Abstract
Incident reports of medication errors are valuable learning resources for improving patient safety. However, pertinent information is often contained within unstructured free text, which prevents automated analysis and limits the usefulness of these data. Natural language processing can structure this free text automatically and retrieve relevant past incidents and learning materials, but to be able to do so requires a large, fully annotated and validated corpus of incident reports. We present a corpus of 58,658 machine-annotated incident reports of medication errors that can be used to advance the development of information extraction models and subsequent incident learning. We report the best F1-scores for the annotated dataset: 0.97 and 0.76 for named entity recognition and intention/factuality analysis, respectively, for the cross-validation exercise. Our dataset contains 478,175 named entities and differentiates between incident types by recognising discrepancies between what was intended and what actually occurred. We explain our annotation workflow and technical validation and provide access to the validation datasets and machine annotator for labelling future incident reports of medication errors.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background & Summary
Natural Language Processing is a promising technique for the extraction of information from medication related records to enhance medication safety1. However, only very few e-health systems can detect and learn from medical incidents. This poses a challenge to the ‘Patient Safety Incident Reporting and Learning Systems’ goal set by the WHO Global Patient Safety Action Plan 2021–20302,3. Medication errors are a common and potentially avoidable cause of patient harm4,5,6 that, as stated in the third WHO Global Challenge on Patient Safety7,8 and the 2022 World Patient Safety Day, must be significantly reduced globally.
To capture clinical near misses and incidents, reporting systems are recommended by the Institute of Medicine and WHO. Millions of reports have been collected by national and institutional reporting systems across the world9,10,11,12. WHO has published the ‘Minimal Information Model for Patient Safety Incident Reporting and Learning Systems’13, a set of guidelines for standardised incident reporting. However, key information is often embedded within unstructured narrative accounts of the incident, e.g., the ‘free text’ section of reports. The number of narrative incident reports is beyond what can feasibly be reviewed and synthesised by humans, necessitating analysis by computers. Synthesising this plethora of varied free text into meaningful, standardised data that can guide learning and reduce medication errors remains challenging2. To guide the development of models capable of structuring this valuable free-text data into a structured format, a large, fully annotated, and validated corpus of incident reports of medication errors would be valuable.
To our knowledge, no such corpus is yet available, perhaps because the data within incident reports are usually proprietary to certain research groups/institutions and not openly accessible. The Japan Council for Quality Health Care (JQ) has collected more than 100,000 reports of adverse events and near misses, as part of their ‘Project to Collect Medical Near-Miss/Adverse Event Information’ (https://www.med-safe.jp/mpsearch/SearchReport.action). These free-text reports, published in the Japanese language and collected on a national scale since 2010, are openly available and could thus be used to create an annotated corpus of incident reports of medication errors.
Natural language processing and artificial intelligence (AI) are used to autonomously retrieve or extract information for the purpose of learning from incident reports12,14,15,16,17,18. Our earlier studies14,19 provided systematic methodologies for annotating and classifying incident reports of medication errors in a structured manner; the resulting high quality gold-standard data has been published20. In this study, we devised a data creation workflow and developed a machine annotator to create a large corpus of machine-annotated incident reports of medication errors, possessing useful drug-related concept and attribute extraction.
In total, we captured 478,175 medication error-related named entities from 58,568 incident reports of medication errors. For each incident report in the corpus, the relevant named entities have been identified, the discrepancy between what was intended and what actually occurred has been determined and what type of incident the report describes has been recognised. This corpus is the world’s largest publicly available body of annotated incident reports covering concepts and attributes (assertions) related to drug errors. Currently, the machine annotator was trained to detect medication error-related named entities within incident reports and was evaluated using incident reports. However, the machine annotator could be rescaled and applied to other document types, e.g., the free text found in electronic health records. This could be done via additional pretraining and finetuning processes and technical validation.
Furthermore, future studies may be conducted to examine how the human factors that contribute to error (e.g., distractions or workload) can be annotated systematically and added to this kind of dataset. Such knowledge would further enhance our potential to learn from incident reports. We envision the structuring of incident reports to ultimately transform incident learning in healthcare and translate recent advances in AI to improved patient safety. The named entities and incident types identified by this model could be used a starting point for exploring natural language processing-based incident reporting and learning systems. For example, learning health systems2 that help medical professionals retrieve key information from vast corpora of incident reports.
