Abstract
Despite exponential growth in ecological data availability, broader interoperability amongst datasets is needed to unlock the potential of open access. Our understanding of the interface of demography and functional traits is well-positioned to benefit from such interoperability. Here, we introduce MOSAIC, an open-access trait database that unlocks the demographic potential stored in the COMADRE, COMPADRE, and PADRINO open-access databases. MOSAIC data were digitised and curated through a combination of existing datasets and new trait records sourced from primary literature. In its first release, MOSAIC (v. 1.0.0) includes 14 trait fields for 300 animal and plant species: biomass, height, growth determination, regeneration, sexual dimorphism, mating system, hermaphrodism, sequential hermaphrodism, dispersal capacity, type of dispersal, mode of dispersal, dispersal classes, volancy, and aquatic habitat dependency. MOSAIC includes species-level phylogenies for 1,359 species and population-specific climate data. We identify how database integration can improve our understanding of traits well-quantified in existing repositories and those that are poorly quantified (e.g., growth determination, modularity). MOSAIC highlights emerging challenges associated with standardising databases and demographic measures.
Similar content being viewed by others
Background & Summary
The ecological sciences have recently joined the open data revolution1,2,3. As a result of initiatives promoting open data, total species distribution records measure in the hundreds of millions4,5. Functional trait data exist for tens of thousands of species across the globe6,7,8. Global distributed networks, remote sensing, and other ecological sensor data networks are feeding information into the open data space, and we are experiencing a rapid increase in the number of ecological databases. The growth of open data is reflected in state-of-the-art climate models (e.g., ERA-5 [https://www.ecmwf.int/en/forecasts/dataset/ecmwf-reanalysis-v5]) becoming available at fine spatial and temporal resolutions suitable for biological analyses9,10, the growth of behavioural trait datasets11, and large population datasets (Living Planet Data, successor to the Global Population Dynamics Database [http://livingplanetindex.org/data_portal]; Human Mortality Database;12 Human Fertility Database;13 AnAge Database;14 DATLife [Max Plank Institute of Demographic Research 2022; https://datlife.org/]). Data access and scaling has extended to biological complexity at the ecological community ecology level (BioTime;15 Web of Life;16 metaCommunity Ecology: Species, Traits, Environment and Space (CESTES17); Environmental Data Initiative [https://environmentaldatainitiative.org/]). The growth of records in ecology datasets and complementary environmental datasets is enabling us to test ecological theory at larger and more complex scales. In line with the expansion of open access data practices, however, there is a need to improve and coordinate data standards to guide the systematic collection and management of data across different trait collection programmes18,19,20.
Despite the increased availability of biological data, synthesizing datasets for analysis is hampered by the lack of complementarity between databases. The rise of data sharing and proliferation of databases can encourage a fragmented and decentralized information landscape unless there is active coordination. Interoperable data systems reflect continuity in the format of data types and structure to allow compatibility across computers and software. Converting datasets into interoperable formats may require the transformation of constituent data types into standardized spatial, temporal, and measurement scales, accounting for known differences/biases in methods21. The need to improve interoperability across datasets is demonstrated by the widescale emergence of data harmonization initiatives across fields of ecology22,23,24. In the past decade, dozens of initiatives have taken shape to both centralize data from existing datasets and to improve data interoperability: standardising units, scales, and terminology for comparative purposes25,26. Unifying data formats and streamlining their integration unlocks the potential for existing datasets to answer questions that cut across levels of biological complexity. Linking together levels of biological complexity is critical for identifying how phenomena emerge and transmit across different levels of biological organisation, upscaling and downscaling through biological systems. For example, the critical linkages between genetics and biochemistry27, biochemistry and physiology28, and physiology and demography29,30 have benefitted from dataset integration.
