Abstract
Dark-operative protochlorophyllide oxidoreductase (DPOR) is a key enzyme to produce chlorophyll in the dark. Among photosynthetic eukaryotes, all three subunits chlL, chlN, and chlB are encoded by plastid genomes. In some gymnosperms, two codons of chlB mRNA are changed by RNA editing to codons encoding evolutionarily conserved amino acid residues. However, the effect of these substitutions on DPOR activity remains unknown. We first prepared cyanobacterial ChlB variants with amino acid substitution(s) to mimic ChlB translated from pre-edited mRNA. Their activities were evaluated by measuring chlorophyll content of dark-grown transformants of a chlB-lacking mutant of the cyanobacterium Leptolyngbya boryana that was complemented with pre-edited mimic chlB variants. The chlorophyll content of the transformant cells expressing the ChlB variant from the fully pre-edited mRNA was only one-fourth of the control cells. Co-purification experiments of ChlB with Strep-ChlN suggested that a stable complex with ChlN is greatly impaired in the substituted ChlB variant. We then confirmed that RNA editing efficiency was markedly greater in the dark than in the light in cotyledons of the black pine Pinus thunbergii. These results indicate that RNA editing on chlB mRNA is important to maintain appropriate DPOR activity in black pine chloroplasts.
Similar content being viewed by others
Introduction
Chlorophyll a (Chl) is an essential tetrapyrrole pigment in photosynthesis that is synthesized from glutamate through a complex pathway consisting of at least 15 enzymes1,2,3. Intermediates of Chl produce reactive oxygen species upon exposure to light. Consequently, Chl biosynthesis is tightly regulated in response to a variety of environmental factors, including light and redox states4. In angiosperms, light is used for photosynthesis and in addition is also a major environmental signal that regulates Chl biosynthesis. In these plants, light is required for reduction of protochlorophyllide (Pchlide) to chlorophyllide a by the enzyme light-dependent Pchlide oxidoreductase (LPOR). LPOR is the sole Pchlide reductase in angiosperms so seedlings grown in the dark are etiolated (lack Chl). Many angiosperms studies on Chl biosynthesis have focused on LPOR, leading to the discovery of the regulatory protein FLU5. Photosynthetic organisms that have a longer evolutionary history, such as gymnosperms, also have an alternative Pchlide reduction system called dark-operative Pchlide oxidoreductase (DPOR) that functions irrespective of light6,7,8,9. Most gymnosperm seedlings have both LPOR and DPOR allowing these plants to produce Chl even in the dark (Fig. 1A)10, 11.
Protochlorophyllide reduction (A). Protochlorophyllide (Pchlide) is reduced by two structurally unrelated enzymes; LPOR and DPOR. DPOR is a nitrogenase-like oxygen-labile enzyme consisting of the three subunits ChlL, ChlN, and ChlB and serves as a key enzyme for Chl biosynthesis in the dark. (B) Partial amino acid sequences of chloroplast DNA encoded ChlB from P. thunbergii (Pth), L. decidua (Lde), P. patens (Ppa), the liverwort Marchantia polymorpha (Mpo), the green alga Chlamydomonas reinhardtii (Cre), Thermosynechococcus elongatus (Tel), Leptolyngbya boryana (Lbo), Synechocystis sp. PCC 6803 (6803), Prochlorococcus marinus SS120 (Pma), and the purple bacterium Rhodobacter capsulatus (Rca). RNA editing derived from amino acid substitutions is shown in bold, and the two amino acid residues from the pre-edited transcript in P. thunbergii and L. decidua are shown in red. The numbers 206 and 213 are from P. thunbergii ChlB.
Even though DPOR and LPOR catalyze the same stereospecific reduction of the C17=C18 double bond of Pchlide, these enzymes are structurally and evolutionarily unrelated9. LPOR is a nuclear-encoded protein that belongs to the short-chain dehydrogenase/reductase (SDR) superfamily, which consists of single polypeptide enzymes that use NADPH as a cofactor to catalyze a variety of redox reactions12. In contrast, DPOR is a nitrogenase-like enzyme encoded by three genes, chlL, chlN and chlB. The three gene products form two separable components, L protein (2ChlL dimer) and NB protein (2ChlN–2ChlB heterotetramer)8. The L protein transfers electrons to the NB protein coupled with ATP hydrolysis13, and the NB protein plays a role as the catalytic component of DPOR. The NB protein binds Pchlide, and the C17=C18 double bond of Pchlide is reduced by electrons transferred from the NB cluster (a [4Fe-4S] cluster). The NB cluster held by three Cys residues from ChlN and one Asp residue from ChlB is placed at the interface between ChlN and ChlB14. Thus, the complex formation between ChlN and ChlB is crucial for the catalytic activity of DPOR.
The por gene encoding LPOR is distributed throughout oxygenic photosynthetic organisms, including cyanobacteria. The three genes, chlL, chlN and chlB, for DPOR are present in photosynthetic bacteria (denoted as bchL, bchN and bchB, respectively), cyanobacteria, green algae, bryophytes, pteridophytes, and gymnosperms. In photosynthetic eukaryotes, the three genes are encoded in chloroplast genomes. However, the three genes coding for subunits of DPOR are absent in angiosperms and various eukaryotic algae15. Nonetheless, the physiological significance of the presence of both LPOR and DPOR in various photosynthetic organisms remains largely unknown, although light and oxygen appear to be key factors in their functional differentiation in cyanobacteria16 and it has been reported that DPOR is required for Chl production under short-day conditions in the liverwort Marchantia polymorpha 17.
