Abstract
Acrossocheilus fasciatus is a stream-dwelling fish species of the Barbinae subfamily. It is valued for its colorfully striped appearance and delicious meat. This species is also characterized by apparent sexual dimorphism and toxic ovum. Biology and aquaculture researches of A. fasciatus are hindered by the lack of a high-quality reference genome. Here, we report chromosome-level genome assemblies of the male and female A. fasciatus. The HiFi-only genome assemblies for both female and male individuals were 899.13 Mb (N50 length of 32.58 Mb) and 885.68 Mb (N50 length of 33.06 Mb), respectively. Notably, a substantial proportion of the assembled sequences, accounting for 96.15% and 98.35% for female and male genomes, respectively, were successfully anchored onto 25 chromosomes utilizing Hi-C data. We annotated the female assembly as a reference genome and identified a total of 400.62 Mb (44.56%) repetitive sequences, 27,392 protein-coding genes, and 35,869 ncRNAs. The high-quality male and female reference genomes will provide genomic resources for developing sex-specific molecular markers, inform single-sex breeding, and elucidate genetic mechanisms of sexual dimorphism.
Similar content being viewed by others
Background & Summary
The Barbinae is a subfamily of the Cyprinidae that is the largest family of freshwater fishes. This subfamily contains the most complex and diverse fish groups within the Cyprinidae1. Their morphologies and habits are highly diverse. For example, Sinocyclocheilus rhinocerous dwells in caves and has evolved relevant traits2. Genome sequences of several Barbinae species, including three species of genus Sinocyclocheilus (S. grahami, S. rhinocerous, and S. anshuiensis), Poropuntius huangchuchieni, Puntigrus tetrazonahas, and Onychostoma macrolepis, have been deciphered, largely due to their phylogeny features and notable evolutionary status2,3,4. Most of the species in the Barbinae had undergone whole genome duplication after the third round of teleost-specific genome duplication (TGD) event that generated tetraploid even hexaploid5. However, some species remain diploids that retain the original chromosome number 2n=50, such as O. macrolepis, P. huangchuchieni and P. tetrazonahas3,4,6. Acrossocheilus fasciatus is also a diploid species in the Barbinae, with chromosome number 2n=507. It is mainly found in streams south of the Yangtze River and is extremely popular with recreational fisheries due to its colorful appearance with six dark stripes. It is a local delicacy and is considered highly nutritious8 by people in southeast China, especially in Zhejiang Province. However, because of its small size and slow growth rate9, this fish is always in short supply and has great market prospects. In addition, A. fasciatus is ichthyootoxic, with toxic ova10. The structures of the toxins remain unknown. Furthermore, it is sexually dimorphic in both body mass and appearance (Fig. 1). The weight of a two-year-old mature female is approximately 1.5 times that of the mature male11. In mature males, the six black transverse stripes gradually faded with the appearance of secondary sex characteristics such as the pearl organs and redness of the abdomen, whereas the females always retain the transverse stripes.
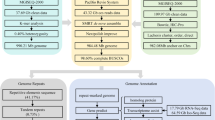
Despite its biological and economic importance, the genomic resources of A. fasciatus are limited. Several studies on A. fasciatus were focused on the mitochondrial DNA or transcriptomes12,13,14,15,16. In this study, we sequenced and annotated the chromosome-scale genome assemblies of the male and female A. fasciatus using PacBio HiFi reads and high-throughput chromosome conformation capture (Hi-C) technologies. The genome size of female A. fasciatus was estimated to be about 880.6 Mb through k-mer frequency distribution analysis with 126.33 Gb (~143 × ) Illumina clean data. The female and male genomes were independently assembled into contigs with PacBio HiFi reads. The female genome assembly spans 899.13 Mb with a contig N50 length of 32.58 Mb using 62.01 Gb (~70 × ) PacBio HiFi clean reads. The male genome spans 885.68 Mb with a contig N50 length of 33.06 Mb using 97.67 Gb (~111 × ) of HiFi clean reads. 96.15% and 98.35% of contig sequences of the female (contigs N50 length = 32.35 Mb; scaffolds N50 length = 33.86 Mb) and male (contigs N50 length = 32.84 Mb; scaffolds N50 length = 33.78 Mb) genomes were anchored onto 25 chromosomes using Hi-C data (Supplementary Table 1). Finally, the female genome was annotated as a reference genome with 44.56% (400.62 Mb) of repetitive sequences, 27,392 protein-coding genes, and 35,869 ncRNAs. The female and male genome assemblies reported here provide genomic resources for development of sex-specific molecular markers and single-sex breeding as well as a better understanding of the mechanisms of sexual dimorphism.
