Abstract
A wide variety of microbial and inflammatory factors induce DNA release from neutrophils as neutrophil extracellular traps (NETs). Consensus on the kinetics and mechanism of NET release has been hindered by the lack of distinctive methods to specifically quantify NET release in time. Here, we validate and refine a semi-automatic live imaging approach for quantification of NET release. Importantly, our approach is able to correct for neutrophil input and distinguishes NET release from neutrophil death by other means, aspects that are lacking in many NET quantification methods. Real time visualization shows that opsonized S. aureus rapidly induces cell death by toxins, while actual NET formation occurs after 90 minutes, similar to the kinetics of NET release by immune complexes and PMA. Inhibition of SYK, PI3K and mTORC2 attenuates NET release upon challenge with physiological stimuli but not with PMA. In contrast, neutrophils from chronic granulomatous disease patients show decreased NET release only in response to PMA. With this refined method, we conclude that NET release in primary human neutrophils is dependent on the SYK-PI3K-mTORC2 pathway and that PMA stimulation should be regarded as mechanistically distinct from NET formation induced by natural triggers.
Similar content being viewed by others
Introduction
The discovery of neutrophil extracellular traps (NETs) has enriched our knowledge on the anti-microbial strategies that neutrophils use to fight invading pathogens. NETs are fibres of decondensed chromatin decorated with granular proteins and are released in the extracellular milieu to kill various pathogens1,2,3,4. Opposite to the beneficial role of NETs to innate immune defence, NET formation has been shown to contribute to inflammation in non-infectious diseases. The formation of NETs in tissue or inside the vasculature could lead to clot forming, metastasis or exposure of autoantigens and thus contribute to the pathogenesis of thrombosis5, cancer6 and autoimmune inflammatory diseases7,8,9,10, respectively. In systemic lupus erythematosus (SLE), the presence of LL-37, human neutrophil peptide (HNP) and autoantibodies against these specific cellular components supports the formation of immune complexes (ICs), which trigger neutrophils to release NETs11. In addition, monosodium urate (MSU) crystals present in joints of gout patients have been described to abundantly induce NET release12.
Many of the current reports on NET release are based on in vitro studies using the non-physiological stimulus phorbol myristate acetate (PMA). PMA is a potent stimulator of protein kinase C (PKC), which in turn activates the nicotinamide adenine dinucleotide phosphate (NADPH)-oxidase complex leading to abundant generation of reactive oxygen species (ROS)13. Although PMA has been used to study the biology of NETs in many publications, it is not a physiological stimulus and thus will differ from NET release induced by microbial and endogenous stimuli.
To experimentally address the release of NETs in vitro, in recent years many strategies have been developed. Microscopic techniques are most often used to determine and quantify NETs (reviewed by14). Microscopy is useful to visualize NET release and to detect specific NET components, but has its limitations. Non-automatic quantification of fixed time point microscopic experiments is rather inaccurate, because the results could easily be biased by the observer and it is impossible to correct for neutrophil input. Another technique combines microscopy with flow cytometry, also called multispectral imaging flow cytometry (MIFC), and quantifies NET release by visualization and side-scatter and is able to identify NET components by multiple fluorescence staining15. MIFC allows unbiased quantitative analysis, however it focuses on neutrophils in an early phase of NET release and may miss neutrophils that actually release NETs.
An important limitation that all these visualization techniques have in common is that they lack the possibility to study NET kinetics, on which there is currently no consensus. Rapid NET release has been shown in response to Staphylococcus aureus (S. aureus)16, Fc receptor (FcR) activation17, 18, MSU19, Candida albicans (C. albicans) hyphea20, and Leishmania 21, while PMA18, ICs22 and C. albicans yeast20 trigger NET formation at later time points. Many of these studies were performed with fluorescence plate reader experiments using DNA binding dye’s (i.e. PicoGreen or Sytox Green), however this will not distinguish DNA expelled as NETs from other forms of DNA release.
Live imaging approaches were shown to enable following single neutrophils and visualize NET release, which makes it the technique of choice to study neutrophil morphology and kinetics23, 24. Recently, a novel semi-automated NET quantification has been described25. This technique is able to detect PMA-induced NETs based on the surface of Sytox Green staining. Because this procedure does not require extensive processing of cells, it has been shown to be reproducible and valid compared to manual counting. Here, we have used live imaging and a validated semi-automatic approach to quantify NET release in response to physiological stimuli in healthy donors (HDs) and in patients with chronic granulomatous disease (CGD). This allowed us to obtain novel information on the kinetics and underlying signalling pathways that result in NET release following non-inflammatory and inflammatory stimuli and it indicates that PMA is not a good model for any of those.
Results
A semi-automatic quantification approach to analyse live imaging NET release
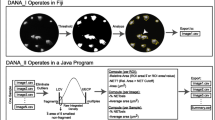
We performed a live imaging assay to monitor NET release over time that uses Hoechst stain at time point zero to control for neutrophil input and validated an earlier described analysis approach that specifically quantifies NET release25. To discriminate NETs from dying, Sytox Green permeable neutrophils (further referred as Sytox Green+ neutrophils), we assessed maximal surface of Sytox Green+ neutrophils and the minimal size of NETs. Using this approach we set thresholds for noise (35 µm2), Sytox Green+ neutrophils (35–68 µm2) and NETs (>68 µm2). The use of Fiji macros ensured a semi-automatic quantification approach (Fig. S1).
To quantify NETs, the Sytox Green images were transferred to binary images and the size of Sytox Green+ neutrophils as well as the NETs was determined. Sytox Green+ neutrophils typically covered a surface of between 40 and 55 µm2 while NETs covered a surface of over 68 µm2 (Fig. 1A). In addition, NET surface increased over time because of diffusion while the size of Sytox Green+ neutrophils remained below 68 µm2.