Methods
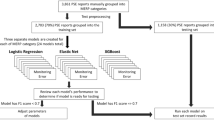
We present our data creation and validation workflow in Fig. 1 and explain the details below.
Collecting free-text incident reports of medication errors from JQ
Our source data was obtained from open-access national-wide incident reports collected by JQ’s Project to Collect Medical Near-Miss/Adverse Event Information’ (https://www.med-safe.jp/index.html)9. The Project collects incident reports from more than 1,000 medical institutions in Japan. The collected incident reports are made up of structured parts (e.g., forms with drop-down menus) and unstructured parts (e.g., free text in ‘comment’ or ‘note’ sections). For details on the JQ structured reporting items and unstructured reporting guidance, please refer to the supplementary information. In our study, we only used the unstructured narrative reports as the source of free text input. The incident reports in this database are categorised according to their type of error, such as errors relating to medication, blood transfusions, treatment, medical devices, surgical drains, examinations, medical care, etc.
There are 121,244 free-text incident reports in total. Our study focuses on annotating incident reports of medication errors; there are 63,856 medication error reports collected from 2010–2020. We only used 58,568 annotatable free-text incident reports (free-text document length > 0) for annotation, though the entire corpus of 121,244 free-text reports was subsequently used to pre-train the annotation model (as described in Section 3 of the methods). The distribution of report lengths is displayed in Fig. 2. All reports were downloaded in.csv format directly from the JQ website. Subsequently, we randomly selected incident reports from the set of incident reports of medication errors for reference labelling in order to perform validation exercises in the next stage. The technical details of how these annotated datasets were created are included in the supplementary information.
Annotation scheme
Our annotation scheme is a framework for annotating the information within incident reports in a way that allows medication errors to be extracted systematically. In developing our annotation scheme19,20,21, we referred to reporting guidelines and methods from the World Health Organization’s Minimal Information Model for Patient Safety13, the European Medicines Agency’s Good Practice Guide22, the NCCMERP index of medication errors23 and other relevant studies24,25. The annotation scheme and our gold-standard ‘Intention and Factuality Annotated Medical Incident Report’ (IFMIR) dataset have been published previously20.
Here, we provide a brief summary of the annotation procedure. First, the drug-related named entities in each report were annotated. Next, each of these named entities was subject to ‘intention/factuality’ analysis, which determined which named entities in the text were intended and which named entities were linked to an event that actually occurred. Lastly, the type of incident for each report was deduced based on associative rules among the identified annotations. As a reference, our system also produces a relation index, which is shared between named entities belonging to the same event. This index was not included in the technical validation. The main steps are described below:
-
Named entity recognition (NER). Named entities are, essentially, the ‘things of interest’ within incident reports – the key units of information that need to be extracted. Named entities that were annotated include ‘drug’, ‘form’, ‘strength’, ‘duration’, ‘timing’, ‘frequency’, ‘date’, ‘dosage’, and ‘route’. Some of these named entities are further separated into subtypes. For example, strength might be described as an amount, e.g., 250 mg, or as a rate, e.g., 250 mg/hr. Note – the current version of the annotation guidelines only covers variables of nouns, proper nouns, compound nouns and numbers.
-
Intention/factuality analysis (I&F). All named entities are labelled as ‘intended and actual’ (IA), ‘intended and not actual’ (IN) or ‘not intended and actual’ (NA). The latter two labels indicate the occurrence of a medication error, as a discrepancy exists between what was intended and what actually happened.
-
Incident type. The incident type, i.e., what kind of medication error occurred, can be determined through an assessment of which named entities were intended and which actually occurred, as well as whether they belong to the primary event. One report might contain more than one incident type. Our pre-defined incidents are as follows: ‘wrong drug’, ‘wrong form’, ‘wrong mode’, ‘wrong strength_amount’, ‘wrong strength_rate’, ‘wrong strength_concentration’, ‘wrong timing’, ‘wrong date’, ‘wrong duration’, ‘wrong frequency’, ‘wrong dosage’ and ‘wrong route’. For errors that fall outside of the above categories, or for free-text inputs that do not contain any error, the incident type is registered as ‘other’.
