33 and Cytochrome c oxidase (COX)34, to initiate nitric oxide synthase33 and active energy metabolism34 for individual development. ZSCAN10 (also named as Zfp206) belongs to the C2H2 zinc fingers family and encodes a zinc finger transcription factor that is specifically expressed in embryonic stem cells (ESCs)35, 36. ZSCAN10 is a regulator of pluripotency and maintains the pluripotent state by interacting with other pluripotency factors, such as Sox2 and Oct437. ZSCAN10-null mice have a reduced weight and mild hypoplasia in the heart, spleen, eyes and long bones38. Consistently, ZSCAN10 expression in ESCs mid-gestation embryos and adults suggests that ZSCAN10 plays a role in maintaining progenitor cell subpopulation and regulating individual development. In the present study, we identified that NRF1 and ZSCAN10 binds to Hap2 and Hap4 and induce a significantly higher transcriptional activity than others based on the luciferase reporter, EMSA and ChIP assays. These results demonstrate that NRF1 and ZSCAN10 play important roles in regulating the transcriptional activity of the SIX1 gene and contributing to better body measurements.
In combination, the H1-H2 and H3-H4 diplotypes showed better BMTs than the H1-H1, H1-H3 and H3-H3 diplotypes. Correlation analysis results were consistent with the interpretation that the transcriptional activities of Hap2 had a higher activity than those Hap1 and Hap3. Notably, the H3-H4 diplotypes showed a lower IFC than the other diplotypes, which is an indicator of lean meat quality characteristics, affects juiciness, flavour, and morphology and is closely related to beef tenderness15. There is significant breed variation in IFC, with the Belgian Blue bulls producing the leanest meat, Limousine producing intermediate levels and Aberdeen Angus producing the highest fat content39, 40. This increased IFC has a positive impact on tenderness, taste and flavour40. Through aggregate selection, based on our findings, we infer that the H1-H2 diplotypes could be used as a molecular marker of combined genotypes for future selection of BMTs in Qinchuan cattle.
In conclusion, our study revealed that bovine SIX1 is highly expressed in the longissimus thoracis and physiological immature individuals. Two SNPs were located within the proximal minimal promoter region from −216 to −28, inducing the binding of transcription factors NRF1 and ZSCAN10 on the promoter region of diplotypes H1-H2 and regulating SIX1 transcriptional activity. Correlation analysis showed the combined haplotypes H1-H2 (-GG-GA-) had significantly greater BMTs than the others. This information may be useful for MAS in cattle breeding.
Materials and Methods
Ethics Statement
All animal procedures were performed according to guidelines laid down by the China Council on Animal Care, and the protocols were approved by the Experimental Animal Manage Committee (EAMC) of Northwest A&F University.
Animal sources, data collection, and genomic DNA isolation
In total, 428 female cows aged 18 to 24 months were randomly selected from the National Beef Cattle Improvement Center’s experimental farm (Yangling, China). The BMTs were measured as described previously41, including BL, WH, CD and CC. ULA, BF and IFC were measured via ultrasound using a Sono-grader model 2 (Renco, USA). Genomic DNA was extracted from blood samples, using standard method42, and stored at −20 °C until subsequent analyses.
Real-time PCR analysis of expression pattern
Fifteen tissues (heart, liver, spleen, kidney, rumen, reticulum, omasum, abomasa, small intestine, large intestine, subcutaneous fat, longissimus thoracis, soleus, psoas and testicle) were obtained from three adult Qinchuan cattle. The longissimus thoracis samples were collected during eight developmental stages from male Qinchuan cattle, including 1, 3, 6, 12, 18, 24, 36 and 48 months after birth, and three parallel individuals were sampled during each period. Total RNA was extracted from the tissues using a Total RNA Kit (Tiangen, Beijing, China) and then reverse-transcribed using a PrimeScript™ RT Reagent Kit (Perfect Real Time) (TaKaRa, Dalian, China). The reaction was performed using a SYBR Green PCR Master Mix Kit (TaKaRa) on a 7500 System SDS V 1.4.0 (Applied Biosystems, USA). All primers used in the real-time PCR experiments are listed in Supplementary Table S1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was invoked as the endogenous control gene. The relative expression levels of the target mRNAs were calculated using the 2−ΔΔCt method43.