Surface-based analysing approach to quantify NET release. Neutrophils from HDs were stimulated with viable opsonized S. aureus and monitored over time for 4 hours using Hoechst 33342 and Sytox Green. (A) Sytox Green images from live imaging were transferred to binary images and the size of Sytox Green+ neutrophils (<53 µm2) and NETs (83–212 µm2) were measured. (B) Fluorescence microscopy using CellMask plasma membrane dye revealed that the DNA of Sytox Green+ neutrophils is intracellular and NETs are extracellular. (C) Citrullinated histone H3 is only present in Sytox Green particles with a surface above 68 µm2, which confirms that these are NETs. Data from 3 independent experiments are presented. Statistical significance (****P < 0.0001) was determined by two-tailed Mann-Whitney test. For quantification, the amount of neutrophils in t = 0 (D), Sytox Green+ neutrophils (blue), and NETs (red) in multiple time points (E) were determined. (F) Indicated time points of live imaging (Supplemental video 1; Hoechst 33342 in magenta and Sytox Green in green) showed the binding of Sytox Green to intracellular DNA (Sytox Green+ neutrophil) and extracellular DNA. For quantification, the amount of Sytox Green+ neutrophils or NETs were divided by the amount of neutrophils in t = 0 and expressed as percentage of Sytox Green+ neutrophils (G) or NETs (H). Data from 5 independent experiments are presented as median ± interquartile range. Statistical significance (**P < 0.01) was determined by two-tailed Mann-Whitney test. The images in A–F are representative of at least 3 experiments with neutrophils from different donors.
We used three separate approaches to confirm the differences between Sytox Green+ neutrophils and NETs. First, we counted all Sytox Green particles at time point 60 min and 240 min after stimulation with viable opsonized S. aureus. After 60 min, the majority of Sytox Green particles covered a surface between 0 and 40 µm2 but remained below 68 µm2. The surface of these Sytox Green particles increased (20–50 µm2) after 240 min of stimulation. Moreover, NETs with a surface above 68 µm2 appeared after 240 min of stimulation (Fig. S2). Second, with CellMask plasma membrane dye we secured that Sytox Green staining of Sytox Green+ neutrophils is intracellular while NETs are extracellular (Fig. 1B). Finally, citrullinated histone H3 was present only in the extracellular DNA while the Sytox Green+ neutrophil lacks citrullinated histone H3 (Fig. 1C). This confirms that the DNA was expelled in the form of NETs and that our NET surface-based threshold of >68 µm2 is valid to genuine NETs.
We determined the number of neutrophils over time and showed similar amount of neutrophils at time point zero (t = 0) and 240 min (Fig. S3). Subsequently, we used t = 0 for correcting neutrophil input. Hoechst images were transferred to binary images and the number of neutrophils was determined (Fig. 1D). Likewise, with Sytox Green images at multiple time points, the number of Sytox Green+ neutrophils (in blue) and NETs (in red) was determined (Fig. 1E).
Upon exposure to viable opsonized S. aureus, permeabilization of the neutrophil plasma membrane occurred within 30 to 60 min where after Sytox Green entered the neutrophils. After 90 min, the neutrophil plasma membrane of part of the cells broke and DNA was released in the extracellular environment, while part of the cells stayed Sytox Green positive but did not release NETs (Fig. 1F and Supplemental video 1).
Quantification demonstrated that within the first 60 min of exposure to viable opsonized S. aureus, neutrophils became Sytox Green+ with ~70% being positive after 240 min (Fig. 1G). The release of NETs occurred after 90 min and ~12% of cells released NETs after 240 min of stimulation with viable opsonized S. aureus (Fig. 1H). Taken together, these data show that we have validated a surface-based semi-automatic analysing approach which is able to specifically quantify physiologically-induced NET release over time and correct for cellular input.
Live imaging distinguishes NET release from toxin-induced cell death
Upon exposure to viable opsonized S. aureus, neutrophils either released NETs or only became permeable, becoming Sytox Green+ without expulsion of their DNA into extracellular space. Even after prolonged incubation, the Sytox Green+ neutrophils did not release NETs (Fig. S4 and Supplemental video 2), suggesting exposure to viable opsonized S. aureus results in two separate mechanisms of cell death. Since S. aureus toxins are known to induce cell death24, we exposed neutrophils to toxins obtained from an overnight culture of S. aureus and monitored neutrophil fate over time. After exposure with toxins, neutrophils became permeable and Sytox Green+ (Fig. 2A). The lack of citrullinated histone H3 confirms that toxin-induced Sytox Green+ neutrophils are not NETs (Fig. 2B).
Distinguishing toxin-induced cell death and apoptosis from NET release. Neutrophils from HDs were stimulated with anti-Fas antibody, toxins from overnight culture of S. aureus or viable and HK opsonized S. aureus and monitored over time for 4 hours. (A) Indicated time points from live imaging (Supplemental video 3) showed that neutrophils release NETs in response to HK opsonized S. aureus while stimulation with toxins alone or a combination of toxins and HK opsonized S. aureus mainly resulted in Sytox Green+ neutrophils. Anti-Fas antibody neither induced Sytox Green+ neutrophils nor NET release. (B) Fluorescence microscopy using CellMask plasma membrane dye and anti-citrullinated histone H3 antibody confirms the presence of extracellular DNA in the form of NETs. Quantification of Sytox Green+ neutrophils (C) and NETs (D) of 2 independent experiments performed with 4 donors. The data are presented as median ± interquartile range. Statistical significance (*P < 0.05; ***P < 0.001) was determined by Kruskal-Wallis test. The images in A and B are representative of at least 3 experiments with neutrophils from different donors.
Our time-lapse NET quantification revealed that the first Sytox Green+ cells appeared after 40 minutes and ~58% of the cells being Sytox Green+ after 240 min without the release of NETs (Fig. 2C,D). Interestingly, not a single neutrophil became Sytox Green+ without also releasing citrullinated histone H3 positive NETs after stimulation with heat-killed (HK) opsonized S. aureus, which cannot produce toxins (Fig. 2B,C,D and Supplemental video 3). To demonstrate the difference between HK and live opsonized S. aureus-induced NET release is because of toxin production, we supplemented HK opsonized S. aureus with toxins. The neutrophil response with HK opsonized S. aureus with exogenous toxins mimicked that of live opsonized S. aureus: neutrophils became Sytox Green+, but the majority of these cells did not produce NETs.