Methods
Sample collection
Two-year-old female and male adults of A. fasciatus were randomly sampled from the second-generation progeny of selective breeding performed in Dingxin Ecological Agriculture Co., Ltd. (Xiuning County, Huangshan City of Anhui Province, China). The sampled fish were euthanized with MS-222 (Sigma-Aldrich, #A5040) and dissected on ice. Eight tissues including the brain, gill, heart, intestine, liver, ovary, muscle, and skin of one female (body length = 16.23 cm, body weight = 43.56 g) were collected and immediately frozen in liquid nitrogen and then stored at −80 °C until DNA and RNA extraction. The blood and muscle tissues of one male (body length = 13.05 cm, body weight = 26.73 g) were collected for DNA extraction.
DNA extraction and sequencing for genomes
The high-molecular weight (HMW) genomic DNA from the female muscle and the male blood of A. fasciatus was extracted using the phenol/chloroform method17. The quality and quantity of the extracted DNA were assessed using 1.0% agarose gel electrophoresis and a Qubit 4.0 fluorometer (Thermo Fisher Scientific, USA).
For PacBio sequencing, the high-quality DNA (main band > 30 kb) was randomly interrupted into 15–18 kb size fragments by a Covaris g-TUBE (Woburn, Massachusetts, USA), and then the SMRTbell libraries were constructed using the PacBio HiFi Express Template Prep Kit 2.0 according to the manufacturer’s instruction18 (Pacific Biosciences, Menlo Park, CA, USA). For the female genome assembly, we generated two cells of HiFi clean reads with 62.01 Gb (~70 × ) data and an N50 read length of 14.12 kb using PacBio Sequel IIe platform. For the male genome assembly, we generated only one cell of HiFi reads with 97.67 Gb (~111 × ) data and an N50 read length of 13.96 kb using PacBio Revio platform (Table 1). For Illumina sequencing, the DNA was randomly interrupted into ~350 bp fragments using the Covaris ultrasonic crusher. Libraries were constructed using NEBNext® UltraTM DNA Library Prep Kit for Illumina (NEB, #E7370L) and sequenced on the Novaseq 6000 platform (Illumina, Inc., San Diego, CA, USA) with paired-end (PE) 150 bp model. We also obtained 126.33 Gb (~143 × ) of Illumina short reads to survey the female genome (Table 1).
For genome scaffolding, Hi-C libraries were prepared using muscle tissues from both female and male individuals for PacBio genome sequencing. The Hi-C library construction, including cell crosslinking, cell lysis, chromatin digestion (MboI), biotin labeling, proximal chromatin DNA ligation and DNA purification, was performed according to the standard protocol described previously19,20. After quality control assessment by Agilent 2100 Bioanalyzer and qPCR test, the resulting Hi-C libraries were subjected to sequencing with PE 150 bp model on Illumina Novaseq. 6000 platform. As a result, a total of 137.24 Gb (~152 × ) and 104.69 Gb (~116 × ) raw read data were generated for the female and male genome, respectively (Table 1).
RNA extraction and transcriptome sequencing
Eight sampled tissues, including the brain, gill, heart, intestine, liver, ovary, muscle, and skin of the female A. fasciatus were each extracted for total RNA using TRIzolTM reagent (Thermo Fisher Scientific, USA). The resulting RNAs were treated with DNase I (NEB, USA) to remove the genomic DNA.