A detailed explanation of our annotation methodology (June 1, 2023; version 1.0.3) is available online26. Figures 3, 4 demonstrate how free-text incident reports are annotated.
Machine-annotation pipeline
Using both rule-based and deep-learning approaches, a three-layer multi-task BERT model performed the abovementioned annotation tasks automatically on the extracted free text detailing medication errors, as shown in Fig. 5. The development of the deep-learning pipeline is briefly described below.
Pre-training with the full JQ corpus
We adopted a BERT model with the SentencePiece tokenizer pre-trained on Japanese Wikipedia and Twitter corpora (https://github.com/yoheikikuta/bert-Japanese); the BERT model was pre-trained with the JQ incident report corpus of 121,244 free-text documents of all incident types.
Fine-tuning with rule-based annotated data
To prepare for the fine-tuning of data in the first phase, we first developed and evaluated an independent rule-based model. We created a list of unique drug names based on the 2022 list of standard drugs published by Japan’s Ministry of Health, Labour and Welfare (https://www.mhlw.go.jp/topics/2021/04/tp20210401-01.html). Furthermore, the lexicons of named entities (including form, mode and route) were obtained from the gold-standard data20. For named entities that were often presented in numerical form, such as strength – amount, strength – rate, strength – concentration, frequency, date, dosage, timing and duration, the ‘regular expressions’ (i.e., text sequence/pattern) in which they often appear were recorded and used to identify and extract them. Using the free-text incident reports of medication errors, the rule-based model incorporated morphological segmentation and annotated the targeted named entities according to the abovementioned rules. The resulting annotated incident reports of medication errors were used to fine-tune the pre-trained model at the named entity recognition layer.
Fine tuning with gold-standard data
In the second phase of fine tuning, we used the IFMIR gold-standard data (522), to further improve the named entity recognition layer of the BERT model. We treated the intention/factuality task as a problem of named entity-based multiclass classification, and used the known labels from the gold-standard data to fine tune the intention/factuality and relation (reference) layers of the BERT model. 5-fold cross validation was conducted, and we reported the best performing cross-validation result.
Data Records
The final multi-task BERT model was used to annotate the free-text incident reports of medication errors; the deposited dataset at the named-entity level is available on figshare27. The corpus is stored as a single.xlsx file. Figure 6 shows the structure of the annotated data, where each row represents a separate named entity. The annotation results for named entity recognition, intention/factuality, and incident type are shown.
We provide a brief explanation of the contents of a typical annotated incident report, as shown in Fig. 6. The first column heading, ‘no’, is the id number of the extracted named entity, ‘id’ refers to the incident report’s ID and ‘year’ refers to the incident reporting year. The heading ‘report’ contains the original free-text report, ‘namedentity’ is the named entity in question, ‘startidx’ is the location index within the report preceding the first character of the named entity and ‘endidx’ is the location index following the final character of the named entity. The column heading ‘entitytype’ refers to the category the named entity belongs to and ‘errorlabel’ is the named entity’s ‘intention/factuality’ status: ‘IA’, ‘IN’ or ‘NA’. The column heading ‘relation’ refers to the index number of the medication event that the named entity belongs to, and ‘incidenttype’ is the inferred incident type described by the report. If multiple types of error occur, this column will display more than one type of incident. In the absence of an error incident, the incident type is indicated as ‘other’. All items in the data that were originally in Japanese text, e.g., ‘report’ or ‘namedentity’, were translated into English using Google Translate (June, 2023); these translated columns are indicated as ‘translated_(heading)’ in the data.
The 58,658 annotated JQ incident reports of medication errors, displayed at the entity level, are available with an English data dictionary at figshare27. The AI/NLP pipeline and the readme file for the machine annotator are also available28, allowing the multi-task BERT model to be applied towards annotating other free-text incident reports of medication errors.