Western blotting
Tissues protein were extracted using the T-PER Tissue Protein Extraction Reagent (Pierce, Thermo Fisher Scientific, USA). The total protein samples were quantified using the Pierce BCA Protein Assay Kit (Thermo Scientific), and 50 μg were electrophoresed on a 10% SDS-polyacrylamide gel, and then transferred to nitrocellulose. After blocking in defatted milk powder, the membranes were incubated with the SIX1 antibody (sc-514441, Santa Cruz, USA) and GAPDH antibody (sc-293335, Santa Cruz). The blots were washed and subsequently treated with a peroxidase labelled secondary antibody. The signals were detected as chemical luminescence to expose X-ray films using the ChemiDoc™ XRS+ System (Bio-Rad, Hercules, CA, USA).
5′-Rapid amplification of cDNA ends (5′-RACE)
To identify the transcriptional start site (TSS) of the bovine SIX1 gene, 5’-RACE was performed on total RNA from the longissimus thoracis muscle using a BD SMARTTM RACE cDNA amplification kit (Clontech Inc., CA, USA) according to the manufacturer’s protocol. PCR was performed using a Universal Primer A Mix (UPM, Clontech Inc., CA, USA) and the nested PCR primers (Table S1) located in exon 1 of the SIX1 gene. The conditions and methods used were as previously described27. For sequencing PCR products were separated by electrophoresis in 2% agarose gels and subsequently cloned into T-Vector pMD19 (simple) (Takara).
Primer design, PCR amplification, and SNPs detection
Three pairs of PCR primers (primer A, B and C) were designed to amplify a 1.8 kb genomic region upstream of the bovine SIX1 gene (AC_000167.1 from 73068130 to 73074697). The primer sequences are reported in Table S1. The PCR amplifications were carried out using pooled genomic DNA from 428 Qinchuan cattle as a template44. The 20 μL PCR reaction volume contained 50 ng of pooled genomic DNA, 0.5 μM of primer, 1 × buffer (including 1.2 mM MgCl2), 200 μM dNTPs, and 0.4 units of KOD DNA polymerase (Toyobo, Osaka, Japan). The PCR was performed using a program of 5 min at 95 °C, 34 cycles of 97 °C 30 s, an annealing temperature for the primers as shown in Table S1 for 30 s and 72 °C for 60 s. All PCR products were sequenced to verify amplification of the intended target. Finally, the sequences were imported into BioXM software (Version 2.6) for the SNP analysis.
Promoter cloning and generation of the luciferase reporter constructs
Fragment primers for −1802, −1488, −1044, −708, −483, −216, −28 and +144 were designed (Table S1) to amplify unidirectional deletions of the bovine SIX1 promoter. Promoter constructs were generated by PCR using specific primers with the sequence of the KpnI and BglII restriction sites incorporated and the wild individuals as DNA templates. Then, all fragments were cloned into Vector pMD19-T (simple) (TaKaRa), and ligated into the luciferase reporter construct pGL3-basic vector digested with the same restriction enzymes KpnI and BglII (TaKaRa). These plasmids were named pGL3-1802, pGL3-1488, pGL3-1044, pGL3-708, pGL3-483, pGL3-216 and pGL3-28. The PCR amplification conditions were similar to the conditions used in the previous step and plasmid DNA was further confirmed by DNA sequencing.
Potential cis-acting elements identification
The Genomatix database (http://www.genomatix.de) was used to search for potential cis-acting elements in the proximal minimal promoter region of the SIX1 gene. These potential cis-acting elements were compared for genotypic differences. Sequences that contained one or two SNPs or being adjacent to SNPs were evaluated.