Thus, bacterial toxins are responsible for the permeabilization of the neutrophil plasma membrane after which Sytox Green enters the neutrophil. Our live imaging approach is able to distinguish NET release from toxin-induced cell death even when both processes are occurring in the same sample. Furthermore, upon stimulation with anti-Fas antibody to induce apoptosis, no Sytox Green+ neutrophils n or NETs were present (Fig. 2A,C,D). This shows that we have developed an assay and method of analysis that distinguishes NETs from other cell death mechanisms.
MSU induces rapid NET release
Also in non-infectious conditions, neutrophils can be triggered by host factors to release NETs. The exposure of neutrophils to MSU or ICs results in abundant NET release22, 26. PMA is a non-physiological stimulus often used for study of NETs in vitro 24. Upon stimulation with MSU, ICs or PMA, neutrophils released NETs (Fig. 3A). NET release in response to ICs and PMA started around 90 min while MSU-induced NET release occurred already within 60 min, starting as soon as 30 min (Fig. 3B). The presence of citrullinated histone H3 in the extracellular DNA of neutrophils stimulated with PMA and MSU confirmed that the DNA was expelled in the form of NETs (Fig. 3C). This shows that NET kinetics is dependent on the type of stimulus, with MSU inducing rapid NET release.
MSU induces rapid NET release. Neutrophils from HDs were stimulated with MSU, ICs and PMA and monitored over time for 4 hours. (A) Indicated time points from live imaging showed MSU-induced NET release within 60 min while NET release in response to ICs and PMA occurred within 120 min. (B) Quantification of NETs of 5 independent experiments. The data are presented as median ± interquartile range and statistical significance (*P < 0.05; **P < 0.01) was determined by Kruskal-Wallis test. (C) Fluorescence microscopic experiments revealed the presence of citrullinated histone H3 in the extracellular DNA after 180 min stimulation with MSU and PMA. The images in A and C are representative of at least 4 experiments with neutrophils from different donors.
PMA and physiological stimuli activate different pathways to induce NETs
To investigate the underlying pathway of NET release, we used chemical inhibitors to target specific downstream signalling molecules. Neutrophils were pre-treated with various inhibitors before exposure to live opsonized S. aureus, MSU, and PMA. We first determined the optimal concentration of the chemical inhibitors and chose the highest concentrations that did not induce Sytox Green signal (dying cells) in otherwise non-stimulated neutrophils (Fig. S5A).
The release of NETs in response to PMA was inhibited with 40–60% in the presence of DPI (NADPH oxidase inhibitor) and BAPTA-AM (chelator of Ca2+) while treatment of neutrophils with Wortmannin (PI3K inhibitor), R406 (SYK inhibitor), MLN0128 (pan-mTORC inhibitor), SB203580 and SB202190 (p38 MAPK inhibitors) did not affect NET release in response to PMA (Fig. 4A). To further study the role of NADPH oxidase in NET release, we used neutrophils from two male CGD patients (twins). These CGD patients contain a homozygous delta GT mutation in the NCF1 gene (gene encoding the p47phox protein; a component of NADPH oxidase), which leads to low levels of ROS production. PMA-induced NET release was abrogated in CGD neutrophils compared to neutrophils from a HD (Fig. 4B,C).
PI3K, SYK and mTORC2 are important players of the underlying signalling pathway of NET release. Neutrophils from HDs were incubated with Wortmannin (PI3K inhibitor), R406 (SYK inhibitor), SB203580 and SB202190 (p38 MAPK inhibitors), MLN0128 (pan-mTORC inhibitor), BAPTA-AM (Chelator of Ca2+), and DPI (NADPH oxidase inhibitor) before stimulation with MSU, PMA and viable opsonized S. aureus. (A) AUC of NET quantification of 3 independent experiments are presented as median ± interquartile range and statistical significance (*P < 0.05; **P < 0.01) was determined by Kruskal-Wallis test. Neutrophils from one HC and two CGD patients were stimulated with MSU, PMA and opsonized S. aureus and monitored over time for 4 hours. (B) Merge images (Hoechst 33342 in magenta and Sytox Green in green) of time point 240 min showed decreased NET release of CGD neutrophils in response to PMA compared to HC neutrophils. (C) Quantification of NET release of 1 experiment with 1 HC and 2 CGD donors.
NET release induced by MSU and viable opsonized S. aureus was reduced with 40–60% when neutrophils were treated with Wortmannin, R406 and MLN0128 (Fig. 4A). Treatment of neutrophils with Rapamycin (mTORC1 inhibitor) did not attenuate NET release in response to these physiological stimuli (Fig. S5B). In addition, Celastrol (NF-κB inhibitor), Bay-11-7082 (IκB phosphorylation inhibitor), GW5074 (cRaf1 kinase inhibitor), U-73122 (PLC inhibitor), Bafilomycin A (vacuolar-type H+ -ATPase inhibitor), JNKi (JNK inhibitor) or Mytoquinone (mitochondrial ROS inhibitor) did not affect NET release (Fig. S5C). This suggests the importance of the SYK-PI3K-mTORC2 signalling pathway in NET release triggered by MSU and viable opsonized S. aureus, since the pan-mTORC inhibitor attenuated NET release and the mTORC1 inhibitor did not.
To prove a distinct underlying signalling mechanism in PMA-induced NET release compared to NET formation induced by MSU and viable opsonized S. aureus, we again used DPI treated neutrophils or neutrophils from CGD patients. Upon stimulation with MSU and viable opsonized S. aureus, DPI-treated neutrophils as well as CGD neutrophils did release NETs (Fig. 4A,B). Quantification revealed that NET release of CGD neutrophils is comparable or even increased compared to NET release from a HD (Fig. 4C). Although we only used one HD in this experiment, the release of NETs in response to PMA, live opsonized S. aureus and MSU of this HD is comparable with other HDs from previous experiments. Together, our data indicate that calcium and NADPH oxidase-induced ROS play an important role in PMA-induced NET release. In contrast, NET release in response to MSU and viable opsonized S. aureus acts via the SYK-PI3K-mTORC2 pathway and is independent of NADPH oxidase-mediated ROS.