Furthermore, we share the manually reviewed datasets used for technical validation on figshare29. These are the randomly sampled labelled incident reports from 2010–2020 (n = 40) for testing/internal validation, randomly sampled labelled incident reports from 2021 (n = 20) for external validation, and error-free reports (n = 10) for error analysis. The procedure for preparing these labelled datasets for technical validation is described in the supplementary information.
Technical Validation
All datasets used for technical validation can be found on figshare29. The supplementary information provides all details of technical validation, which comprised cross-validation, internal validation, external validation and error-analysis. We briefly summarise the main results of the cross-validation, internal and external validations in Fig. 7. 5-fold cross validation was conducted to search for model parameters using the IMFIR gold standard data. In the internal validation phrase, we randomly selected 40 incident reports from the 58,568 incident reports of medication errors from 2010–2020 and manually reviewed the reference labels of the selected incident reports. In total, this internal validation dataset contains 263 identified named entities. Since this set of validation data was within the produced annotated dataset, this validation exercise revealed the extent to which the labels are correctly identified within the deposited dataset. Furthermore, we conducted external validation using 20 external reports extracted from 2021 and this set of data contains 110 named entities identified by manual review. The performance evaluation metrics of precision, recall and F1-score are reported in these exercises. The best performing F1-scores at cross-validation for NER and I&F were 97%, and 76%, respectively. In the internal validation exercise, the macro-average of the F1-score across all named entities was 83% and 57% for I&F tasks. In the external validation exercise, these same two metrics were 83% and 50%, respectively. In error analysis, the machine annotator achieved 90% accuracy (false positive rate of 10%) in correctly identifying reports without error.
Usage Notes
This deposited dataset characterises medication incident reports in a structured and machine analysable format. In this way, the dataset serve as a library of similar incidents, connecting users with a wide variety of relevant past cases. Search queries can draw upon combinations of annotations for named entities, intention/factuality and incident types, facilitating the efficient retrieval of similar or relevant cases, particularly high-risk scenarios. For instance, using the identified drug named entity, one can retrieve past incidents of multiple-drug consumption situations, such as polypharmacy and look-alike-sound-alike medications. Furthermore, among the incidents involving incorrect dosages, one can further pinpoint opioid-based drugs to better understand instances of opioid misuse. Furthermore, the machine annotator is capable of on-demand annotation of other free-text incident reports of medication errors in the Japanese language, such as new JQ incident reports, or those collected by individual hospitals.
Digital health system designers can use the machine annotator and the annotated corpus for developing incident report systems, providing function to automated knowledge extraction and retrieval of relevant past cases. We previously demonstrated a proof-of-concept system online (https://www.aiforpatientsafety.com/). The machine annotator can also be further fine tuned and repurposed for other applications related to medication errors, e.g., the free text found in electronic health records.
Incident reports are often proprietary data and are not shared publicly. This open, annotated dataset, generated using ‘real life’ incident reports, has been translated into English for users in other countries. This serves as a valuable and unique resource for systematic studies of medication errors around the world. The workflow and annotation guidelines were designed to be usable in other languages and have been experimentally tested using English incident reports. This large dataset of real-world reports can serve as standard annotated data for natural language processing challenges, examples of which include drug–drug interaction challenges30,31, medication errors/adverse drug challenges32 and n2c2 challenges33.
Annotated data associated with this Data Descriptor are available at figshare27,29 and are released under CC-BY 4.0 to maximise reuse and further study. The original data used to create these reports were collected as part of JQ’s ‘Project to Collect Medical Near-Miss/Adverse Event Information’. When using this deposited dataset, one should also cite the original data source from JQ (https://www.med-safe.jp/index.html).
As described in the section on technical validation, we used samples of randomly selected reports that were manually annotated to validate our model. Despite our best efforts to make the delivered datasets as accurate as possible, some errors might remain due to the high variability in content across reports and the inability to thoroughly validate every label. Please email the corresponding author of this paper to report any errors or provide other comments.
Code availability
The machine annotator code is written in Python 3.9, using libraries for pandas, NumPy, multiprocessing, tqdm, transformers, SentencePiece, neologdn, etc. and can be operated on Google Colab or a local integrated development environment. The annotation guidelines26, deposited dataset27 and machine annotator28 are available at figshare. The manually reviewed and machine-annotated datasets used for technical validation are also available at figshare29.