Cell culture and transfection
QCMCs were isolated from Qinchuan foetal bovine as described previously45, 46. The QCMCs were maintained in Dulbecco’s Modified Eagle Medium (DMEM-F12) and supplemented with 20% newborn calf serum (NBCS, Invitrogen, USA) and antibiotics (100 IU/mL penicillin; 100 µg/mL streptomycin) at 37 °C and 5% CO2 in a normal atmosphere incubator. Cells were grown overnight to 80–90% confluence at a density of 1.2 × 105 cells in the growth medium without antibiotics in 24-well plates. In each well, 800 ng of the each series of reporter plasmid (pGL3 −1802, −1488, −1044, −708, −483, −216 and −28) were co-transfected with pRL-TK normalizing reporter plasmid (Promega, USA) into QCMCs with 3 μL X-tremeGENE HP DNA transfection reagent (Roche, USA). The pGL3-Basic vector served as a negative control. At 5 h after the transfection, we replaced the media with DMEM with 2% horse serum (HS) (GIBCO, Invitrogen) and incubated for 40 h to induce the differentiation of the QCMC myoblasts into myotubes. We performed all remaining steps as previously described28. Firefly luciferase activity and Renilla luciferase activity were measured according to the dual-luciferase reporter assay standard protocol in three independent experiments. The relative luciferase activities were determined using a NanoQuant Plate™ (TECAN, infinite M200PRO).
Statistical analysis
Statistical analysis was performed for allelic frequencies, genotype frequencies, He, Hardy-Weinberg equilibriums and PIC parameters according to Nei’s methods17. Linkage Disequilibrium (LD) and haplotype distributions of the SNPs were analyzed using the expectation maximization algorithm with Haploview software47. The association between single SNPs and body measurements traits were analyzed using the general linear models (GLM) procedure in SPSS (version 13.0). The linear model was used:
where Yijkl were the traits measured on each individual cow, μ was the overall population mean for the traits, Gi was the fixed genotype effect, Aj was the fixed effect of age, Ak was the fixed effect due to the age of dam, Sl was the fixed effect due to the season of sampling (spring vs. fall) and Eijkl was the standard error.
EMSA assays
Nuclear extracts from the QCMCs were prepared using the Nuclear Extract Kit (Active Motif Corp., Carlsbad, CA, USA) according to the manufacturer’s protocol. The Bradford dye assay (Bio-Rad Corp., Richmond, CA, USA) was used to adjust the concentration of nuclear fraction protein. The LightShift Chemiluminescent EMSA Kit (Thermo Fisher Corp., Waltham, MA, USA) was used for the EMSA assays according to the manufacturer’s protocol. Briefly, 200 fmol of the 5′ end with biotin labelled probes (listed in Table S1) were incubated room temperature for 20 min with a reaction mixture containing 2 μL 10 × binding buffer, 1 μL poly (dI.dC) and 10 μg of nuclear protein extract in a volume of 20 μL. For the competition assay, unlabelled or mutated DNA probes were added to the reaction mixture and incubated for 15 min; then, 200 fmol labeled probes in a volume of 20 μL were added and incubated at room temperature for 20 min. For the super-shift assay, 10 μg of the NRF1 (ab86516, Abcam, USA) or ZSCAN10 (ab45344, Abcam) antibodies were added to the reaction mixture and then incubated on ice for 30 min; then, 200 fmol of the labelled probes in a volume of 20 μL were added and incubated at room temperature for 20 min. Finally, the main complexes were resolved on 6% non-denaturing polyacrylamide gel electrophoresis (PAGE) using 0.5 × TBE buffer for 1 h and image with ChemiDoc™ XRS+ System molecular imager (Bio-Rad).