Discussion
NETs serve as an innate immune defence mechanism to protect the host against invading pathogens, while on the other hand uncontrolled NET release can contribute to disease development and -progression. To visualize and quantify NET release in vitro, many experimental techniques are described, including fixed time point microscopy27, live imaging23, 24, 28, 29, image-based flow cytometry15, fluorescence plate reader assay using DNA binding dye’s16, 30, 31 and ELISA based assays30. These techniques are useful for studying NET components, morphology and kinetics. However, it is important to be aware of the limitations of each of these techniques. In this study, we use a semi-automatic approach to quantify NET release in a live imaging assay, which is based on previous described methods25, 32. Our approach was able to correct for neutrophil input and distinguishes NET release from other mechanisms of cell death, crucial aspects that are lacking in many NET quantification methods.
A unique mechanism of NET release in response to S. aureus was previously described: rapid (within minutes) and without breaching the neutrophil plasma membrane16. Consistent with this study, we previously have shown that, when using a fluorescence plate reader assay, Sytox Green fluorescence appears within 30 min when neutrophils were exposed to viable S. aureus 16, 19. We concluded at the time that neutrophils are able to rapidly release NETs in response to these stimuli. However, S. aureus secrete pore-forming toxins that contribute to bacterial virulence and are able to permeabilize neutrophils for DNA dyes that are normally cell membrane impermeable33,34,35. We now show that cytolytic toxins from viable S. aureus induce rapid permeabilization of the neutrophil plasma membrane allowing for Sytox Green influx and nuclear DNA staining without histone H3 citrullination. In strong contrast, actual NET release occurs at a later time point (Supplemental video 2). Indeed, HK S. aureus that cannot secrete toxins, are not able to permeabilize neutrophils and only trigger NET release. Without imaging, i.e. with a fluorescence plate reader, it is not possible to discriminate between nuclear DNA and NETs. Thus, we conclude that imaging is essential for measuring NET release.
Visualization of NET release in a live imaging assay shows that expelled DNA from the neutrophil diffuses around the neutrophil itself23,24,25. In contrast, fixed time point microscopic experiments show DNA structures that seem to be pulled apart and cover a large surface25, 36. Similar structures of NETs were shown in vivo where blood flow and moving cells result in stretched NETs37,38,39. In vitro, this stretched morphology is induced by pipetting (Fig. S6). This shows that quantification of live imaging NET release is more accurate than quantification of NET release visualized with fixed time point microscopy.
Neutrophils rapidly kill pathogens by phagocytosis or by the secretion of proteases to protect the host against severe infection40, 41. NETs have also been shown to eliminate microbes1,2,3,4. So far, it is not known which signal triggers the neutrophil to release their nuclear content in the extracellular milieu. The release of NETs could act as a cell death mechanism in which the neutrophil sacrifices itself to protect others. This suggests that NET release is a late response, which is induced when other anti-microbial strategies have failed. In addition, neutrophils sense microbe size and release NETs in response to large pathogens42, arguing for an active process that occurs directly after neutrophil contact with the microbe. In contrast with fluorescence plate reader data from our previous study19, we here show that only MSU triggers rapid NET release while NET formation in response to opsonized S. aureus, PMA, and ICs occurs at a later phase. Others have also shown that PMA18 and ICs22 induce late NET release. Although these experiments were performed with a fluorescence plate reader assay and therefore unable to discriminate between bona fide NETs and permeable cells, we now confirm late phase NET release in response to PMA and ICs. Similarly, fluorescence plate reader experiments revealed that FcR activation17, 18, Leishmania 21, and C. albicans hyphea20 induce rapid NET release while C. albicans yeast20 triggers NET formation at a later time point. Our current data underline that these kinetics should be interpreted carefully and analysed in an assay like ours, to determine the difference between cellular permeability and actual NET release.
There is no consensus of the requirement of ROS production for NET formation and current data on NET release suggests there are two differential signalling pathways that induce the release of NET. Direct stimulation of PKC, i.e. by PMA, provokes ROS-dependent NET formation, which diminishes in the presence of DPI. Similarly, DPI was published to attenuate NET release upon stimulation with ICs22, 43, anti-LL3744 and non-opsonized S. aureus 24. Indeed, CGD neutrophils do not release NETs upon PMA23, 24, 45,46,47 or non-opsonized S. aureus 24 stimulation. This demonstrates that some stimuli require NADPH-induced ROS to release NETs. On the contrary, we and others showed that DPI does not affect NET release when neutrophils are stimulated with MSU19, ICs48, uric acid49, Leishmania 21, ionomycin50 and opsonized S. aureus 16, 19. In addition, it was shown that CGD neutrophils are still able to release NETs when exposed to uric acid49, which suggests the presence of a NADPH-oxidase independent NET signalling pathway. We now show that CGD neutrophils as well as DPI-treated neutrophils release NETs upon exposure of MSU and viable opsonized S. aureus while PMA-induced NET release is low. So far, data obtained with neutrophils from CGD patients confirm that the role of NADPH oxidase-dependent ROS in NET release is stimulus dependent. However, the discrepancies about the role of ROS in NET release in studies that use DPI are challenging to interpret. We propose two distinct explanations: 1) The effect of DPI is concentration dependent and/or non-specific or; 2) inappropriate NET measuring techniques and quantification methods that lack experimental sensitivity are masking the true effect of DPI on NET release. Thus, the use of a live imaging assay that allows visualization and quantification of NET release and discriminates between actual NETs and other cell death mechanisms is essential for future NET studies.