References
Wong, A., Plasek, J. M., Montecalvo, S. P. & Zhou, L. Natural language processing and its implications for the future of medication safety: a narrative review of recent advances and challenges. Pharmacotherapy 38, 822–841, https://doi.org/10.1002/phar.2151 (2018).
Patient safety incident reporting and learning systems: technical report and guidance. (World Health Organization, 2020).
Global patient safety action plan 2021–2030: towards eliminating avoidable harm in health care. (World Health Organization, 2022).
Keers, R. N., Williams, S. D., Cooke, J. & Ashcroft, D. M. Prevalence and nature of medication administration errors in health care settings: a systematic review of direct observational evidence. Ann. Pharmacother. 47, 237–256, https://doi.org/10.1345/aph.1R147 (2013).
Makary, M. A. & Daniel, M. Medical error—the third leading cause of death in the US. BMJ 353, i2139, https://doi.org/10.1136/bmj.i2139 (2016).
Morimoto, T., Gandhi, T., Seger, A., Hsieh, T. & Bates, D. Adverse drug events and medication errors: detection and classification methods. BMJ Qual. Saf. 13, 306–314, https://doi.org/10.1136/qshc.2004.010611 (2004).
World Health Organization. Medication without harm. World Health Organization https://www.who.int/initiatives/medication-without-harm (2017).
Donaldson, L. J., Kelley, E. T., Dhingra-Kumar, N., Kieny, M. P. & Sheikh, A. Medication without harm: WHO’s third global patient safety challenge. Lancet 389, 1680–1681, https://doi.org/10.1016/s0140-6736(17)31047-4 (2017).
Project to collect medical near-miss/adverse event information: project details and how to participate [Iryō jiko jōhō shūshū-tō jigyō jigyō no naiyō to sanka hōhō]. (The Japan Council for Quality Health Care, 2022).
Cooper, J. et al. Nature of blame in patient safety incident reports: mixed methods analysis of a national database. Ann. Fam. Med. 15, 455–461, https://doi.org/10.1370/afm.2123 (2017).
AIRS (Advanced Incident Reporting System): framework for HA wide incident reporting. (Hong Kong Hospital Authority, 2017).
Wang, Y., Coiera, E., Runciman, W. & Magrabi, F. Using multiclass classification to automate the identification of patient safety incident reports by type and severity. BMC Med. Inform. Decis. Mak. 17, 84, https://doi.org/10.1186/s12911-017-0483-8 (2017).
Minimal information model for patient safety incident reporting and learning systems: user guide. (World Health Organization, 2016).
Liu, J., Wong, Z. S. Y., So, H. Y. & Tsui, K. L. Evaluating resampling methods and structured features to improve fall incident report identification by the severity level. J. Am. Med. Inform. Assoc. 28, 1756–1764, https://doi.org/10.1093/jamia/ocab048 (2021).
Gong, Y., Song, H.-Y., Wu, X. & Hua, L. Identifying barriers and benefits of patient safety event reporting toward user-centered design. Saf. Health 1, 7, https://doi.org/10.1186/2056-5917-1-7 (2015).
Wang, Y., Coiera, E. & Magrabi, F. Can unified medical language system–based semantic representation improve automated identification of patient safety incident reports by type and severity? J. Am. Med. Inform. Assoc. 27, 1502–1509, https://doi.org/10.1093/jamia/ocaa082 (2020).
Wong, Z. S. Y. Statistical classification of drug incidents due to look-alike sound-alike mix-ups. Health Inform. J. 22, 1–17, https://doi.org/10.1177/1460458214555040 (2014).
Wong, Z. S. Y., So, H. Y., Kwok, B. S., Lai, M. W. & Sun, D. T. Medication-rights detection using incident reports: A natural language processing and deep neural network approach. Health Inform. J. 26, 1460458219889798, https://doi.org/10.1177/1460458219889798 (2019).
Wong, Z. S. Y. et al. Annotation guidelines for medication errors in incident reports: validation through a mixed methods approach. Stud. Health Technol. Inform. 290, 354–358, https://doi.org/10.3233/shti220095 (2022).