RNA editing is a post-transcriptional modification that introduces changes in RNA sequences18. RNA editing in plant mitochondria and plastids modifies various codons to encode different amino acid residues as well as a few cases where initiation and stop codons are generated19, 20. In the black pine Pinus thunbergii, chlN and chlB in the chloroplast genome have been reported to have one and two RNA editing sites, respectively11, 21, 22. The site in chlN undergoes the conversion from the codon CCU (Pro285) to UCU (Ser), and the two sites in chlB undergo the conversion from codons CCG (Pro206) and CGG (Arg213) to CUG (Leu) and UGG (Trp), respectively. These three amino acids in ChlN and ChlB are highly conserved among many oxygenic phototrophs23 (Fig. 1B). The two C-to-U changes in chlB have also been found in the larch Larix decidua by Demko et al.11 who showed that the RNA editing efficiency of chlB mRNA correlates with the accumulation of Chl in the dark suggesting that RNA editing in chlB affects Chl biosynthesis in these gymnosperms. However, no direct causal links among these amino acid substitutions and DPOR activity has been identified, in part, because DPOR is extremely sensitive to oxygen and can be assayed only under anaerobic conditions in vitro 16, 24, 25.
To evaluate DPOR activity in vivo, we developed a complementation system in the cyanobacterium Leptolyngbya boryana 24, 25. In this system, a cyanobacterial chlB mutant, which lacks the ability to produce Chl in the dark, is used to express an exogenous chlB query gene. DPOR activity of the query gene is then evaluated by measuring the Chl content in transformant cells grown in the dark. In a previous study, we used this system to successfully evaluate chloroplast DNA-encoded DPOR activity from the moss Physcomitrella patens 24. In the present study, we focused on the activity of DPOR using cyanobacterial ChlB subunit variants carrying amino acid substitutions that correspond to those of pre-edited chlB mRNA in P. thunbergii. In addition, we analyzed changes in the RNA editing efficiency in chlB mRNA in response to light conditions in seedlings of P. thunbergii. These results suggest that RNA editing in chlB serves as a regulatory system for DPOR activity in response to environmental light conditions in black pine chloroplasts.
Results
Pre-edited mimic variants of cyanobacterial ChlB
Comparisons of ChlB amino acid sequences among various photosynthetic organisms that lack RNA editing processes indicated that the two amino acid residues in black pine ChlB that are generated by RNA editing (Leu206 and Trp213) are highly conserved among plants and cyanobacteria (Fig. 1B). The only significant divergence occurs at the second site that diverges in a group of marine cyanobacteria (Asp) and in proteobacteria (His) (Fig. 1B, Pma and Rca)23. To investigate whether these two amino acid residues are functionally important for DPOR activity, and to evaluate the activity of the edited and pre-edited forms of ChlB, we tried to express chlB together with chlN encoding the other subunit of the NB protein of DPOR from P. thunbergii in Escherichia coli. However, all portions of the expressed protein were recovered in the insoluble fraction of E. coli. We also tried to evaluate the activity of edited and pre-edited ChlB proteins in vivo using the in vivo complementation system of the cyanobacterium L. boryana 24, 25. However, Chl biosynthesis in the dark was not restored even with the edited ChlB from P. thunbergii, owing to very low expression of the black pine ChlN and ChlB proteins and probable incompatibility between the cyanobacterial L protein and the black pine NB protein.
Given that the Leu and Trp residues present in P. thunbergii ChlB are also conserved in ChlB from the cyanobacterium L. boryana (Figs 1B and 2A), we subsequently decided to evaluate the effects of Leu209 (CTA) to Pro209 (CCA) and Trp216 (TGG) to Arg216 (CGG) amino acid substitutions on DPOR activity in this cyanobacterial species. For in vivo analysis, we expressed ChlB variants bearing pre-edited codons Pro209 (ChlB_P) and Arg216 (ChlB_R) as well as a doubly pre-edited variant (ChlB_PR) bearing both pre-edited codons of Pro209 and Arg216 (Fig. 2A). Shuttle vectors carrying pre-edited mimic chlB variants with strep-chlN as an artificial operon strep-chlN–chlB as previously reported24, 25 were introduced into the chlB-lacking L. boryana mutant YFB1426. These transformants, YFB14/NB2, YFB14/NB2_P, YFB14/NB2_R, and YFB14/NB2_PR express the wild-type ChlB, ChlB_P, ChlB_R, and ChlB_PR variants respectively, as well as Strep-ChlN.
Effects of pre-edited mimic substitutions of ChlB in cyanobacterial cells. (A) The partial ChlB amino acid sequence from L. boryana (WT) and the pre-edited mimic sequences from pHBNB2_P (P), pHBNB2_R (R), and pHBNB2_PR (PR) are shown. The two codons (two T-to-C substitutions shown in red) were altered in the three plasmids. (B) (a) Chl and Pchlide contents of dark-grown transformants of YFB14 harboring the shuttle vectors pHBNB2 (WT, lane 1), pHBNB2_P (P, lane 2), pHBNB2_R (R, lane 3), pHBNB2_PR (PR, lane 4), and empty vector (Control, lane 5) (b) The presence of ChlB was confirmed by Western blot analysis of total cell extract of transformant cells using the anti-ChlB antiserum. (c, d) Strep-ChlN (c) and ChlB (d) in the soluble fractions were detected by anti-ChlN and anti-ChlB antisera, respectively. (e, f) Affinity purified proteins from these crude extracts were detected by anti-ChlN (e) and anti-ChlB (f) antisera, respectively.