To facilitate genome annotation, both Iso-Seq and RNA-Seq were performed. For PacBio Iso-Seq, the RNAs were mixed equimolarly and subjected to sequencing. Specifically, the concentration, integrity, and purity of the RNA isolated from each tissue of the female were confirmed using Qubit, Agilent 2100 and Nanodrop, then pooled together at an equimolar concentration. A double-stranded cDNA library was prepared with SMARTer® PCR cDNA Synthesis Kit (Clontech, USA). Subsequently, the cDNA library was sequenced using the PacBio Sequel IIe platform. After filtering and treating using SMRTlink v11.0 (https://www.pacb.com/support/software-downloads/) with parameters–minLength=50, a total of 20.25 Gb of subreads data were generated (Table 1). For Illumina RNA-seq, eight cDNA libraries from the aforementioned tissues were constructed independently and sequenced using Illumina NovaSeq 6000. A total of 56.32 Gb clean data were generated after removing reads containing adapters, reads with more than 10% unknown nucleotides (Ns) or low-quality bases (more than 20% bases with Phred quality < 5) (Table 1).
De novo genome assembly with PacBio HiFi reads and Hi-C technologies
Before de novo assembly, the size of the female genome was estimated with k-mer analysis of Illumina reads. The Illumina clean reads were filtered to remove redundancy with in-house script redup.v2 developed by Novogene (Beijing, China), and utilized to calculate the k-mer frequency with k=17 using Jellyfish v2.2.721,22. Based on the formula: genome size = k-mer number/peak depth, the female genome size of A. fasciatus was estimated to be 880.6 Mb, with a heterozygous ratio of 0.53% and repeat rate of 47.82% (Supplemental Fig. 1).
PacBio HiFi reads from the female and the male individuals were assembled into the female contigs and the male contigs using Hifasm v0.16.123 with default parameters. A total of 110 female contigs were built with a total length of 899,126,031 bp and an N50 length of 32.58 Mb. And a total of 174 male contigs were built with a total length of 885,680,593 bp and an N50 length of 33.06 Mb.
The Hi-C raw reads were processed to remove paired reads that contain adapters or low-quality bases (more than 20% bases with Phred quality <5), and quality-controlled by HiCUP24. Subsequently, the contigs were anchored into 25 pseudo-chromosomes using ALLHiC pipeline25 with the clean Hi-C data (Fig. 2a). Juicebox software was used to correct chromosome interaction strength artificially (Supplemental Fig. 2)26. As a result, 84 scaffolds of the female genome were generated with a total length of 899,129,631 bp and an N50 length of 33.86 Mb, of which 96.15% (864,515,734 bp) was anchored onto 25 chromosomes (Tables 2, 3). 167 scaffolds of the male genome were generated with a total length of 885,681,293 bp and an N50 length of 33.78 Mb, of which 98.35% (871,084,321 bp) was anchored onto 25 chromosomes (Tables 2, 3). Finally, we obtained the high-quality chromosome-level male and female reference genomes with Hi-C technologies20 for genome characters analysis (Fig. 2b).
Genome assembly and chromosome synteny analysis. (a) Hi-C interactive heatmap of the female genome. (b) Genomic landscape of female A. fasciatus. The Circos plot illustrates from outside to inside (I) gene density, (II) TE protein density, (III) transposon amount, (IV) GC ratio, and (V) GCskew. (c) Chromosome synteny analyses. Chromosome synteny between Danio rerio and female A. fasciatus (I), and between female and male A. fasciatus (II).