The limitations of global-scale datasets are shifting away from data availability and toward data interoperability. For functional traits, momentum toward data integration is evidenced by recent database initiatives31 and global networks that, like the Open Trait Network (https://opentraits.org/), aim to standardize and integrate trait data across taxa18. Despite major improvements in the consolidation and accessibility of trait data, there is not yet a single-source centralized database spanning behaviour, physiology, habitat, and other trait data for a wide range of species. Existing databases are often linked by taxonomy (e.g., FishBase32, CoralTraits33, MammalBase34, AmphiBio35); trait type (e.g., Tree of Sex36, TreeBase37, Xylum Functional Traits38); data type (e.g., GBIF (https://www.gbif.org/), MOL39, TetraDensity40); or a combination of the taxonomies and traits (WooDiv41, CarniDiet42). A number of other databases take a more general approach in their thematic scope, but are still constrained to a limited set of traits and taxonomy (e.g., Amniote43, Pantheria44, BIEN7,45, TRY23). MOSAIC offers a platform that integrates databases in the remit of species with structured population models across their ecological traits.
Here, we introduce MOSAIC, a centralized database of trait data across the Tree of Life. MOSAIC is an open-access database that complements the existing demographic data available in the COMPADRE Plant Matrix Database46, COMADRE Animal Matrix Database47, and the new PADRINO IPM Database48. MOSAIC v. 1.0.0 includes 14 frequently used traits that encompass morphological, reproductive, dispersal, and habitat type attributes for some 300 species of animals and plants. Additional traits will be added in the future (see Future Direction, below). MOSAIC allows users to integrate structured population data to probe larger questions through the collection, curation, and complementarity of relevant contextual data.
Methods
Scope and coverage of MOSAIC
The MOSAIC database (v. 1.0.0) includes 14 key trait records across 300 species (Fig. 1). MOSAIC is designed to provide complementary data for analysis in connection with structured population models: matrix population models (MPMs49), where state variables are discrete (e.g., age50, ontogeny/development51, discrete classes of size52), and integral projection models (IPMs53), where the state variables are continuous (e.g., size54, mass55, parasite load56). The traits included in MOSAIC 1.0.0 were identified as a set of physical, physiological, geographic, and behavioural attributes of most immediate relevance to demographic research (see Table 1; more information at https://mosaicdatabase.web.ox.ac.uk). Traits were also selected in consideration of the lack of standardized and centralized databases for certain traits (e.g., volancy, modularity, and growth indeterminacy; see Fig. 2 for trait variance and taxonomic structure of select traits excluded from existing databases). Importantly, we note that MOSAIC is not a general dataset for analysis of functional traits, as this is available through other extensive repositories (e.g., TRY23, BIEN7,45). Instead, the focus of MOSAIC is on providing taxonomic coverage to species with open-access structured population models available in the COMADRE47, COMPADRE46, and PADRINO48 databases (See Figs. 3, 4 for spatial scope and taxonomic scope with respect to structured population databases, respectively). MOSAIC provides a much-needed interoperability between existing databases that are relevant to demography. By doing so, MOSAIC helps to fill critical data needs of population ecologists and functional trait ecologists (see Fig. 5 for relevant covariance structure).
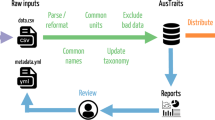
Structure of the MOSAIC database (v1.0.0). The MOSAIC database comprises a combination of existing trait records centralised from data servers, databases, and datasets and new records collected from the primary literature by the MOSAIC team. Existing records are labelled, MOSAIC-A, in the mosaic metadata attribute field and include provenance in the metadata. New data records that adhere to existing data standards and that fit into the remit of an existing database (i.e., a data gap filled by the MOSAIC team) are labelled MOSAIC-B. New records that do not have an existing data standard or for which there is not an existing database are labelled MOSAIC-C.
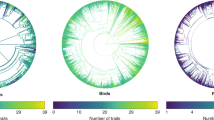
Trait fields in the MOSAIC database (v1.0.0). The MOSAIC database contains a combination of continuous and discrete trait fields. Six of the 14 trait fields are illustrated here for animals, and organised by trait level (all discrete fields) and taxonomic classification. Trait distributions vary strongly by trait and taxonomy. Observed variation in trait values by taxonomic group is a prerequisite condition to their potential value toward explaining reported variation in vital rates and resulting demographic properties across the Tree of Life. Species included in the MOSAIC database are limited to those for which stage-structured population data exist in the COMADRE, COMPADRE, and PADRINO databases.