In YFB14, Chl synthesis in the dark is arrested at the Pchlide reduction step caused by a disruption of chlB 26. The Chl content of YFB14/NB2_P and YFB14/NB2_R cells grown in the dark was almost identical to that observed in the positive control YFB14/NB2 indicating that these mutations are functionally active (Fig. 2B,a). In contrast, the Chl content of YFB14/NB2_PR cells grown in the dark was significantly lower than that of the positive control. In addition, while only traces of Pchlide were detected in YFB14/NB, YFB14/NB2_P, and YFB14/NB2_R cells, the YFB14/NB2_PR cells contained markedly higher concentrations of Pchlide, reaching approximately 60% of that of the negative control (YFB14 with an empty pPBH202 vector25) (Fig. 2B,a). The Pchlide accumulations were also well correlated with decreased Chl contents. The contents of ChlB proteins in transformant cells were subsequently assessed by Western blot analysis (Fig. 2B,b). In all three YFB14/NB2 mutant variants, ChlB proteins were present at levels comparable to the amount of wild type ChlB present in the control strain YFB14/NB2, suggesting that only the PR mutant of ChlB is functionally impaired as a DPOR subunit in cyanobacterial cells.
Co-purification of ChlB variants with Strep-ChlN
We checked the amount of ChlB in soluble fractions and observed that ChlB_R was present at same level as WT ChlB. This is contrasted by ChlB_P and ChlB_PR variants that were present at less than half the wild type level (Fig. 2B,d). This indicates that the Pro substitution reduced solubility of ChlB. In addition, the amount of Strep-ChlN, which does not have any amino acid substitutions, also largely decreased in the soluble fractions that contained ChlB_P and ChlB_PR variants (Fig. 2B,c).
We also addressed whether ChlB variants can form a stable complex with ChlN by performing a co-purification assay. For this assay, ChlB was co-purified with Strep-ChlN from cyanobacterial soluble extracts using a Strep-tag affinity column and the amount of co-purified ChlB was evaluated by Western blot analysis (Fig. 2B,f). This analysis showed that ChlB was indeed co-purified with Strep-ChlN from extracts that expressed ChlB_R as well as from extracts that expressed WT ChlB. However, the yield of Strep-ChlN from extracts that expressed ChlB_P was very low. Interestingly, equivalent low amounts of ChlB_P was co-purified with these low yields of Strep-ChlN. Little amount of Strep-ChlN was also obtained from extracts that expressed ChlB_PR. However in these extracts, no detectable ChlB_PR was observed (Fig. 2B,e,f).
We also used an E. coli expression system to assay the interaction between the cyanobacterial ChlN and ChlB in heterologous cells (Fig. 3). In this E. coli expression assay, the amount of ChlB variants in the total cell free crude extract was about the same as observed with WT ChlB (Fig. 3,a). However, when assaying amount in a clarified soluble fraction there was significantly less soluble ChlB_PR than observed with the ChlB_R and ChlB_P variants (Fig. 3,b). In spite of the decreased amount of soluble ChlB proteins, a significant amount of ChlB was co-purified with Strep-ChlN from the extracts that expressed ChlB_R variant as well as WT control (Fig. 3,c). However, quantitation of the yields of Strep-ChlN and ChlB showed that while WT ChlB formed a stable ~1:1 interaction with Strep-ChlN, ChlB_R co-purified with Strep-ChlN at ~60% lower amounts. The observed ~60% reduction in co-purification of ChlB_R is similar to the observed ~60% reduction in relative activity (Fig. 3,d). On the other hand, ChlB_P and ChlB_PR both showed no detectable co-purification with Strep-ChlN (Fig. 3,c), which was consistent with no detectable activity of these proteins (Fig. 3,d). These results with E. coli extracts indicate that the formation of stable ChlN-ChlB complex is indeed inhibited by the Pro substitution with the Arg substitution enhancing this inhibitory effect.
Effects of pre-edited mimic substitutions of ChlB expressed in E. coli. Purified NB protein with pre-edited mimic ChlB variants from E. coli. Western blot analysis of total extracts (a, 0.1 µg per lane) and soluble fractions (b, 1.25 µg per lane) of E. coli using the anti-ChlB antiserum, and SDS-PAGE profiles of affinity-purified NB protein variants (c, 3.0 µg per lane); Proteins were stained using Coomassie Brilliant Blue (c). A nonspecific signal detected just above the ChlB signal in the soluble fractions is shown by an asterisk (b). Relative DPOR activity was measured using Strep-purified proteins with ChlL (d). The 100% activity of the NB protein was 22.0 nmol min−1 mgprotein −1. N.D. denotes “not detected”.
RNA editing of chlB transcripts in pine seedlings
Black pine (P. thunbergii) seedlings were cultivated under continuous light or dark conditions for 10 and 14 days (Fig. 4A). The dark cultivated black pine seedlings produced Chl as indicated by a yellow–green color (Fig. 4A) with Chl content assays showing that the dark-grown seedlings produced 20%–25% of the amount of Chl observed with light-grown seedlings (Fig. 4B). To investigate the correlation between light-independent Chl production and DPOR gene expression, transcript levels of DPOR subunits were semi-quantified using reverse transcriptase (RT)-PCR (Fig. 4C). The transcript levels of psbA, psbB, and rbcL, which are encoded by the chloroplast genome, were almost constant under both light and dark conditions. This result is consistent with a previous report27. However, the transcript levels of all three DPOR genes were markedly higher in dark-grown seedlings than those in light-grown seedlings. This tendency was more pronounced after 14 days, with transcript levels in light-grown seedlings markedly decreasing from 10 days to 14 days. This result suggests that the expression of three DPOR genes is down-regulated in the light and/or up-regulated in the dark.