ChIP assay
The ChIP assays were performed using the SimpleChIP® Enzymatic Chromatin IP Kit (CST, Massachusetts, USA) according to the manufacturer’s protocol. The samples (n = 3) from the Hap2 and Hap4 Qinchuan bovine were used. The protein-DNA complexes were cross-linked with 37% formaldehyde and neutralized with glycine. After digesting the DNA with micrococcal nuclease into fragments of approximately 150–900 bp in length, the fragmented chromatin samples were suspended in the ChIP dilution buffer. The cross-linked chromatin samples were immunoprecipitated with 4 μg of the NRF1 or ZSCAN10 antibodies and normal rabbit IgG overnight at 4 °C. The immunoprecipitated products were isolated with protein G agarose beads, and the bound chromatin was then collected with salt washes. The eluted ChIP in the Elution Buffer was then digested with proteinase K and purified for a quantitative PCR analysis. The ChIP primers used in the RT-PCR experiment are listed in Table S1. Percent input was calculated as follows: % Input = 2[−ΔCt(Ct[ChIP] − (Ct[Input]−Log2(Input Dilution Factor)))]48. We used the immunoprecipitated products from the normal rabbit IgG group as a negative control.
Change history
13 May 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41598-022-12159-8
References
Pedersen, L. D., Sørensen, A. C. & Berg, P. Marker-assisted selection can reduce true as well as pedigree-estimated inbreeding. J Dairy Sci.92, 2214–23, doi:10.3168/jds.2008-1616 (2009).
Boukha, A. et al. Genetic parameters of carcass and meat quality traits of double muscled Piemontese cattle. Meat Sci.89, 84–90, doi:10.1016/j.meatsci.2011.03.024 (2011).
Hirway, C.D. et al. Genes Related to Economically Important Traits in Beef Cattle. Asian J Anim sci. 1, doi:10.3923/ajas.2011.34.45 (2011).
Kawakami, K., Sato, S., Ozaki, H. & Ikeda, K. Six family genes–structure and function as transcription factors and their roles in development. Bioessays22, 616–626, doi:10.1002/1521-1878 (2000).
Li, X. et al. Eya protein phosphatase activity regulates SIX1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature426, 247–254, doi:10.1038/nature02083 (2003).
Laclef, C. et al. Altered myogenesis in six1-deficient mice. Development130, 2239–2252, doi:10.1242/dev.00440 (2003).
Xu, P. X. et al. Six1 is required for the early organogenesis of mammalian kidney. Development130, 3085–3094, doi:10.1242/dev.00536 (2003).
Ikeda, K. et al. SIX1 is essential for early neurogenesis in the development of olfactory epithelium. Dev Biol.311, 53–68, doi:10.1016/j.ydbio.2007.08.020 (2007).
Grand, F. L. et al. SIX1 regulates stem cell repair potential and self-renewal during skeletal muscle regeneration. J Cell Biol.198, 815–832, doi:10.1083/jcb.201201050 (2012).
Liu, Y. et al. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res.38, 6857–6871, doi:10.1093/nar/gkq585 (2010).
Chakroun, I. et al. Genome-wide association between Six4, MyoD, and the histone demethylase Utx during myogenesis. FASEB J.29, 11, doi:10.1096/fj.15-277053 (2015).
Suzuki, Y., Ikeda, K. & Kawakami, K. Development of gustatory papillae in the absence of Six1 and Six4. J Anat.219, 710–721, doi:10.1111/j.1469-7580.2011.01435.x (2011).
Grifone, R. et al. SIX1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol Cell Biol.24, 6253–6267, doi:10.1128/MCB.24.14.6253-6267.2004 (2004).
Hetzler, K. L. et al. The homoeobox gene SIX1 alters myosin heavy chain isoform expression in mouse skeletal muscle. Acta Physiologica210, 415–428, doi:10.1111/apha.12168 (2014).
Choe, J. H. et al. The relation between glycogen, lactate content and muscle fiber type composition, and their influence on postmortem glycolytic rate and pork quality. Meat Sci.80, 355–362, doi:10.1016/j.meatsci.2007.12.019 (2008).
Pastinen, T. & Hudson, T. J. Cis-acting regulatory variation in the human genome. Science306, 647–650, doi:10.1126/science.1101659 (2004).
Nei, M. Genetic distance between populations. Am Nat.106, 223–229, doi:10.1086/282771 (1972).
Wang, J. et al. Haplotypes in the promoter region of the CIDEC gene associated with growth traits in Nanyang cattle. Sci Rep.5, 12075, doi:10.1038/srep12075 (2015).