The spatial distribution of MOSAIC (v1.0.0) records relative to the (a) COMADRE and (b) COMPADRE databases. The COMADRE and COMPADRE dataset include animal and plant demographic data, respectively, on all continents except Antarctica, with a substantial bias to North America and Central Europe. MOSAIC introduces trait records that include models in all major geographies of COMADRE and COMPADRE. The map shows the density of matrix population models globally (color graded by density per area in ca. 150 km2 hexagonal cells). Cells bordered by white represent localities of population models for which there are trait records included in the first release of the MOSAIC trait database. Species prioritised in v1.0.0 of MOSAIC did not include the PADRINO dataset, so no distribution figure is shown for integral projection models.
Phylogenetic mapping of traits included in MOSAIC (v1.0.0). (a) Phylogenetic distribution of seven major traits for animals: (1) growth determination; (2) sexual dimorphism; (3) monogamy; (4) gonochory; (5) volancy; (6) dispersal; and (7) biomass. (b) Phylogenetic distribution of four traits in plants: (1) dispersal capability; (2) dispersal type; (3) dispersal mode; and (4) dispersal class. Figures developed in R using the ggtree package.
Trait covariance in the MOSAIC database (v1.0.0). Some traits exhibit high correlation in the MOSAIC database, such as between growth determination and between growth regeneration. Trait associations are expected to occur in the MOSAIC dataset and may reflect widespread constraints or statistical anomalies, particularly when dealing with small samples or specific taxonomic subgroups. Trait covariation can be symptomatic of biomechanical constraints (e.g., flight and biomass), major growth characteristics (e.g., modularity and growth determination), or other past or presently compelled associations (e.g., height and vessel density).
Data sources
MOSAIC is both a meta-database (a database of databases) and a database in its own right, containing new trait records from primary literature (Fig. 1). The MOSAIC database contains records centralized from existing datasets where functional traits relevant to population ecologists can be openly accessed and redistributed (e.g., BIEN7,45). Licensing terms of these databases are included in the supplement (Appendix S1: Database Licensing Terms for Constituent Databases of MOSAIC). The records reflected in the MOSAIC database do not encompass the entirety of the source databases but are instead partial facsimiles of those databases that reflect records relevant to demographic databases (COMADRE, COMPADRE, PADRINO). MOSAIC has three major components: (1) trait records sourced from existing databases (22%); (2) trait records newly procured through searching the primary literature (71%); and (3) trait record markers indicating the presence of records in non-open-access database (7%). MOSAIC trait markers exist for one of two reasons: the database containing the records of interest does not allow records to be accessed or limits redistribution rights behind individual registration and/or specific use applications; or records in other databases contain multiple records for a species, which do not currently fit within the data structure of version 1.0.0 of MOSAIC (see Future Targets for MOSAIC).
In addition to identifying whether a trait record is new, the “MOSAIC” attribute field also identifies whether the attribute field (i.e., trait name) is part of an existing database. For example, the MOSAIC field might indicate that a record for specific leaf area is new for a specific species, and also that the attribute is part of databases such as TRY23 or BIEN7,45. By contrast, a new species record for volancy would indicate that there are currently no databases that systematically collect data on volancy attributes and therefore that all volancy records in the MOSAIC database are new.
Organisation of sources in MOSAIC
The MOSAIC attribute field is a factorial variable with three levels: MOSAIC-A, MOSAIC-B, and MOSAIC-C. The first of these levels, MOSAIC-A (Fig. 1), labels only records that reflect existing databases (i.e., provenance of an existing data acquisition service); the second, MOSAIC-B, labels new records collected by the MOSAIC team that are in a trait field within the scope of an existing database initiative (e.g., specific leaf area in BIEN); and the third, MOSAIC-C, labels new records collected by the MOSAIC team on traits for which there is not currently a database initiative centralising records. If a datum has been adopted from another dataset or database, then the relevant source is referenced in the Database attribute column. Note that this value will be “NA” for all MOSAIC-C records, logically. Over time, data sharing will move records in MOSAIC-B to MOSAIC-A as the MOSAIC-B traits are assimilated in the database networks that specialize on an existing trait (i.e., data feedback; see Fig. 1).