Efficiency of RNA editing of chlB mRNA in cotyledons of black pine (A). Seedlings of the black pine P. thunbergii were grown for 10 and 14 days under light (L) and dark (D) conditions (Scale bars = 10 mm). (B) Chl (including Chl a and Chl b) content (µg cotyledon−1) in the cotyledons at 10 and 14 days under light (L) and dark (D) conditions. (C) Semi-quantification of the transcripts of several chloroplast genes in cotyledons of seedlings grown in light (lanes 1 and 3) and dark (lanes 2 and 4) for 10 (lanes 1 and 2) and 14 days (lanes 3 and 4). Three chlB, chlL, and chlN genes encode the DPOR subunits. The psbA, psbB, and rbcL genes were treated as genes representative of photosynthesis in chloroplast genomes. rrn23 was used as an internal standard. Cycle numbers are shown in parentheses. (D) The RNA editing efficiency of chlB transcript is expressed as the ratio of the number of clones with edited codons to the total number of examined clones. Ratios among clones (Supplementary Table S1) are color-coded; both edited codons (CTA/TGG), green; singly edited codons (CCA/TGG, CTA/CGG), yellow and blue, respectively; and pre-edited codons (CCA/CGG), light gray.
Two codons CCA (Pro206) and CGG (Arg213) of chloroplast DNA-encoded chlB are post-transcriptionally changed to CUA (Leu206) and UGG (Trp213) by RNA editing11. To determine the RNA editing efficiency of chlB mRNA, partial chlB fragments covering the two editing sites were amplified from cDNA samples using PCR and cloned into the vector. The nucleotide sequences of these chlB fragments were determined in more than 30 clones for each condition, and the RNA editing efficiency was estimated as the ratio of the number of edited clones (sequenced as CTA and TGG) to the total number of clones (Fig. 4D, Supplementary Table S1). In dark-grown seedlings, over 80%–90% of chlB transcripts were correctly edited in both codons while singly edited clones (CTA/CGG or CCA/TGG) present less than 10% of the clones. This result suggested that the two RNA editing reactions are tightly coupled or proceed very rapidly. In addition, the total numbers of each singly edited codon were almost identical, suggesting no site preference in the two editing sites. In contrast to the dark-grown seedlings, only 40%–60% of cDNA clones were edited at both sites in light-grown seedlings. The RNA editing efficiency for both sites in 10-day seedlings was slightly higher than that in 14-day seedlings grown in the dark, whereas that in 10-day light-grown seedlings was approximately 60% and decreased to approximately 40% in 14-day light-grown seedlings. These results indicate that light not only suppresses the transcription of DPOR genes but also reduces the efficiency of RNA editing of chlB mRNA.
Discussion
In this study, we showed that a cyanobacterial ChlB variant (ChlB_PR) that mimics pre-edited plastid ChlB significantly decreases DPOR activity relative to wild-type ChlB. We also showed that the transcript levels of three plastid genes coding for DPOR subunits are up- and down-regulated in dark and light conditions, respectively, in seedlings of P. thunbergii. In addition, the RNA editing efficiency of chlB transcripts correlated well with transcript levels, being markedly higher in dark-grown than in light-grown seedlings. If the activity observed in the cyanobacterial ChlB variant is applicable to black pine chloroplast ChlB then this suggests that DPOR activity is also regulated at the post-transcriptional stage via RNA editing.
The single Pro substitution on the cyanobacterial ChlB (ChlB_P) at the same position as that which occurs by RNA editing of the eukaryotic homologs has a negative effect on formation of a complex with ChlN. In addition, this mutation also causes a decrease in solubility. These negative effects are also manifest by expression in E. coli cells in which no complex formation with ChlN was observed (Fig. 3,c). Interestingly, ChlB_P did complement Chl biosynthesis in chlB deletion strain (YFB14/NB2_P; Fig. 2B,a) indicating that this mutation can form enough of a functional NB complex in cyanobacterial cells to maintain Chl synthesize in the dark. These results also indicate that cyanobacterial cells may contain a stabilizing factor(s), such as Pchlide that allows the ChlB Pro substitution to form a NB complex. The additional Arg substitution (ChlB_PR) caused more severe effect on complex formation with ChlN with this negative effect observed even in cyanobacterial cells.
Interestingly, a significant decrease in the amount of ChlN was observed in YFB14/NB2_P and YFB14/NB2_PR strains that express the Pro substituted ChlB variants (Fig. 2B,c). Given that the Pro substitution affects formation of a stable NB protein complex, it is possible that a large part of ChlN may exist as a monomer in YFB14/NB2_P and YFB14/NB2_PR and that monomeric ChlN is unstable. Previous work on nitrogenase reported that NifD (corresponding to ChlN on DPOR) was rapidly degraded and accumulated as an insoluble aggregate without its partner NifK (corresponding to ChlB), while NifK was stable alone and formed homo tetramer28. Taken together, the results in L. boryana and E. coli suggest that the Pro substitution affects NB complex formation and the drastic decrease of DPOR activity observed with the pre-editing mimic ChlB variant (ChlB_PR) is caused by defect in complex formation with ChlN.