Genomic synteny analysis
To assign the chromosome ID of A. fasciatus genomes and assess the accuracy of genome assemblies, we performed the genomic synteny analysis between zebrafish Danio rerio, and the female and male A. fasciatus. For synteny analysis between the assemblies of zebrafish and female A. fasciatus, Mummer27 (v4.0.0beta2) was used to match the maximal unique sequences between the genomes with parameter “–mincluster 500”. The matched sequence sets were filtered by removing the sets with sequence similarity less than 80%. For synteny analysis between the female and the male assemblies of A. fasciatus, the matched sequence sets were filtered by removing the sets with sequence similarity of less than 95% and length less than 10 kb. Genomic synteny graphs were generated with the matched sets using RectChr v1.36 (https://github.com/BGI-shenzhen/RectChr) (Fig. 2c). The synteny graphs indicated a moderate level of collinearity with minor rearrangements between the genomes of zebrafish and A. fasciatus, and the genome assemblies of the female and male A. fasciatus are remarkably accurate. No obvious chromosome structure variation was observed between female and male genomes through synteny analysis.
Repeat annotation of the female genome
The repeat sequences mainly consisted of interspersed repeats (mainly transposable elements, TEs) and tandem repeats. The repeat sequences of TEs in the female A. fasciatus genome were identified using a strategy combing homology alignment and ab initio search. Tandem repeats were predicted ab initio using TRF28. Firstly, the homolog prediction of TEs was based on Repbase29 database employing RepeatMasker and RepeatProteinMask30 (https://www.repeatmasker.org/) with default parameters. Secondly, de novo repetitive elements were identified by LTR_FINDER31, RepeatScout32, and RepeatModeler33 with the default parameters. All repeat sequences with length > 100 bp and a gap ‘N’ less than 5% constituted the de novo TE library. Finally, a customized library (combination of homolog and de novo TE library without redundancy) was subjected to homology search using RepeatMasker to identify TEs. As a result, extensive repeat sequences including tandem repeats and interspersed repeats were detected in the genome, accounting for approximately 44.56% (400.62 Mb) of the genome (Table 4), which is close to the repeat rate of 47.82% estimated by the genome survey. The tandem repeat sequences were 57.51 Mb in length, accounting for 6.40% of the genome (Table 4).
Gene prediction and functional annotation
Three strategies were used to predict gene structures in the female genome: homology searching, ab initio prediction, and transcriptome-assisted prediction. For homology searching, the homologous protein sequences of Danio rerio, Ctenopharyngodon idella, Megalobrama amblycephala, Poropuntius huangchuchieni, Puntigrus tetrazona, Onychostoma macrolepis, and Oryzias latipes were downloaded from NCBI database (https://ftp.ncbi.nlm.nih.gov/genomes/refseq). Protein sequences were aligned to the genome using TBLASTN (v2.2.26; E-value ≤1e−5)34, and then the matched proteins were aligned to the homologous genome sequences for accurate spliced alignments with GeneWise (v2.4.1)35 which was used to predict gene structure contained in each protein region. For gene predication ab initio, AUGUSTUS36 (v3.2.3), GeneID37 (v1.4), GENSCAN38 (v1.0) and GlimmerHMM39 (v3.04) and SNAP40 (2013-11-29) were used in an automated gene prediction pipeline. For RNA-sequencing-assisted prediction, transcriptome read assemblies were generated with Trinity (v2.1.1) for the genome annotation41. To optimize the genome annotation, the RNA-Seq reads from different tissues were aligned to genome sequences using HISAT (v2.0.4) with default parameters to identify exon regions and splice positions42. The alignment results were then used as the input for Cufflinks (v2.2.1) with default parameters for genome-based transcript assembly43. The non-redundant reference gene set was generated by merging genes predicted by three methods with EvidenceModeler (EVM, v1.1.1) and then further annotated with PASA (Program to Assemble Spliced Alignment)44. As a result, we identified 27,392 protein-coding genes in the female reference genome (Table 5, Supplemental Fig. 3a).