Zhang, Y. R. et al. Molecular Characterization of Bovine SMO Gene and Effects of Its Genetic Variations on Body Size Traits in Qinchuan Cattle (Bos taurus). Int J Mol Sci.16, 16966–16980, doi:10.3390/ijms160816966 (2015).
Yajima, H. et al. Six family genes control the proliferation and differentiation of muscle satellite cells. Exp Cell Res.316, 2932–2944, doi:10.1016/j.yexcr.2010.08.001 (2010).
Grifone, R. et al. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development132, 2235–2249, doi:10.1242/dev.01773 (2005).
Boucher, C. A. et al. Cloning of the Human SIX1 Gene and Its Assignment to Chromosome 14. Genomics33, 140–142, doi:10.1006/geno.1996.0172 (1996).
Wu, W. et al. Molecular characterization, expression patterns and polymorphism analysis of porcine SIX1 gene. Mol Biol Rep.38, 2619–2632, doi:10.1007/s11033-010-0403-9 (2011).
Wang, H. et al. Molecular cloning and expression pattern of duck SIX1 and its preliminary functional analysis in myoblasts transfected with eukaryotic expression vector. Indian J Biochem Bio.51, 271–281 (2014).
Brown, M. Change in fibre size, not number, in ageing skeletal muscle. Age Ageing16, 244–248, doi:10.1093/ageing/16.4.244 (1987).
Rehfeldt, C., Fiedler, I., Dietl, G. & Ender, K. Myogenesis and postnatal skeletal muscle cell growth as influenced by selection. Livest Prod Sci.66, 177–188, doi:10.1016/S0301-6226(00)00225-6 (2000).
Zhao, Z. D. et al. Characterization of the promoter region of the bovine long-chain acyl-CoA synthetase 1 gene: Roles of E2F1, Sp1, KLF15, and E2F4. Sci Rep.6, 19661, doi:10.1038/srep19661 (2016).
Li, A., Zhang, Y., Zhao, Z., Wang, M. & Zan, L. Molecular Characterization and Transcriptional Regulation Analysis of the Bovine PDHB Gene. Plos One.11, e0157445, doi:10.1371/journal.pone.0157445 (2016).
Chan, J. Y., Han, X. L. & Kan, Y. W. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci USA90, 11371–11375 (1993).
APABaar, K. et al. Skeletal muscle overexpression of nuclear respiratory factor 1 increases glucose transport capacity. Faseb J.17, 1666–1673, doi:10.1096/fj.03-0049com (2003).
Fukatsu, Y. et al. Muscle-specific overexpression of heparin-binding epidermal growth factor-like growth factor increases peripheral glucose disposal and insulin sensitivity. Endocrinology.150, 2683–2691, doi:10.1210/en.2008-1647 (2009).
Herzig, R. P., Andersson, U. & Scarpulla, R. C. Dynein light chain interacts with NRF-1 and EWG, structurally and functionally related transcription factors from humans and drosophila. J Cell Sci.113, 4263–73, http://jcs.biologists.org/content/113/23/4263.long (2001).
Wu, Z. et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell98, 115–124, doi:10.1016/S0092-8674(00)80611-X (1999).
APADhar, S. S., Ongwijitwat, S. & Wong-Riley, M. T. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem.283, 3120–3129, doi:10.1074/jbc.M707587200 (2008).
Wang, Z. X. et al. Zfp206 is a transcription factor that controls pluripotency of embryonic stem cells. Stem Cells25, 2173–2182, doi:10.1634/stemcells.2007-0085 (2007).
Liang, Y. et al. Structural analysis and dimerization profile of the SCAN domain of the pluripotency factor Zfp206. Nucleic Acids Res, 40, 8721–8732, doi:10.1093/nar/gks611 (2012).
Yu, H. B., Kunarso, G., Hong, F. H. & Stanton, L. W. Zfp206, Oct4, and Sox2 are integrated components of a transcriptional regulatory network in embryonic stem cells. J Biol Chem.284, 31327–31335, doi:10.1074/jbc.M109.016162 (2009).