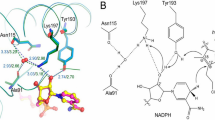
Crystal structures of the DPOR complex of L protein and NB protein from Prochlorococcus marinus has been recently reported as well as crystal structures of NB protein from Rhodobacter capsulatus and Thermosynechococcus elongatus 14, 29, 30. Although sequence similarity among these three ChlB is not high (the most distant pair: about 35% between T. elongatus and P. marinus), the overall structure of the NB protein is conserved very well. The spatial arrangement of these two edited amino acid residues, Leu and Trp (Asp/His), is almost identical in the structures of the three NB proteins (Fig. 5). Given that black pine ChlB shows high similarity to cyanobacterial ChlB (68% to T. elongatus and 65% to L. boryana), the observed effects of the PR substitution in L. boryana ChlB likely reflect the actual effects of amino acid substitutions by RNA editing in P. thunbergii.
Spatial arrangement of the two editing amino acid residues corresponding to the edited codons in the crystal structures of NB proteins from three photosynthetic organisms. Close-up view of the two amino acid residues corresponding to editing sites in crystal structures of NB proteins from T. elongatus (A; PDB code 2XDQ), P. marinus (B; PDB code 2YNM), and R. capsulatus (C; PDB code 4AEK); ChlN/BchN and ChlB/BchB subunits are colored green and blue, respectively. The two amino acid residues corresponding to the editing sites are shown in red. The [4Fe-4S] clusters shown in a CPK model (yellow and orange) and Pchlide molecules are shown as stick models (pink). The crystal structure from T. elongatus is of the Pchlide-free form.
The first editing site (Leu209 in T. elongates ChlB, Fig. 5A; Leu213 in P. marinus ChlB, Fig. 5B; and Leu199 in R. capsulatus BchB, Fig. 5C) is commonly present as a short α-helix consisting of nine amino acid residues that may stabilize the adjacent two α-helices via a hydrophobic interaction. The second editing site (Trp216 of the cyanobacterial ChlB, Fig. 5A), which is not conserved in R. capsulatus (His206, Fig. 5C) or P. marinus (Asp220, Fig. 5B), is commonly located in a very short loop connecting the α-helix that contains the first editing site and the short β-sheet. Common spatial positions of two residues among the three crystal structures are distal from the catalytic Pchlide-binding cavity, [4Fe-4S] cluster, and subunit-interaction region between ChlN/BchN and ChlB/BchB (Fig. 5). Although the editing site is distant from the interface between ChlN, our results indicate that the Pro substitution has a big impact on ChlN-ChlB complex formation. Consequently the Leu to Pro substitution in the α-helix likely has a significant impact on the overall tertiary structure that manifests a defect in this subunit interaction. Even though a single Arg substitution exerts only slight negative effects on the complex formation, our results also imply that the double substitution affects the activity of NB protein with an additive destabilization effect.
In a previous study using L. decidua, production of Chl was largely arrested after 7 days in cotyledons grown in the dark, despite maintenance of DPOR mRNA levels11. These authors also reported a significant decrease in the RNA editing efficiency of chlB mRNA from 7 to 14 days, suggesting a strong correlation between Chl production and RNA editing in dark-grown L. decidua seedlings. Our evaluation of ChlB variants in the present study indicates a causal relationship between RNA editing of chlB mRNA and DPOR activity.
Our results suggested that the DPOR activity is maintained at a high level by up-regulated transcription of the chlLNB genes and high efficient RNA editing under the dark conditions, and under the light conditions the DPOR activity decreases by down-regulated transcription and low efficiency of RNA editing. One consequence of RNA editing of the chlB mRNA may be to provide an additional regulatory layer of the DPOR activity in addition to the main transcriptional regulation in black pine chloroplasts.
It would be reasonable that the editing efficiency of chlB is higher in the dark, where DPOR is critical for Chl supply, than in the light in P. thunbergii. Under light conditions, LPOR is a better enzyme to reduce Pchlide than DPOR. LPOR, which is a small protein of approximately 30 kD, has a tolerance to oxygen and requires light and NADPH for its activity31. In contrast, DPOR is a relatively large protein complex (approximately 360 kD; consisting one NB protein and two L protein30) and requires reduced ferredoxin and ATP8, 32 with an oxygen-labile FeS cluster25. In addition, DPOR may compete for Pchlide with LPOR if both enzymes are present in the chloroplast. Given that the Pchlide binding site is located at the interface between ChlB and ChlN14, 30, the dissociation of NB protein may contribute to prevent competition for Pchlide between LPOR.
There are significant amounts of ChlB_PR left as a soluble protein in YFB14/NB2_PR strain, while soluble ChlN protein decreased obviously in the cell (Fig. 2B,c,d). This result implies that ChlB variant from pre-edited mRNA would be present as a homomeric protein in chloroplasts. Transgenic tobacco overexpressing pre-edited chlB of P. thunbergii in chloroplasts showed stimulation of root development, suggesting alternative functions of the ChlB variant derived from pre-edited mRNA33. Pre-edited mRNAs of rps2 encoded by maize mitochondrial genome are also translated as well as edited rps2 mRNA34. In animal nerve cells, proteins translated from pre-edited mRNA are likely to have different functions from those of corresponding proteins from edited mRNA35. If a homomeric ChlB protein translated from pre-edited mRNA also has a different function, RNA editing in chlB might work as a switch to change the function of ChlB in black pine. Further studies are needed to identify such additional functions of ChlB in photosynthetic organisms.