Gene functions were assigned according to the best match by aligning the protein sequences to the Swiss-Prot45 (http://www.uniprot.org/) using BLASTP (E-value ≤ 1e-5). The motifs and domains were annotated using InterProScan7046 (v5.31) (https://www.ebi.ac.uk/interpro/). The Gene Ontology (GO) IDs for each gene were assigned according to the corresponding InterPro entry. We predicted the protein function by transferring annotations from the closest BLAST hit (E-value ≤ 1e-5) in the Swiss-Prot database and DIAMOND (v0.8.22)/BLAST hit (E-value < 10-5) in the NR database (ftp://ftp.ncbi.nih.gov/blast/db). We also mapped the gene set to a KEGG pathway and identified the best match for each gene47. As a result, 96.1% of the predicted 27,392 protein-coding genes have functional annotations (Supplementary Fig. 3b).
For non-coding RNA (ncRNA) annotation, the tRNAs were predicted using the program tRNAscan-SE48. Since rRNAs are highly conserved, the rRNA sequences of Homo sapiens were chosen as references, and rRNA sequences were predicted using BLASTN (E-value ≤ 1e-5). Other ncRNAs, including miRNAs and snRNAs were identified by searching against the Rfam database with default parameters using the infernal software49. Finally, a total of 35,869 ncRNAs were identified including 2,588 miRNAs, 18,386 tRNAs, 12,709 rRNAs, and 2,186 snRNAs (Supplementary Table 2).
Furthermore, the male genome of A. fasciatus was also annotated using the annotation result of the female genome as a reference with the liftoff50 software, an accurate gene annotation mapping tool, capable of mapping genes from a reference genome to a target genome.
Data Records
All the raw sequencing data for genome assembly have been deposited in the NCBI database (https://www.ncbi.nlm.nih.gov/bioproject). Specifically, for the female genome, the Illumina WGS data (SRR2699340851-SRR2699340952), PacBio WGS data (SRR2699339353-SRR2699339454), transcriptome data (SRR26993400-SRR26993400755,56,57,58,59,60,61,62,SRR2699339263) and Hi-C data (SRR26993395-SRR2699339964,65,66,67,68) were deposited under the BioProject accession number PRJNA1045882. For the male genome, the PacBio WGS data (SRR2712617969) and Hi-C data (SRR2758855370) were deposited under the BioProject accession number PRJNA1049304. The final files of the assembled genome of A. fasciatus have been deposited at GenBank under the accession number JAXUIB000000000 (female)71 and JAZDCR000000000 (male)72. Meanwhile, all the data including the male and female genome sequences and annotation files are accessible through the Figshare73.
Technical Validation
Benchmarking Universal Single-Copy Orthologues (BUSCO)74, Core Eukaryotic Genes Mapping Approach (CEGMA)75, and Merqury software76 were used to evaluate the genome assemblies. The BUSCO (v5.2.2) was used to evaluate the completeness of the genome assemblies with the vertebrata database (vertebrata_odb10). Out of the 3,354 orthologous genes, 3,304 (98.5%) genes were identified as complete genes, 16 (0.5%) genes were identified as fragmented genes, and 34 (1%) genes were missing from the female genome assembly (Fig. 3a). On the other hand, 3,301 (98.5%) genes were identified as complete genes, 19 (0.5%) genes were identified as fragmented genes, and 34 (1%) genes were missing from the male genome assembly (Fig. 3b). Meanwhile, CEGMA (v2.5) evaluation was also considered for genome completeness evaluation. Out of the 248 Eukaryotic core genes, 235 (94.76%) genes and 233 (93.95%) were identified in the female and male genomes, respectively (Supplementary Table 3). To further assess the completeness of genome assemblies, we identified telomeric repeats in both female and male genomes using tidk (v0.2.41) (https://zenodo.org/records/10091385) with Cypriniformes-specific telomeric repeat sequences. The results demonstrated telomeric repeat sequences could be identified in almost all of the chromosome ends (Supplementary Fig. 4). These results indicate an extremely high level of completeness of the genome assemblies.