Sources and provenance of records
Because of existing limitations on data access, some datasets cannot be transferred into MOSAIC. Where data exist outside of the MOSAIC platform, but have restricted access, the MOSAIC database directs users toward the appropriate database on a trait and taxa-specific level (see metaMOSAIC). The MOSAIC User Guide (Appendix S2) explains differences between data gaps that are yet to be reviewed and those that are true gaps (e.g., volancy/flight capability of plants). Data were obtained through searching peer-reviewed records and PhD dissertations of ISI Web of Science, Scopus, and Google Scholar using key words pertinent to the species name and trait field in question (see Appendix S3 for a list of keywords queried). The archives of data repositories, including the Figshare digital repository57 (https://figshare.com/articles/dataset/MOSAIC_trait_database/21035857; see Appendix S3 for a complete list of repositories reviewed – to be maintained hereafter on the MOSAIC portal) and journal archives that have a high occurrence of data publishing, including Nature Scientific Data, Methods in Ecology and Evolution, and Journal of Ecology were carefully reviewed. MOSAIC also reflects a review of data from data aggregating servers, such as the open traits network (https://opentraits.org/), the ecological data wiki (https://ecologicaldata.org/), environmental data initiative (https://environmentaldatainitiative.org/), and databases that aggregate other databases (e.g., BIEN7,45 and TRY23). The complete set of key words used in this review is detailed in a supplement to the User Guide (Appendix S3). A current list of databases reviewed in the development of MOSAIC is included in Appendix S4. Suggested data sources and key terms can be submitted through the MOSAIC data portal.
metaMOSAIC: licensed data, access limitations, and restricted redistribution of existing records and databases
Not all datasets permit open use, dissemination, and redistribution of their trait data. Where limitations on the data collation and redistribution apply, there may be application procedures, registration, and other actions necessary for an individual to obtain access to specific trait records for analysis (e.g., TRY). MOSAIC centralizes the metadata for datasets that do not allow data to be redistributed to help navigate to relevant data resources outside the scope of open access. Records in limited access databases can be searched in MOSAIC by taxonomic group and by trait. MOSAIC links researchers to application materials for requesting access to those limited-access databases. The data access of licensed or non-open-access databases is stored in a data object called metaMOSAIC that is an extension of the MOSAIC database. Thus, the MOSAIC dataset provides data where it is accessible and metaMOSAIC guides researchers to where data exist with registration. When searching fields in the MOSAIC database, the metaMOSAIC adjunct dataset indicates if data are available in these ancillary sources (see User Guide in Appendix S2 for more information) and provide links to pertinent sources and data through the provider. metaMOSAIC is part of the main database object accessible through the MOSAIC portal.
Database updates
The MOSAIC database will be updated as new data are added to the COMADRE, COMPADRE, and PADRINO databases. Updates of the MOSAIC database will account for newly discovered data sources and new literature that adds to or changes the species-level traits in the database, as well as correct errors from earlier versions. New MOSAIC versions will be released periodically with a notice published on the website, in the data object metadata, the mosaic GitHub page (https://github.com/mosaicdatabase), and through updates in associated packages in conformance with standard semantic versioning (a three-part version code reflecting major, minor, and patch updates, in respective positions). Updates will be published to the mosaic portal (https://mosaicdatabase.web.ox.ac.uk), social media (Twitter: @mosaicdatabase), and GitHub (https://github.com/mosaicdatabase). The MOSAIC portal includes a location for submission of recommendations for additions or suggested changes to the database.
Future targets of MOSAIC
Versioning
MOSAIC database updates and version history will be posted to the MOSAIC portal in the future.
Taxonomic scope
Future versions of MOSAIC are scheduled for expansion to include 50 core traits across some 1,500 species (the current entirety of COMPADRE, COMADRE, and PADRINO; MOSAIC target traits; see User Guide [Appendix S2]). The MOSAIC core traits list will guide the future roll-out of attributes gathered and stewarded by the MOSAIC team, but additional trait suggestions can be submitted through the online portal.