Zhang, H. K., Sasano, R., Takeda, K. & Wong, Z. S. Y. Development of a medical incident report corpus with intention and factuality annotation. LREC 2020 4578–4584 (2020).
Shiima, Y. & Wong, Z. S. Y. Classification scheme for incident reports of medication errors. Stud. Health Technol. Inform. 265, 113–118, https://doi.org/10.3233/shti190148 (2019).
Good practice guide on recording, coding, reporting and assessment of medication errors. (European Medicines Agency, 2015).
NCC MERP index for categorizing medication errors. (National Coordinating Council for Medication Error Reporting and Prevention, 2022).
Westbrook, J. I., Woods, A., Rob, M. I., Dunsmuir, W. T. & Day, R. O. Association of interruptions with an increased risk and severity of medication administration errors. Arch. Intern. Med. 170, 683–690, https://doi.org/10.1001/archinternmed.2010.65 (2010).
Westbrook, J. I. et al. What are incident reports telling us? A comparative study at two Australian hospitals of medication errors identified at audit, detected by staff and reported to an incident system. Int. J. Qual. Health Care 27, 1–9, https://doi.org/10.1093/intqhc/mzu098 (2015).
Wong, Z. S. Y. Annotation guidelines for incident reports of medication errors. figshare https://doi.org/10.6084/m9.figshare.25025984.v1 (2024).
Wong, Z. S. Y. A corpus of machine-annotated incident reports of medication errors. figshare https://doi.org/10.6084/m9.figshare.21541650.v3 (2023).
Wong, Z. S. Y. Machine annotator. figshare https://doi.org/10.6084/m9.figshare.25026059.v2 (2024).
Wong, Z. S. Y. Gold standard/manual reviewed annotated datasets for technical validation. figshare https://doi.org/10.6084/m9.figshare.23504922.v1 (2023).
Segura-Bedmar, I., Martínez, P. & Herrero-Zazo, M. SemEval-2013 task 9: extraction of drug-drug interactions from biomedical texts (DDIExtraction 2013). SemEval 2013 341–350 (2013).
Segura-Bedmar, I., Martinez, P. & Sanchez-Cisneros, D. The 1st DDIExtraction-2011 challenge task: extraction of drug-drug interactions from biomedical texts. SEPLN 2011 1–9 (2011).
University of Massachusetts, Lowell. NLP challenges for detecting medication and adverse drug events from electronic health records (MADE1.0). MADE1.0 Project https://bio-nlp.org/index.php/projects/39-nlp-challenges (2018).
Uzuner, Ö., Stubbs, A. & Lenert, L. Advancing the state of the art in automatic extraction of adverse drug events from narratives. J. Am. Med. Inform. Assoc. 27, 1–2, https://doi.org/10.1093/jamia/ocz206 (2019).
Acknowledgements
This research was supported by the Japan Society for the Promotion of Science KAKENHI (Grant No. 18H03336) and the publication fee is supported by the President’s Special Fund from St. Luke’s International University. Jiaxing LIU is supported by the Fundamental Research Funds for the Central Universities (No. 2722023BQ053) and Hubei Province Postdoctoral Funding Project. We acknowledge the contributions of Dentsu Digital, Japan, and Dentsu Data Artist Mongol, Mongolia, for program development. The team members include Amarsanaa Agchbayar, Batmunkh Batsaikhan, Telmuun Enkhbold, Khatanbold Batzorig and Od Ganzorig. The authors wish to express their gratitude to Dr. Ryohei Sasano and Mr. Hongkuan Zhang for their early contributions in developing the annotation guidelines and the Gold Standard IFMIR dataset, which subsequently enabled this annotated incident reports study.
Author information
Authors and Affiliations
Contributions
The study was conceived by Z.W. and U.S. Z.W. and J.L. contributed to the study design and implementation. The data described were retrieved and interpreted by Z.W., J.L. and N.W. Z.W. and N.W. led the writing of the paper and all authors approved the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wong, Z.S.Y., Waters, N., Liu, J. et al. A large dataset of annotated incident reports on medication errors. Sci Data 11, 260 (2024). https://doi.org/10.1038/s41597-024-03036-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03036-2
- Springer Nature Limited