A wide variety of cell surface receptors on neutrophils act via SYK and PI3K to induce neutrophil activation51, 52. FcRs, Toll-like receptors (TLRs) and the P2Y6 receptor are amongst them and could be triggered upon exposure with ICs, opsonized S. aureus and MSU53, respectively. We show that the SYK-PI3K-mTORC2 pathway is involved in NET release upon physiological stimuli but not in PMA-induced NET release, which is consistent with previous data22, 54. In addition, PMA provokes NET release that is morphologically different from other NETs (Supplemental video 4). This indicates that PMA-induced NETs should be regarded as mechanistically distinct from NET formation induced by natural triggers. Therefore we recommend to interpret statements from NET studies performed with PMA with care.
This study highlights the potency of a live imaging technique to study NET release. Using a validated semi-automatic quantification approach, we are able to correct for neutrophil input and distinguish between NETs and other cell death mechanisms, two aspects that increase the accuracy of NET quantification. At the same time, we reveal the limitations of fluorescence plate reader assays using cell-impermeable DNA binding dyes for measurement of NET release. The use of PMA to induce NET release should be avoided or interpreted with great care because it induces NET release completely different from all physiological stimuli.
Materials and Methods
Isolation of human neutrophils
Peripheral blood from HDs and from two CGD patients (twin boys) was collected in sodium-heparin tubes (Greiner Bio-One). All blood donors gave informed consent. Patients with CGD were recruited in the outpatient clinic of the department of Paediatric Immunology and Infectious Diseases of the UMC Utrecht. Diagnosis of CGD was determined functionally by absence of respiratory burst upon stimulation in neutrophil (Phagoburst, BD Bioscience) and genetically by Sanger sequencing of the NCF1 gene showing a pathogenic homozygous mutation (delta GT). Neutrophils were isolated by Ficoll-Paque (GE Healthcare) density gradient centrifugation, after which erythrocytes were lysed in ammonium chloride buffer (155 mM NH4Cl; 10 mM KHCO3; 0.1 mM EDTA in double-distilled H2O; pH = 7.2). Cells were resuspended in RPMI 1640 (Life Technologies) supplemented with 10% (v/v) heat-inactivated (HI) foetal bovine serum (FBS) (Biowest) and 50 U/ml Penicillin-Streptomycin (referred to as RPMI 10% hereafter). Purity of isolated neutrophils was analysed using the CELL-DYN Emerald and was >88%. All experiments were performed in accordance with relevant guidelines and regulations approved by the Medical Ethical Committee of the University Medical Centre Utrecht.
Bacterial culture and preparation
S. aureus Wood 46 was grown up to exponential phase at 37 °C under aerobic conditions in Todd-Hewitt Broth (THB) containing 1% (v/w) yeast extract. After quantification of bacteria by measuring OD600 (2 × 108 CFU with OD600 = 0.4), bacteria were washed twice with chilled PBS and opsonized for 30 min at 37 °C with 10% HI human pooled serum in PBS. In some experiments, bacteria were HK for 60 min at 70 °C before opsonisation.
Chemical inhibitors
To study the underlying signalling NET pathway, several chemical inhibitors were used. Before stimulation, neutrophils were pre-treated for 30 min at 37 °C in RPMI 10% with DMSO (vehicle control), 1 µM Wortmannin (PI3K inhibitor; Sigma-Aldrich), 1 µM R406 (SYK inhibitor; Selleckchem), 10 µM SB203580 (p38 MAPK inhibitor; Selleckchem), 10 µM SB202190 (p38 MAPK inhibitor; Santa Cruz Biotechnology) 10 µM MLN0128 (pan-mTORC inhibitor; Selleckchem), 10 µM BAPTA-AM (chelator of Ca2+;Focus Biomolecules), 1 µM DPI (NADPH oxidase inhibitor; Sigma-Aldrich), 1 µM Rapamycin (mTORC1 inhibitor; Selleckchem), 1 µM Celastrol (NF-κB inhibitor; InvivoGen), 0.1 µM Bay-11-7082 (IκB phosphorylation inhibitor; InvivoGen), 0.1 µM GW5074 (cRaf1 kinase inhibitor; Santa Cruz Biotechnology), 0.1 µM U-73122 hydrate (PLC inhibitor; Sigma-Aldrich), 1 nM Bafilomycin A (vacuolar-type H+ -ATPase inhibitor; Sigma-Aldrich), 0.1 µM JNKi II 420128 (JNK inhibitor; Merck Millipore) or 0.1 µM Mytoquinone (mitochondrial ROS inhibitor; BIOTREND).
Preparation of ICs
Insoluble ICs were formed by using human serum albumin (HSA) (Sanquin) and rabbit polyclonal anti-HSA IgG (Sigma Aldrich) as described previously55. Briefly, a mix of 5 µg HSA and 45 µg rabbit-anti-HSA antibody was made in a final volume of 50 µl PBS. After 1 h incubation at 37 °C, insoluble ICs were formed and used for stimulation of neutrophils.
Microscopic live imaging NET assay
Neutrophils were incubated in RPMI 10% containing 20 μM Hoechst 33342 for 30 min at 37 °C, in the presence or absence of chemical inhibitors, and washed twice with RPMI 1640 (without phenol red) supplemented with 2% FBS, 50 U/ml Penicillin-Streptomycin, and 10 mM HEPES (referred to as RPMI-pr 2% hereafter). A total of 5 × 104 neutrophils were seeded on 0.001% poly-L-lysine (Sigma Aldrich) pre-coated wells of a clear bottom 96-wells plate (Ibidi) and challenged with 25 ng/ml PMA (Sigma Aldrich), 100 μg/ml MSU, 10 µg/ml ICs, opsonized S. aureus at a MOI of 10. All stimuli were resuspended in RPMI-pr 2% containing 4 nM Sytox Green (Life Technologies). NET release was recorded at 37 °C and 5% CO2 on the Pathway 855 bioimaging system (BD Biosciences) with a 10x objective during a period of 240 min. Every 30 min, a set of two images (Exc/Em: 350/461 nm (Hoechst) and 504/523 nm (Sytox Green)) was taken with an Orca high-resolution CCD camera and four fields of view (each 415 × 317 μm) per condition were captured. The system was controlled by the AttoVision software (version 1.7/855). To make movies, images were taken every 2 min.