Interspecific variation
Future versions of the MOSAIC database will adopt a file structure that will accommodate multiple records per species. Once records for COMADRE, COMPADRE, and PADRINO are fully populated across the MOSAIC traits with mean, pooled, or other representative quantities (e.g., mean leaf size for all plants or adult bodymass of animals), secondary records will be added. Existing trait databases may contain multiple records per species (see, for example, structure of COMADRE47, COMPADRE46, BIEN7,45, TRY23), although some databases host a single record per species’ trait, such as age and growth rate for animals in AnAge14. To facilitate research into intra-specific trait variation58,59,60, MOSAIC provides provenance of records, whether records were subject to selection or merger (means or pooling), and fields that identify whether multiple records are known to exist. Where records for a given species were isolated from existing databases, mean values are often retrieved, and the database sourcing additional data is noted in the database under the attributed field (“Additional Trait Data Available” field).
citMOSAIC: Citizen science
In addition to metaMosaic, which guides users to licenced data not reported in the MOSAIC database, MOSAIC plans to roll out a database of identical structure to MOSAIC that gathers information from citizen science datasets. Like MOSAIC, citMOSAIC will have three components (citMOSAIC-A, citMOSAIC-B, citMOSAIC-C), reflecting the same relationship of databases and fields to MOSAIC. citMOSAIC will be kept independent of the main MOSAIC database to avoid conflation of peer reviewed literature and PhD dissertations from citizen science data. Where appropriate to use datasets together, the metadata, query functionality, and design of citMOSAIC will mirror that of MOSAIC to promote interoperation of databases. citMOSAIC will be downloadable from the MOSAIC portal website.
Data Records
The current version of MOSAIC (v. 1.0.0) is deposited as series of flat data files (comma separated values and text files) in Figshare (https://figshare.com/articles/dataset/MOSAIC_trait_database/21035857). The data in this paper and complete database are also published on the MOSAIC website https://mosaicdatabase.web.ox.ac.uk and associated GitHub repository https://github.com/mosaicdatabase/mosaicdatabase. All data is open access without restrictions on access.
The MOSAIC database can be downloaded directly into R as a series of flat tabular data (comma separated values and text files) on Figshare (https://figshare.com/articles/dataset/MOSAIC_trait_database/21035857) and, optionally, as an S4 object (see Usage Notes—Accessing MOSAIC). Future versions of the MOSAIC datasets with any corrections and new data records will be updated to the GitHub page (github.com/mosaicdatabase/mosaicdatabase) linked with the direct download in R and updated to Figshare. Data use and redistribution is covered by a Creative Commons CC-BY license, with unrestricted use and modification with attribution, consistent with MOSAIC’s constituent databases (Appendix S1).
Description of the individual trait fields are included in Table 1. Detailed discussion of each data field, units, precision, and cautionary notes included in the MOSAIC User Guide (Appendix S2). In its first version (v. 1.0.0), MOSAIC data has 41% density coverage across 14 core trait fields of approximately 300 species. MOSAIC provides 100% density of climate data for all species in COMADRE, COMPADRE, and PADRINO for which there are GPS locations (86% of all records across the databases). 71.9% of species in COMADRE, COMPADRE, and PADRINO had records in the Online Tree of Life phylogeny included in MOSAIC. Data completeness varies greatly amongst trait fields, as does the sourcing of data from primary and secondary datasets (Fig. 2).
Across formats, each tabular row or vector element represents a single species. Columns are representative of attributes or metadata associated with attributes. Unique identifiers associated with the matrix population models and integral population models in COM(P)ADRE and PADRINO databases, respectively, to link demographic models with trait records. In the current MOSAIC version (v. 1.0.0), only one record per species is released, but future versions will incorporate multiple records per species.
Technical Validation
The technical validity of records in the MOSAIC database is based on three levels of review. First, all records in MOSAIC, regardless of whether they are collected from primary or secondary data sources, are obtained from either peer reviewed journals or scholarly equivalent documents (e.g., PhD dissertation). Second, all data sources in MOSAIC were reviewed to ensure that they are reasonably representative of the trait record for the species (e.g., screened for sample size, geography relative to the pertinent population record(s), and methods suitable to the trait). Third, MOSAIC will be periodically reviewed, added to, and amended, additive to any revisions that might come about through the feedback portal. As a result, MOSAIC will be adaptively managed to ensure that all records are of suitably high-quality and grounded in peer-reviewed data. MOSAIC also contains metadata for all records, ensuring provenance to original records and secondary standardisation.