Materials and Methods
Cyanobacterial strains, mutants, and culture conditions
The cyanobacterial mutant YFB14 lacking chlB 26 was derived from Leptolyngbya boryana (formerly Plectonema boryanum) strain dg5 36, which was used as the host strain to overexpress chlB variants. YFB14 was cultivated in BG-11 medium with 15 µg ml−1 kanamycin under medium-intensity light conditions (40 µmolphoton m−2 s−1) using fluorescent lamps as described previously16. Transformants harboring the overexpression plasmids were cultivated as above in medium supplemented with 10 µg ml−1 chloramphenicol for plasmid maintenance. The medium was supplemented with glucose (30 mM) for heterotrophic growth in the dark.
Plasmid construction
To introduce pre-editing amino acid substitutions into ChlB from L. boryana, pHANB2_P, pHANB2_R, and pHANB2_PR were constructed from pHANB2 (Supplementary Tables S2 and S3), which have chlN–chlB fragments in pASK-IBA5plus as the template25. The T-to-C substitution in the Leu209 codon (CTA) produced the Pro codon (CCA) and was introduced by PCR using primers PbNf1 and PbchlBedit-r1 (fragment II; Supplementary Table S2). The corresponding fragment without the substitution was amplified using primers PbNf1 and PbchlBWT-r1 (fragment I; Supplementary Table S2). The second mutation, which substituted the Trp216 codon (TGG) with the Arg codon (CGG), was introduced as a partial chlB fragment and was amplified using primers PbchlBedit-f1 and PbBr1 (fragment IV; Supplementary Table S2). The corresponding fragment without the substitution was amplified using primers PbchlBWT-f1 and PbBr1 (fragment III; Supplementary Table S2). Fragments I (II) and III (IV) have an identical 13-bp sequence that was used to overlap PCR37. The entire chlN–chlB fragment containing the L209P substitution (using fragments II and III), W216R substitution (using fragments I and IV), and both substitutions (using fragments II and IV) were then produced. These connected fragments were cloned into BsaI sites in pASK-IBA5plus to yield pHANB2_P (L209P), pHANB2_R (W216R), and pHANB2_PR (both substitutions), respectively (Supplementary Table S3). Shuttle vectors expressing these chlB variants with chlN in L. boryana were constructed as described previously25. The chlN–chlB fragments were amplified by PCR using the primers PBHLI18-f1 and PBHchlNBr1 (Supplementary Table S2), with pHANB2_P, pHANB2_R, and pHANB2_PR as templates, and the PCR fragment was introduced into the SphI-BamHI sites of pPBHLI1825 to yield pHBNB2_P, pHANB2_R, and pHANB2_PR (Supplementary Table S3). These plasmids were then introduced into cyanobacterial cells by electroporation38.
Determination of Chl and Pchlide
Pigments were extracted from the dark-grown cyanobacteria in 90% (v/v) methanol as described previously16. Chl and Pchlide contents in methanol extracts were determined by HPLC. For HPLC analyses, aliquots of methanol extracts were applied to a 4.6 × 150 mm Symmetry C8 3.5-µm column (Waters), and pigments were separated as described previously39, 40. Pchlide and Chl were eluted at 7.9 and 22.2 min, respectively, and their contents were determined using standard pigments.
ChlB co-purification
Pre-edited mimic ChlB variants were co-purified with Strep-ChlN from soluble fractions of L. boryana 25, 41 and E. coli 25, 41 using a Strep-Tactin column as described previously42. The purification procedure was performed in an anaerobic chamber as described previously16. DPOR activity assay were then performed as described previously24.
Western blot analysis
Crude and soluble fractions of E. coli were prepared as described previously41. Crude extracts of cyanobacterial cells were prepared and Western blot analyses were performed as described previously16. After SDS-PAGE, proteins were transferred onto PVDF membranes and were incubated with an antiserum against ChlB or ChlN from L. boryana 15, 26, followed by goat anti-rabbit IgG-horseradish peroxidase conjugate (Bio-Rad). Protein bands were visualized using a chemiluminescent substrate (ECL Western Blotting Analysis System; GE Healthcare).
Pine cultivation and RNA extraction from cotyledons
Seedlings of the black pine P. thunbergii were grown under continuous light or continuous dark at 25 °C. Cotyledons were harvested at 10 and 14 days of age. RNA was extracted from cotyledons using RNeasy Plant Mini Kit (Qiagen) and DNase (RQ1 RNase-Free DNase; Promega) and was used for RT-PCR analyses and sub-cloning of chlB fragments. After preparation of cDNA from the extracted RNA, PCR was carried out using the gene-specific primers (Supplementary Table S2) with 25 cycles for chlB, chlL, psbA, rbcL, and rrn23 genes and 30 cycles for chlN and psbB genes. Amplified chlB fragments were cloned into pGEM-T Easy Vector (Promega), and nucleotide sequences were determined in both directions using the primer pair M13 forward and M13 reverse (3730XI, ABI). For each condition 33 or 36 clones were sequenced (Supplementary Table S1). RNA editing efficiency was defined as the number of edited clones/total number of clones.
References
Tanaka, R. & Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58, 321–346 (2007).
Masuda, T. & Fujita, Y. Regulation and evolution of chlorophyll metabolism. Photochem Photobiol Sci 7, 1131–1149 (2008).
Stenbaek, A. & Jensen, P. E. Redox regulation of chlorophyll biosynthesis. Phytochemistry 71, 853–859 (2010).
Czarnecki, O. & Grimm, B. Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J Exp Bot 63, 1675–1687 (2012).