Semi-automatic NET quantification approach
This NET quantification approach is based previously described methods25, 32. For quantification of NETs, images were processed with Fiji software (version 2.0.0-rc-43/1.51d). Image stacks were created and converted in 8-bit greyscale. To count neutrophils, thresholding of t = 0 Hoechst image was done with “Moments” logarithm followed by watershed segmentation to separate particles that touch. To count NETs, thresholding of Sytox Green stacks were performed with “Li” logarithm followed by watershed segmentation. The amount of Sytox Green+ neutrophils (35–68 μm2) or NETs (>68 μm2) were divided by the total number of neutrophils in t = 0 to calculated percentages. For semi-automatic quantification analysis, three macros were created for: 1) making stacks, 2) counting neutrophils, and 3) counting Sytox Green+ neutrophils and NETs (Fig. S1).
Induction of neutrophil apoptosis and toxin-mediated cell death
Neutrophil apoptosis and toxin-mediated cell death was induced as described before24. Briefly, neutrophils were incubated with 20 ng/ml anti-human CD95 (Fas) antibody clone EOS9.1 (BioLegend) to induce apoptosis and 25 µg/ml toxins from S. aureus to induce cell death. Toxins were obtained by growing S. aureus Wood 46 overnight in THB containing 1% yeast. Bacteria were pelleted, supernatant was collected and sterilized with a 0.20 µm filter. Protein concentration was determined with BCA Protein Assay (Thermo Fisher Scientific).
Immunofluorescence staining of citrullinated histone H3
Neutrophils were resuspended in RPMI-pr 2% and a total of 5 × 105 neutrophils were seeded on 0.001% poly-L-lysine coated glass coverslips. Neutrophils were challenged as described before for 180 min at 37 °C and 5% CO2. Medium was gently discarded and neutrophils were fixed with 2% paraformaldehyde (Electron Microscopy Sciences) for 15 min at room temperature (RT). After blocking with 20% normal goat serum (Cell Signaling Technologies) in PBS containing 2% bovine serum albumin (BSA) (Roche) for 30 min at RT, 0.25 µg/ml rabbit-anti-citrullinated histone H3 antibody (Abcam) in PBS/2% BSA was used for incubation overnight at 4 °C. Next day, neutrophils were incubated with 2.5 µg/ml goat-anti-rabbit antibody conjugated with Alexa Fluor 594 (Life Technologies) for 120 min at RT, followed by 30 min of incubation at RT with 5 µM Hoechst 33342 in PBS to stain neutrophil nucleus. Coverslips were mounted in Fluoromount-G (Southern Biotech) and analysed with Pathway 855 bioimaging system with a 20x objective.
For immunofluorescence staining of citrullinated histone H3 in the live imaging NET assay, neutrophils were incubated in RPMI-pr 2% containing Hoechst 33342 and 1x CellMaskTM Deep Red Plasma membrane stain (Thermo Fisher Scientific) for 30 min at 37 °C. After washing with RPMI-pr 2%, neutrophils were seeded and challenged with toxins, viable and HK opsonized S. aureus. All stimuli were resuspended in RPMI-pr 2% containing Sytox Green. After 180 min incubation at 37 °C and 5% CO2, 2 µg/ml rabbit-anti-citrullinated histone H3 antibody was added and incubated for 30 min. A mix of 0.67 µg/ml donkey-anti-rabbit (Life Technologies) and 0.67 µg/ml goat-anti-rabbit antibody, both conjugated with Alexa Fluor 594, was added and incubated for 15 min before NET release was recorded with a 40x objective. Sets of four images (Hoechst, Sytox Green, Exc/Em: 590/617 nm (AF594) and 649/666 nm (CellMask Deep Red)) were taken.
Statistical analysis
Data were analysed in GraphPad Prism 6. The comparison of two samples was performed by two-tailed Mann-Whitney test and multiple comparisons were established by Kruskal-Wallis test. Each experiment was performed at least three times on independent occasions. P < 0.05 was considered significant.
References
Brinkmann, V. et al. Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535, doi:10.1126/science.1092385 (2004).
Urban, C. F., Reichard, U., Brinkmann, V. & Zychlinsky, A. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol 8, 668–676, doi:10.1111/j.1462-5822.2005.00659.x (2006).
Guimaraes-Costa, A. B. et al. Leishmania amazonensis promastigotes induce and are killed by neutrophil extracellular traps. Proc Natl Acad Sci USA 106, 6748–6753, doi:10.1073/pnas.0900226106 (2009).
Saitoh, T. et al. Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe 12, 109–116, doi:10.1016/j.chom.2012.05.015 (2012).
Fuchs, T. A. et al. Extracellular DNA traps promote thrombosis. Proc Natl Acad Sci USA 107, 15880–15885, doi:10.1073/pnas.1005743107 (2010).
Cools-Lartigue, J. et al. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J Clin Invest. doi:10.1172/JCI67484 (2013).
Hakkim, A. et al. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci USA 107, 9813–9818, doi:10.1073/pnas.0909927107 (2010).
Kessenbrock, K. et al. Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med 15, 623–625, doi:10.1038/nm.1959 (2009).
Khandpur, R. et al. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med 5, 178ra140, doi:10.1126/scitranslmed.3005580 (2013).
Mitroulis, I. et al. Neutrophil extracellular trap formation is associated with IL-1beta and autophagy-related signaling in gout. PLoS One 6, e29318, doi:10.1371/journal.pone.0029318 (2011).
Lande, R. et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med 3, 73ra19, doi:10.1126/scitranslmed.3001180 (2011).
Schauer, C. et al. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat Med 20, 511–517, doi:10.1038/nm.3547 (2014).