Peer-reviewed data
All data in MOSAIC are either published in a peer reviewed academic journal, PhD dissertation, or other peer reviewed source (e.g., Oxford Bibliographies). Unique identifiers for the publications that source the data is provided for every record. Prior to the review of the record by the MOSAIC team, all trait records were technically scrutinized and reviewed by more than one subject area expert. In addition to peer review of the publication record, many MOSAIC records are sourced from existing databases that impose technical standards beyond those imposed by the initial publication and have technically assessed the quality of underlying data.
Record review and selection
The MOSAIC team evaluated whether records were consistent across the literature. Where multiple records were identified containing different numeric values for a continuous trait, the MOSAIC team selected a focal record based on: (1) sample size, (2) geographic extent, (3) geographic proximity to the site, and (4) method of measurement. If multiple records were identified for factorial traits that were in conflict with one another, then those records were excluded from the initial release of MOSAIC v.1.0.0. Only unanimously consistent factorial trait values were included in the first version of MOSAIC. At least two members of the MOSAIC team assessed records in every case to ensure that there was agreement in the sourcing of the data.
Quality checks
The MOSAIC team reviewed the data for any outliers. In addition to reviewing for general anomalies from the data variance structure, as part of the data collection protocol, the MOSAIC team screened data for bounds applicable to each trait (see User Guide, Appendix S2). These value bounds ensured that all reported data were within the range of biologically realistic values as an additional safeguard against spurious values. Beyond confirming the general data structural integrity, the MOSAIC team compared the overlay of records in COMADRE, COMPADRE, and PADRINO with the source data for MOSAIC to ensure congruence of the data transferred from source databases. All automated data overlays were manually checked for their integrity against source data.
Ongoing development & growth
The MOSAIC database will continue to grow and, in the future, will accommodate multiple trait records per species. This trait record redundancy will provide another level of protection against spurious records, in addition to providing quantification of trait variance. Future database development plans include a schedule to incorporate GPS coordinates to formalize the distance between each trait record and the corresponding population records.
Metadata
The MOSAIC database contains the source data for each trait record, including (1) Author(s), (2) Year of Publication, (3) Journal of Publication, (4) DOI/ISBN of the publication, and (5) Inclusion in other databases. Future versions of the database may contain additional metadata for the trait data.
Usage Notes
Accessing MOSAIC
A guide for downloading the MOSAIC database can be accessed in the Supplementary Material (Appendix S2). The MOSAIC database is open access and can be downloaded into R as a csv using the below line of code:
read.csv(“https://raw.githubusercontent.com/mosaicdatabase/mosaicdatabase/main/MOSAIC_v1.0.0.csv”)
The MOSAIC database is a comma separated value (csv) format that is publicly archived under a CC-BY-4.0 license accessible from Figshare (https://figshare.com/articles/dataset/MOSAIC_trait_database/21035857), GitHub (https://github.com/mosaicdatabase/mosaicdatabase), and the MOSAIC portal (https://mosaicdatabase.web.ox.ac.uk). Phylogenetic data is formatted for use and manipulation using the open-source R-package ape (Analyses of Phylogenetics and Evolution; Schliep & Paradis 2019) which can be installed by CRAN.
Mosaic User Guide, Vignette, and Data Collection Protocols are all included in the Supplemental materials (Appendix S2, S3, and S5). The User Guide specifies the classifications, precision, and data types for each trait field. The MOSAIC User Guide details the metadata on the structure of data for each trait (e.g., species, genus, mosaic index). An updated list of the databases that are directly or indirectly addressed by the MOSAIC database is maintained on the MOSAIC portal.
Field values
MOSAIC records contain one of three values: “NA,” “NDY,” or a trait value that can be numeric, factorial, or a character string, as discussed further in the User Guide. NAs apply to fields that have been reviewed and which do not apply to the species of interest. For example, plants do not have flight capacity, and thus NAs apply to them for this trait. Likewise, height and canopy size are key morphological dimension of plants that may not transfer meaningfully to most vertebrates, where adult biomass is a more relevant61,62 and oftentimes used trait. By contrast, NDYs represent “Not Digitized Yet”, indicating fields/species that have not been reviewed for potential records. All other values will reflect the units described in the User Guide.