Meskauskiene, R. et al. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98, 12826–12831 (2001).
Fujita, Y. Protochlorophyllide reduction: a key step in the greening of plants. Plant Cell Physiol 37, 411–421 (1996).
Armstrong, G. A. Greening in the dark: Light-independent chlorophyll biosynthesis from anoxygenic photosynthetic bacteria to gymnosperms. J. Photochem. Photobiol. B. 43, 87–100 (1998).
Fujita, Y. & Bauer, C. E. In Chlorophylls and Bilins: Biosynthesis, Synthesis, and Degradation, 109–156 (Academic Press, 2003).
Reinbothe, C. et al. Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci. 15, 614–622 (2010).
Breznenová, K. et al. Light-independent accumulation of essential chlorophyll biosynthesis- and photosynthesis-related proteins in Pinus mugo and Pinus sylvestris seedlings. Photosynthetica 48, 16–22 (2010).
Demko, V. et al. A novel insight into the regulation of light-independent chlorophyll biosynthesis in Larix decidua and Picea abies seedlings. Planta 230, 165–176 (2009).
Masuda, T. & Takamiya, K.-I. Novel insights into the enzymology, regulation and physiological functions of light-dependent protochlorophyllide oxidoreductase in angiosperms. Photosynth Res 81 (2004).
Nomata, J., Terauchi, K. & Fujita, Y. Stoichiometry of ATP hydrolysis and chlorophyllide formation of dark-operative protochlorophyllide oxidoreductase from Rhodobacter capsulatus. Biochem Biophys Res Commun 470, 704–709 (2016).
Muraki, N. et al. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465, 110–114 (2010).
Hunsperger, H. M., Randhawa, T. & Cattolico, R. A. Extensive horizontal gene transfer, duplication, and loss of chlorophyll synthesis genes in the algae. BMC Evolutionary Biology 15, 1–19 (2015).
Yamazaki, S., Nomata, J. & Fujita, Y. Differential operation of dual protochlorophyllide reductases for chlorophyll biosynthesis in response to environmental oxygen levels in the cyanobacterium Leptolyngbya boryana. Plant Physiol 142, 911–922 (2006).
Ueda, M., Tanaka, A., Sugimoto, K., Shikanai, T. & Nishimura, Y. chlB requirement for chlorophyll biosynthesis under short photoperiod in Marchantia polymorpha L. Genome Biol Evol 6, 620–628 (2014).
Knoop, V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci 68, 567–586 (2011).
Chateigner-Boutin, A. L. & Small, I. Plant RNA editing. RNA Biol 7, 213–219 (2010).
Castandet, B. & Araya, A. RNA editing in plant organelles. Why make it easy? Biochemistry (Mosc) 76, 924–931 (2011).
Karpinska, B., Karpinski, S. & Hallgren, J. The chlB gene encoding a subunit of light-independent protochlorophyllide reductase is edited in chloroplasts of conifers. Curr Genet 31, 343–347 (1997).
Kusumi, J., Sato, A. & Tachida, H. Relaxation of functional constraint on light-independent protochlorophyllide oxidoreductase in Thuja. Mol Biol Evol 23, 941–948 (2006).
Gupta, R. S. Origin and spread of photosynthesis based upon conserved sequence features in key bacteriochlorophyll biosynthesis proteins. Mol Biol Evol 29, 3397–3412 (2012).
Yamamoto, H., Kurumiya, S., Ohashi, R. & Fujita, Y. Functional evaluation of a nitrogenase-like protochlorophyllide reductase encoded by the chloroplast DNA of Physcomitrella patens in the cyanobacterium Leptolyngbya boryana. Plant Cell Physiol 52, 1983–1993 (2011).
Yamamoto, H., Kurumiya, S., Ohashi, R. & Fujita, Y. Oxygen sensitivity of a nitrogenase-like protochlorophyllide reductase from the cyanobacterium Leptolyngbya boryana. Plant Cell Physiol 50, 1663–1673 (2009).
Fujita, Y., Takagi, H. & Hase, T. Identification of the chlB gene and the gene product essential for the light-independent chlorophyll biosynthesis in the cyanobacterium Plectonema boryanum. Plant Cell Physiol 37, 313–323 (1996).
Mukai, Y., Yamamoto, N. & Koshiba, T. Light-independent and tissue-specific accumulation of light-harvesting chlorophyll a/b binding protein and ribulose bisphosphate carboxylase in dark-grown pine seedlings. Plant Cell Physiol 32, 1303–1306 (1991).
Li, J. G., Tal, S., Robinson, A. C., Dang, V. & Burgess, B. K. Analysis of Azotobacter vinelandii strains containing defined deletions in the nifD and nifK genes. J Bacteriol 172, 5884–5891 (1990).
Bröcker, M. J. et al. Crystal structure of the nitrogenase-like dark operative protochlorophyllide oxidoreductase catalytic complex (ChlN/ChlB)2. J Biol Chem 285, 27336–27345 (2010).
Moser, J. et al. Structure of ADP-aluminium fluoride-stabilized protochlorophyllide oxidoreductase complex. Proc Natl Acad Sci USA 110, 2094–2098 (2013).
Heyes, D. J., Martin, G. E., Reid, R. J., Hunter, C. N. & Wilks, H. M. NADPH:protochlorophyllide oxidoreductase from Synechocystis: overexpression, purification and preliminary characterisation. FEBS Lett 483, 47–51 (2000).