Kraus, P. et al. Pleiotropic Functions for Transcription Factor Zscan10. PLoS ONE9, e104568, doi:10.1371/journal.pone.0104568 (2014).
Cuvelier, C. et al. Comparison of composition and quality traits of meat from young finishing bulls from Belgian Blue, Limousin and Aberdeen Angus breeds. Meat Sci.74, 522–531, doi:10.1016/j.meatsci.2006.04.032 (2006).
Razminowicz, R. H., Kreuzer, M. & Scheeder, M. R. L. Quality of retail beef from two grass-based production systems in comparison with conventional beef. Meat Sci.73, 351–361, doi:10.1016/j.meatsci.2005.12.013 (2006).
Gui, L. S., Zhang, Y. R., Liu, G. Y. & Zan, L. S. Expression of the SIRT2 gene and its relationship with body size traits in Qinchuan cattle (Bos taurus). Int J Mol Sci.16, 2458–2471, doi:10.3390/ijms16022458 (2015).
Sambrock, J. & Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA (2001).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods25, 402–408, doi:10.1006/meth.2001.1262 (2001).
Sham, P. et al. DNA Pooling: a tool for large-scale association studies. Nat Rev Genet.3, 862–871, doi:10.1038/nrg930 (2002).
Kamanga-Sollo, E. et al. Roles of igf-i and the estrogen, androgen and igf-i receptors in estradiol-17beta- and trenbolone acetate-stimulated proliferation of cultured bovine satellite cells. Domest Anim Endocrin.35, 88–97, doi:10.1016/j.domaniend.2008.02.003 (2008).
Kamanga-Sollo, E. et al. Effect of estradiol-17β on protein synthesis and degradation rates in fused bovine satellite cell cultures. Domest Anim Endocrin.39, 54–62, doi:10.1016/j.domaniend.2010.08.007 (2010).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics21, 263–265, doi:10.1093/bioinformatics/bth457 (2005).
Chakrabarti, S. K., James, J. C. & Mirmira, R. G. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J Biol Chem.277, 13286–93, doi:10.1074/jbc.M111857200 (2002).
Acknowledgements
This research was supported by the National Modern Agricultural Industry Special Program (No. CARS-38), the National 863 Program of China (No. 2013AA102505), the National Science and Technology Support Projects (No. 2015BAD03B04) and the Shaanxi Technological Innovation Engineering Program (No. 2014KTZB02-02-01).
Author information
Authors and Affiliations
Contributions
Lin-Sen Zan and Da-Wei Wei conceived and designed the experiments. Da-Wei Wei performed the experiments and wrote the manuscript. Lin-Sheng Gui mainly assisted in analyzing the data. Rajwali Khan and Sayed Haidar Abbas Raza provided constructive suggestions for the discussion and a language modification. Song-Zhang and Li-Wang helped to collect the samples and data, Hong-Fang Guo disposal the data.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wei, DW., Gui, LS., Raza, S.H.A. et al. NRF1 and ZSCAN10 bind to the promoter region of the SIX1 gene and their effects body measurements in Qinchuan cattle. Sci Rep 7, 7867 (2017). https://doi.org/10.1038/s41598-017-08384-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-08384-1
- Springer Nature Limited
This article is cited by
-
The polymorphism of the ovine insulin like growth factor-2 (IGF2) gene and their associations with growth related traits in Tibetan sheep
Tropical Animal Health and Production (2024)
-
Association of polymorphism in the promotor area of the caprine BMPR1B gene with litter size and body measurement traits in Damani goats
Tropical Animal Health and Production (2024)
-
Exosomes from human urine-derived stem cells carry NRF1 to alleviate bladder fibrosis via regulating miR-301b-3p/TGFβR1 pathway
Molecular and Cellular Biochemistry (2023)
-
Polymorphism and association study of lactoferrin (LF) gene with milk yield, milk composition, and somatic cell count in Beetal goats
Tropical Animal Health and Production (2023)
-
Genetic variants in the SIRT6 transcriptional regulatory region affect gene activity and carcass quality traits in indigenous Chinese beef cattle (Bos taurus)
BMC Genomics (2018)