Database navigation
The User Guide contains detailed information on navigating the MOSAIC R data object. The primary MOSAIC object contains species-specific attribute values for 14 traits. Climate data and phylogeny are independent files accessible in the same locations described above (csv and phylo formatted, respective). Climate is an independent file because it is based solely on model-specific coordinates (and therefore has multiple values per species). Phylogeny is an independent file because it is a phylo object (a special kind of list object in R accessed through ape) and therefore is not in a format amenable to species-specific csv summary.
The primary MOSAIC file contains 14 fields and can be queried through S3 syntax. Rows represent species and are included in the column titled: species_accepted. Columns represent attributes that are either the trait value or trait metadata. The base format for accessing data is: mosaic$trait.name. In the aforementioned syntax, the trait.name should reflect one trait (e.g., volancy). Metadata can be accessed with the syntax: mosaic$trait.name_metadata, where metadata is replaced by the name of the metadata field (e.g., journal, doi, author). Species names are specified in conformance with the Catalogue of Life (www.catalogueoflife.org), also consistent with the COMPADRE, COMADRE, and PADRINO databases. The User Guide provides specific guidelines for querying fields within the data object in R and for negotiating the dataset in finer detail (Appendix S2).
Error reporting
Users can submit errors for correction by email to: mosaicdatabase@biology.ox.ac.uk. The MOSAIC portal also has an Error Report page for reporting potentially erroneous records, or to query additional questions (https://mosaicdatabase.web.ox.ac.uk/suggested-additions-error-reporting; but see also FAQ: https://mosaicdatabase.web.ox.ac.uk/frequently-asked-questions). Potential errors can be reported anonymously or with contact information (e.g., name, email). Decisions on reported errors will be disclosed on the Error Report page (https://mosaicdatabase.web.ox.ac.uk/suggested-additions-error-reporting) and to the reporting party if contact information is included in the request.
Recommended records
Users can submit records by email to: mosaicdatabase@biology.ox.ac.uk. The MOSAIC portal also has a Recommended Records page (https://mosaicdatabase.web.ox.ac.uk/suggestion-additions) for reporting suggested records that are not included in the MOSAIC database. Recommendations can be made with or without contact information. Contact information will be used exclusively for clarifying questions and updating the commenter when records are included. MOSAIC will report decisions on the Recommendations Incorporated page (https://mosaicdatabase.web.ox.ac.uk/suggestion-additions). Users may also request new data fields to be prioritized in future rollouts. Given the realities of limiting resources, The MOSAIC team will do their best to include the requested records in future versions.
Cautionary notes
Records in the MOSAIC database are gathered and standardized under the protocols detailed in the User Guide (Appendix S2; also available through the MOSAIC portal). Users should be attentive to the precision, levels, and context of data in MOSAIC when used for analysis. For those records in the dataset that come from multiple individuals, we present them as statistical components (e.g., minimum, maximum, or mean trait values). Functional traits in MOSAIC may be estimated from populations studied in COMADRE, COMPADRE, PADRINO, or other databases (Table S4). The potential temporal and spatial mismatch between databases that are linked in an analysis merits close attention63. The studies in the MOSAIC database also include research conducted by different investigators using independent tools, technologies, sample designs, and study methods. The influence of research methods and instruments on the error values in the dataset may require additional consideration for potential bias, noise, and imprecision. Where more than one life history trait value exists for a given species, MOSAIC users will need to determine whether averaging or selective filtering to one study is most appropriate in view of the specifics of the given research question. In certain cases, trait values for a species might only be available for a single st/age and therefore may not provide a complete picture of the trait variation amongst st/ages. Users are encouraged to be cautious when contextualising the scope of representation of the values in the database and their analyses.