Nomata, J., Swem, L. R., Bauer, C. E. & Fujita, Y. Overexpression and characterization of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. Biochim Biophys Acta 1708, 229–237 (2005).
Nazir, S. & Khan, M. S. Chloroplast-encoded chlB gene from Pinus thunbergii promotes root and early chlorophyll pigment development in Nicotiana tabaccum. Mol Biol Rep 39, 10637–10646 (2012).
Phreaner, C. G., Williams, M. A. & Mulligan, R. M. Incomplete editing of rps12 transcripts results in the synthesis of polymorphic polypeptides in plant mitochondria. Plant Cell 8, 107–117 (1996).
Colina, C., Palavicini, J. P., Srikumar, D., Holmgren, M. & Rosenthal, J. J. Regulation of Na+/K+ ATPase transport velocity by RNA editing. PLoS Biol 8 (2010).
Hiraide, Y. et al. Loss of cytochrome c M stimulates cyanobacterial heterotrophic growth in the dark. Plant Cell Physiol 56, 334–345 (2015).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual, Third edition. (Cold Spring Harbor Laboratory Press, 2001).
Fujita, Y., Takahashi, Y., Chuganji, M. & Matsubara, H. The nifH-like (frxC) gene is involved in the biosynthesis of chlorophyll in the filamentous cyanobacterium Plectonema boryanum. Plant Cell Physiol 33, 81–92 (1992).
Goto, T., Aoki, R., Minamizaki, K. & Fujita, Y. Functional differentiation of two analogous coproporphyrinogen III oxidases for heme and chlorophyll biosynthesis pathways in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 51, 650–663 (2010).
Aoki, R., Goto, T. & Fujita, Y. A heme oxygenase isoform is essential for aerobic growth in the cyanobacterium Synechocystis sp. PCC 6803: modes of differential operation of two isoforms/enzymes to adapt to low oxygen environments in cyanobacteria. Plant Cell Physiol 52, 1744–1756 (2011).
Yamamoto, H., Nomata, J. & Fuita, Y. Functional expression of nitrogenase-like protochlorophyllide reductase from Rhodobacter capsulatus In Escherichia coli. Photochem Photobiol Sci 7, 1238–1242 (2008).
Nomata, J., Ogawa, T., Kitashima, M., Inoue, K. & Fujita, Y. NB-protein (BchN-BchB) of dark-operative protochlorophyllide reductase is the catalytic component containing oxygen-tolerant Fe-S clusters. FEBS Lett. 582, 1346–1350 (2008).
Acknowledgements
We thank Yoshihiko Tsumura (University of Tsukuba) for providing seeds of P. thunbergii, Makiko Aichi (Chubu University) and Yoshitake Ohashi (Nagoya University) for technical assistance with cultivation of cyanobacteria, Mamoru Sugita (Nagoya University) for valuable suggestions on RNA editing, and Carl Bauer (Indiana University), Tatsuo Omata (Nagoya University), and Kazuki Terauchi (Ritsumeikan University) for valuable discussions. We also thank Hidenori Takagi and Toshiharu Hase (Osaka University) for their previous works on the chlB mutant of L. boryana and preparation of the antisera against ChlN and ChlB. This work was supported by the Japan Society for the Promotion of Science (JSPS) [Grants-in-Aid for Scientific Research Nos. 19570036, 20200063, and 23370020 (to Y.F.), a Fellowship from JSPS for Japanese Junior Scientists No. 25-2354 (to H.Y.)]; the Ministry of Education, Culture, Sports, Science and Technology of Japan [Global COE program “Advanced System-Biology”]; the Circle for the Promotion of Science and Engineering (to H.Y.); the Japan Science Society [Sasagawa Scientific Research Grant to H.Y.]; and the Japan Science and Technology Agency (JST) [Precursory Research for Embryonic Science and Technology (PRESTO) (to Y.F.)] and [the Advanced Low Carbon Technology Research and Development Program (ALCA) (to Y.F.)].
Author information
Authors and Affiliations
Contributions
Haruki Y., J.K. and Y.F. designed this research. J.K. cultivated pine cotyledons and extracted mRNA from the cotyledons to qPCR analysis, and also J.K. sequenced sub-cloned chlB cDNA fragments. Haruki Y. prepared the cyanobacterial mutants expressing ChlB variants and performed pigment analysis. Haruki Y. and Hisanori Y. cultivated cyanobacteria and detected immunologically the ChlB and ChlN proteins. Haruki Y. purified DPOR subunits from E. coli and assayed DPOR activity. Haruki Y. and Y.F. analyzed the data and wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yamamoto, H., Kusumi, J., Yamakawa, H. et al. The Effect of Two Amino acid Residue Substitutions via RNA Editing on Dark-operative Protochlorophyllide Oxidoreductase in the Black Pine Chloroplasts. Sci Rep 7, 2377 (2017). https://doi.org/10.1038/s41598-017-02630-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-02630-2
- Springer Nature Limited
This article is cited by
-
Expression of Genes in New Sprouts of Cunninghamia lanceolata Grown Under Dark and Light Conditions
Journal of Plant Growth Regulation (2020)
-
Evolution of light-independent protochlorophyllide oxidoreductase
Protoplasma (2019)
-
Functional expression of an oxygen-labile nitrogenase in an oxygenic photosynthetic organism
Scientific Reports (2018)
-
Dark chlorophyll synthesis may provide a potential for shade tolerance as shown by a comparative study with seedlings of European larch (Larix decidua) and Norway spruce (Picea abies)
Trees (2018)