Saito, T., Takahashi, H., Doken, H., Koyama, H. & Aratani, Y. Phorbol myristate acetate induces neutrophil death through activation of p38 mitogen-activated protein kinase that requires endogenous reactive oxygen species other than HOCl. Biosci Biotechnol Biochem 69, 2207–2212, doi:10.1271/bbb.69.2207 (2005).
de Buhr, N. & von Kockritz-Blickwede, M. How Neutrophil Extracellular Traps Become Visible. J Immunol Res 2016, 4604713, doi:10.1155/2016/4604713 (2016).
Zhao, W., Fogg, D. K. & Kaplan, M. J. A novel image-based quantitative method for the characterization of NETosis. J Immunol Methods 423, 104–110, doi:10.1016/j.jim.2015.04.027 (2015).
Pilsczek, F. H. et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol 185, 7413–7425, doi:10.4049/jimmunol.1000675 (2010).
Aleyd, E. et al. IgA enhances NETosis and release of neutrophil extracellular traps by polymorphonuclear cells via Fcalpha receptor I. J Immunol 192, 2374–2383, doi:10.4049/jimmunol.1300261 (2014).
Aleman, O. R., Mora, N., Cortes-Vieyra, R., Uribe-Querol, E. & Rosales, C. Transforming Growth Factor-beta-Activated Kinase 1 Is Required for Human FcgammaRIIIb-Induced Neutrophil Extracellular Trap Formation. Front Immunol 7, 277, doi:10.3389/fimmu.2016.00277 (2016).
Van Avondt, K., van der Linden, M., Naccache, P. H., Egan, D. A. & Meyaard, L. Signal Inhibitory Receptor on Leukocytes-1 Limits the Formation of Neutrophil Extracellular Traps, but Preserves Intracellular Bacterial Killing. J Immunol 196, 3686–3694, doi:10.4049/jimmunol.1501650 (2016).
Kenno, S., Perito, S., Mosci, P., Vecchiarelli, A. & Monari, C. Autophagy and Reactive Oxygen Species Are Involved in Neutrophil Extracellular Traps Release Induced by C. albicans Morphotypes. Front Microbiol 7, 879, doi:10.3389/fmicb.2016.00879 (2016).
Rochael, N. C. et al. Classical ROS-dependent and early/rapid ROS-independent release of Neutrophil Extracellular Traps triggered by Leishmania parasites. Sci Rep 5, 18302, doi:10.1038/srep18302 (2015).
Behnen, M. et al. Immobilized immune complexes induce neutrophil extracellular trap release by human neutrophil granulocytes via FcgammaRIIIB and Mac-1. J Immunol 193, 1954–1965, doi:10.4049/jimmunol.1400478 (2014).
Remijsen, Q. et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res 21, 290–304, doi:10.1038/cr.2010.150 (2011).
Fuchs, T. A. et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol 176, 231–241, doi:10.1083/jcb.200606027 (2007).
Hoffmann, J. H., Schaekel, K., Gaiser, M. R., Enk, A. H. & Hadaschik, E. N. Interindividual variation of NETosis in healthy donors: introduction and application of a refined method for extracellular trap quantification. Exp Dermatol 25, 895–900, doi:10.1111/exd.13125 (2016).
Schorn, C. et al. Monosodium urate crystals induce extracellular DNA traps in neutrophils, eosinophils, and basophils but not in mononuclear cells. Front Immunol 3, 277, doi:10.3389/fimmu.2012.00277 (2012).
Brinkmann, V., Laube, B., Abu Abed, U., Goosmann, C. & Zychlinsky, A. Neutrophil extracellular traps: how to generate and visualize them. J Vis Exp. doi:10.3791/1724 (2010).
Bruns, S. et al. Production of extracellular traps against Aspergillus fumigatus in vitro and in infected lung tissue is dependent on invading neutrophils and influenced by hydrophobin RodA. PLoS Pathog 6, e1000873, doi:10.1371/journal.ppat.1000873 (2010).
Brinkmann, V. & Zychlinsky, A. Neutrophil extracellular traps: is immunity the second function of chromatin? J Cell Biol 198, 773–783, doi:10.1083/jcb.201203170 (2012).
Sil, P., Yoo, D. G., Floyd, M., Gingerich, A. & Rada, B. High Throughput Measurement of Extracellular DNA Release and Quantitative NET Formation in Human Neutrophils In Vitro. J Vis Exp. doi:10.3791/52779 (2016).
Seper, A. et al. Vibrio cholerae evades neutrophil extracellular traps by the activity of two extracellular nucleases. PLoS Pathog 9, e1003614, doi:10.1371/journal.ppat.1003614 (2013).
Brinkmann, V., Goosmann, C., Kuhn, L. I. & Zychlinsky, A. Automatic quantification of in vitro NET formation. Front Immunol 3, 413, doi:10.3389/fimmu.2012.00413 (2012).
Wang, R. et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13, 1510–1514, doi:10.1038/nm1656 (2007).
Konig, B., Prevost, G. & Konig, W. Composition of staphylococcal bi-component toxins determines pathophysiological reactions. J Med Microbiol 46, 479–485, doi:10.1099/00222615-46-6-479 (1997).
Kobayashi, S. D. et al. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. J Innate Immun 2, 560–575, doi:10.1159/000317134 (2010).
Parker, H., Dragunow, M., Hampton, M. B., Kettle, A. J. & Winterbourn, C. C. Requirements for NADPH oxidase and myeloperoxidase in neutrophil extracellular trap formation differ depending on the stimulus. J Leukoc Biol 92, 841–849, doi:10.1189/jlb.1211601 (2012).
Yipp, B. G. et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med 18, 1386–1393, doi:10.1038/nm.2847 (2012).
McDonald, B., Urrutia, R., Yipp, B. G., Jenne, C. N. & Kubes, P. Intravascular neutrophil extracellular traps capture bacteria from the bloodstream during sepsis. Cell Host Microbe 12, 324–333, doi:10.1016/j.chom.2012.06.011 (2012).
Kolaczkowska, E. et al. Molecular mechanisms of NET formation and degradation revealed by intravital imaging in the liver vasculature. Nat Commun 6, 6673, doi:10.1038/ncomms7673 (2015).