To evaluate the quality and accuracy of the female genome assembly, we employed a three-step validation process. Firstly, the Illumina short-reads for the genome survey were mapped to genome assembly using BWA-MEM (v0.7.8)77 with default parameters, and then SAMtools77 was used for SNP calling. As a result, 99.30% of reads were mapped to the genome with approximately 99.95% coverage. Subsequently, the base quality value (QV) of genome sequences was quantified using Merqury software, resulting in a QV score of 52.22. All these results indicate a high-quality genome assembly. The GC skew of genome assembly was calculated with a 10 kb slide window using SOAP.coverage (v2.7.7)78. GC content was 37.49% with no obvious separation, indicating no foreign contamination in the genome (Supplementary Fig. 5).
Code availability
There were no custom software codes developed. The tools used for reads quality control are non-open scripts developed by the Novogene (Beijing, China). All bioinformatics tools and pipelines were performed following the instructions of the manuals and protocols. The versions of the software used, along with their corresponding parameters, have been thoroughly described in the Methods section.
References
Zheng, L. P., Yang, J. X. & Chen, X. Y. Molecular phylogeny and systematics of the Barbinae (Teleostei: Cyprinidae) in China inferred from mitochondrial DNA sequences. Biochem. Syst. Ecol. 68, 250–259 (2016).
Yang, J. X. et al. The Sinocyclocheilus cavefish genome provides insights into cave adaptation. BMC Biol. 14, (1) (2016).
Chen, L. et al. Chromosome-level genome of Poropuntius huangchuchieni provides a diploid progenitor-like reference genome for the allotetraploid Cyprinus carpio. Mol. Ecol. Resour. 21, 1658–1669 (2021).
Li, J. T. et al. Parallel subgenome structure and divergent expression evolution of allo-tetraploid common carp and goldfish. Nat. Genet. 53, 1493–1503 (2021).
Xu, M. R. X. et al. Maternal dominance contributes to subgenome differentiation in allopolyploid fishes. Nat. Commun. 14 (2023).
Cui, W. Y. et al. Embryonic development and phylogenetic analysis of Puntius tetrazona. Journal of Fisheries of China (in Chinese) 44, 1286–1295 (2020).
Jiang, J., Li, M. Y. & Wu, E. M. Chromosome karyotyping of Acrossocheilus fasciatus. Freshwater Fisheries of China (in Chinese) 39, 77–79 (2009).
Yu, Y. Y., Zhou, J. B., Zhang, Y. M. & Li, M. Y. The nutritional compositions and evalution of wild and cultured Acrossocheilus fasciatus. Journal of Fishery Sciences of China (in Chinese) 31, 207–210 (2012).
Yan, Y. Z. et al. Life-history strategies of Acrossocheilus fasciatus (Barbinae, Cyprinidae) in the Huishui Stream of the Qingyi watershed, China. Ichthyol. Res. 59, 202–211 (2012).
Wu, H. L. New records of toxic and medicinal fishes in China. (China Agriculture Press, 2002).
Zhang, Y. M., Cheng, S., Jiang, J. H., Lei, S. Y. & Yang, L. J. Primary study on the growth of Acrossocheilus fasciatus in cultivation. Journal of Shanghai Ocean University (in Chinese) 21, 542–548 (2012).
Zhou, M. Y. et al. Historical landscape evolution shaped the phylogeography and population history of the cyprinid fishes of Acrossocheilus (Cypriniformes: Cyprinidae) according to mitochondrial DNA in Zhejiang Province, China. Diversity (Basel) 15 (2023).
Wei, Z. Z., Fang, Y., Shi, W., Chu, Z. J. & Zhao, B. Transcriptional modulation reveals physiological responses to temperature adaptation in Acrossocheilus fasciatus. Int. J. Mol. Sci. 24 (2023).
Wei, W. B. et al. Integrated mRNA and miRNA expression profile analysis of female and male gonads in Acrossocheilus fasciatus. Biology 11 (2022).