DeLeo, F. R., Allen, L. A., Apicella, M. & Nauseef, W. M. NADPH oxidase activation and assembly during phagocytosis. J Immunol 163, 6732–6740 (1999).
Hirsch, J. G. & Cohn, Z. A. Degranulation of polymorphonuclear leucocytes following phagocytosis of microorganisms. J Exp Med 112, 1005–1014 (1960).
Branzk, N. et al. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol 15, 1017–1025, doi:10.1038/ni.2987 (2014).
Lood, C. et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med 22, 146–153, doi:10.1038/nm.4027 (2016).
Van Avondt, K., Fritsch-Stork, R., Derksen, R. H. & Meyaard, L. Ligation of signal inhibitory receptor on leukocytes-1 suppresses the release of neutrophil extracellular traps in systemic lupus erythematosus. PLoS One 8, e78459, doi:10.1371/journal.pone.0078459 (2013).
Dreyer, A. K. et al. TALEN-mediated functional correction of X-linked chronic granulomatous disease in patient-derived induced pluripotent stem cells. Biomaterials 69, 191–200, doi:10.1016/j.biomaterials.2015.07.057 (2015).
Nishinaka, Y., Arai, T., Adachi, S., Takaori-Kondo, A. & Yamashita, K. Singlet oxygen is essential for neutrophil extracellular trap formation. Biochem Biophys Res Commun 413, 75–79, doi:10.1016/j.bbrc.2011.08.052 (2011).
Bianchi, M. et al. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood 114, 2619–2622, doi:10.1182/blood-2009-05-221606 (2009).
Kraaij, T. et al. A novel method for high-throughput detection and quantification of neutrophil extracellular traps reveals ROS-independent NET release with immune complexes. Autoimmun Rev 15, 577–584, doi:10.1016/j.autrev.2016.02.018 (2016).
Arai, Y. et al. Uric acid induces NADPH oxidase-independent neutrophil extracellular trap formation. Biochem Biophys Res Commun 443, 556–561, doi:10.1016/j.bbrc.2013.12.007 (2014).
Douda, D. N., Khan, M. A., Grasemann, H. & Palaniyar, N. SK3 channel and mitochondrial ROS mediate NADPH oxidase-independent NETosis induced by calcium influx. Proc Natl Acad Sci USA 112, 2817–2822, doi:10.1073/pnas.1414055112 (2015).
Futosi, K. & Mocsai, A. Tyrosine kinase signaling pathways in neutrophils. Immunol Rev 273, 121–139, doi:10.1111/imr.12455 (2016).
Hawkins, P. T., Stephens, L. R., Suire, S. & Wilson, M. PI3K signaling in neutrophils. Curr Top Microbiol Immunol 346, 183–202, doi:10.1007/82_2010_40 (2010).
Uratsuji, H. et al. P2Y6 receptor signaling pathway mediates inflammatory responses induced by monosodium urate crystals. J Immunol 188, 436–444, doi:10.4049/jimmunol.1003746 (2012).
DeSouza-Vieira, T. et al. Neutrophil extracellular traps release induced by Leishmania: role of PI3Kgamma, ERK, PI3Ksigma, PKC, and [Ca2+]. J Leukoc Biol 100, 801–810, doi:10.1189/jlb.4A0615-261RR (2016).
Crockett-Torabi, E. & Fantone, J. C. Soluble and insoluble immune complexes activate human neutrophil NADPH oxidase by distinct Fc gamma receptor-specific mechanisms. J Immunol 145, 3026–3032 (1990).
Acknowledgements
This work was supported by the Dutch Arthritis Foundation (Grant 12-2-406). M.V. is supported by the Netherlands Organization for Scientific Research (NWO), division Earth and Life Sciences (ALW; Grant 863.14.016). L.M. is supported by a Vici grant (91815608) from Netherlands Organization for Scientific Research (NWO). The authors thank Florianne Hafkamp and Casper Schouls for the development of the ImageJ macros. The authors thank Dr. Nina van Sorge (Medical Microbiology, University Medical Centre Utrecht, Utrecht, the Netherlands) and Dr. Paul Naccache (Department of Microbiology-Infection and Immunology, Faculty of Medicine, Laval University, Quebec, Canada) for providing bacterial strains and MSU, respectively. The authors thank Tineke Kraaij, Dr. Onno Teng and Prof. Dr. Cees van Kooten (Department of Nephrology, Leiden University Medical Centre, Leiden, the Netherlands) for helpful discussions and sharing expertise. The authors thank Julia Drylewicz for performing statistical analysis (Laboratory of Translational Immunology, Department of Immunology, University Medical Centre Utrecht, Utrecht, The Netherlands).
Author information
Authors and Affiliations
Contributions
M.L. and L.M. conceived and designed the experiments. M.L. and M.W. performed the experiments and analysed the data. M.V. brought useful input to the study and critically read the manuscript. J.M. collected patient samples and critically read the manuscript. L.M. supervised the research and revised and edited the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van der Linden, M., Westerlaken, G.H.A., van der Vlist, M. et al. Differential Signalling and Kinetics of Neutrophil Extracellular Trap Release Revealed by Quantitative Live Imaging. Sci Rep 7, 6529 (2017). https://doi.org/10.1038/s41598-017-06901-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-06901-w
- Springer Nature Limited
This article is cited by
-
Vascular smooth muscle cells in response to cholesterol crystals modulates inflammatory cytokines release and promotes neutrophil extracellular trap formation
Molecular Medicine (2024)
-
Gap junction-mediated contraction of myoepithelial cells induces the peristaltic transport of sweat in human eccrine glands
Communications Biology (2023)
-
Neutrophil metabolomics in severe COVID-19 reveal GAPDH as a suppressor of neutrophil extracellular trap formation
Nature Communications (2023)
-
Aging hampers neutrophil extracellular traps (NETs) efficacy
Aging Clinical and Experimental Research (2022)
-
C-type lectins and extracellular vesicles in virus-induced NETosis
Journal of Biomedical Science (2021)