Wang, L. et al. Influences of chronic copper exposure on intestinal histology, antioxidative and immune status, and transcriptomic response in freshwater grouper (Acrossocheilus fasciatus). Fish Shellfish Immunol. 139 (2023).
Wang, L. et al. Dietary berberine against intestinal oxidative stress, inflammation response, and microbiota disturbance caused by chronic copper exposure in freshwater grouper (Acrossocheilus fasciatus). Fish Shellfish Immunol. 139 (2023).
Green, M. R. & Sambrook, J. Isolation of High-Molecular-Weight DNA using organic solvents. Cold Spring Harb. Protoc. 2017, pdb.prot093450 (2017).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Belton, J. M. et al. Hi-C: A comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 (2012).
Rao, S. S. P. et al. A 3D Map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Marcais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Li, R. Q. et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272 (2010).
Cheng, H. Y., Concepcion, G. T., Feng, X. W., Zhang, H. W. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods. 18, 170–175 (2021).
Wingett, S. et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Research 4, 1310 (2015).
Zhang, X. T., Zhang, S. C., Zhao, Q., Ming, R. & Tang, H. B. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants 5, 833–845 (2019).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Delcher, A. L., Phillippy, A., Carlton, J. & Salzberg, S. L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30, 2478–2483 (2002).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573–580 (1999).
Jurka, J. et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005).
Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. Chapter 4, Unit 4.10 (2004).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21, I351–I358 (2005).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 117, 9451–9457 (2020).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Birney, E., Clamp, M. & Durbin, R. GeneWise and genomewise. Genome Res. 14, 988–995 (2004).
Stanke, M. & Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19, II215–II225 (2003).
Parra, G., Blanco, E. & Guigó, R. GeneID in Drosophila. Genome Res. 10, 511–515 (2000).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J.Mol. Biol. 268, 78–94 (1997).
Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004).
Korf, I. Gene finding in novel genomes. BMC Bioinform. 5 (2004).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12, 357–360 (2015).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–U174 (2010).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to assemble spliced alignments. Genome Biol. 9, R7 (2008).
Bairoch, A. & Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48 (2000).
Mulder, N. & Apweiler, R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol. Biol. (Clifton, N.J.) 396, 59–70 (2007).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Shumate, A. & Salzberg, S. L. Liftoff: accurate mapping of gene annotations. Bioinformatics 37, 1639–1643 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993408 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993409 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993393 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993394 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993400 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993401 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993402 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993403 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993404 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993405 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993406 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993407 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993392 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993395 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993396 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993397 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993398 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR26993399 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR27126179 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR27588553 (2023).
NCBI GenBank https://identifiers.org/ncbi/insdc:JAXUIB000000000 (2023).
NCBI GenBank https://identifiers.org/ncbi/insdc:JAZDCR000000000 (2023).
Yuan, Y. X. The genome annotations of Acrossocheilus fasciatus. figshare https://doi.org/10.6084/m9.figshare.24995825 (2023).
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A. & Zdobnov, E. M. BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654 (2021).
Parra, G., Bradnam, K. & Korf, I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21 (2020).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (No.2022YFD2400102) and the National Natural Science Foundation of China (No. 31872207).
Author information
Authors and Affiliations
Contributions
J.F.R., M.Y.L. and J.L.L. conceived and supervised the study. T.X.Z., Y.F.W. and J.K.X. collected the samples. Y.X.Y., T.X.Z. and J.F.R. performed the bioinformatics analysis. Y.X.Y., T.X.Z. and J.F.R. drafted the manuscript. J.Q.Y., L.G., Y.B.S., J.J.Z., Y.-W.C.-D. and W.M.L. provided review comments and modification of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yuan, Y., Zhong, T., Wang, Y. et al. Chromosome-scale genome assemblies of sexually dimorphic male and female Acrossocheilus fasciatus. Sci Data 11, 653 (2024). https://doi.org/10.1038/s41597-024-03504-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03504-9
- Springer Nature Limited