Representation, variance, and Bias in MOSAIC v1.0.0
MOSAIC has the potential to help identify macroecological patterns and guide targeted experimental studies that can mechanistically examine the causes and correlates of demographic variability. MOSAIC leverages thousands of animal and plant species housed in COMPADRE, COMADRE, and PADRINO and offers promise for evaluating general hypotheses and identifying novel ones from newly discovered patterns. Despite the inductive value and generality of macroecological inference64,65,66, caution is required in inferring process and cause from trait-demographic patterns using MOSAIC. MOSAIC is a starting point and contextualising instrument, not a stand-alone tool for inferring how traits determine demography and/or how demographic processes may shape traits. In its version 1.0.0, MOSAIC contains a high degree of variance in trait values known to shape demographic outcomes across major taxonomic groups. For example, determinant growth is present for 0% of Amphibians and Birds and 100% of Bivalves, while volancy is present for 0% in Reptiles, Amphibians, and Bony Fish and 77% in Birds (Fig. 2). Animal adult biomass and plant height follow lognormal trait distributions (see Appendix S5). 11% of mammals are indeterminate growers vs. 54% of reptiles and 0% birds, and 95% of mammals are monogamous compared to 100% of reptiles and 20% of birds in MOSAIC. Recent studies have examined how different vital rates are explained by functional traits67,68,69,70. However, understanding how trait variation across taxa translates to demographically influential properties remains underdeveloped.
MOSAIC’s initial release (v1.0.0) includes records for all major regions of the globe for which we have structured population models (Fig. 3). Nevertheless, species trait values are not necessarily gathered from the same localities as population models (see future directions for more information on systematising spatial mismatch). This is an important consideration for users of MOSAIC (and more generally of trait-based approaches) wishing to bring together functional traits and demographic rates, as traits and vital rates are known to vary considerably within the same species across spatial scales71,72,73. Moreover, while there is at least one trait for each of these locations, the data density remains variable. Thus, records are not necessarily representative of the global spread or the full spatial scope of MOSAIC. For example, the highest complete coverage for MOSAIC traits is concentrated toward localities with the with the longest-term demographic models (see COMPADRE locations associated with MOSAIC records).
Phylogenetically, the initial release of MOSAIC is somewhat limited. Version 1.0.0 covers 300 of the 1,400 species currently available in COMPADRE, COMADRE and PADRINO. However, MOSAIC trait data are well distributed across clades (Fig. 4). While there is not a highly skewed phylogenetic concentration with respect to the existing structured population models or clustering of records into small groups across the Tree of Life, phylogenetic density of MOSAIC records remains low. Therefore, information from the MOSAIC database may be limited for a given genus or order and, as such, should be approached with caution. In future versions of MOSAIC, the phylogenetic bias is expected to diminish with more samples and stronger phylogenetic representation.
Covariance across traits is also an important source of confound in existing analyses linking traits and vital rates. Positive and negative correlations across traits that have independent influences on vital rates can create apparent associations of demographic properties with traits, spuriously functionalising non-functional traits74. Disentangling the relevance of key axes of trait variation for their demographic influences demands a clear quantification of the direction and strength networks of trait associations, trade-offs, and demographic consequences. Population biologists seek to understand not only how individual traits relate to different aspects of demographic performance (e.g., population growth rate, risk of quasi-extinction, etc.), but also understanding how trait syndromes shape those demographic outcomes. MOSAIC presents a highly varied covariance structure in trait values for the examined 300 species. For example, without a priori expectation, indeterminate growth and regeneration traits show strong correlation (r = 0.51; Fig. 5), which could influence each other’s effects on vital rates. The same could be argued for the correlation between volancy and reproductive strategy (r = 0.28 with monogamy; Fig. 5).
The MOSAIC database can be used as a platform to showcase the lack of overlap between trait and vital rate data for the same species. This picture calls for a more systematic way address global biases in ecological data quantification/collection. Even where we have complete information about species in the COMADRE, COMPADRE, and PADRINO databases, we are subject to the constraints and biases of those datasets, such as spatial bias toward high-GDP countries and the phylogenetic bias toward temperate regional perennial plants75,76. The compounding of error and density across datasets highlights the need to prioritize stronger representation of functional traits linked with demography. The standardized framework of MOSAIC is an ideal platform to work from to achieve this goal.
In view of potential biases introduced by low sampling density and the patchiness of cover in traits, users of the database are advised to consult the literature about the representativeness and congruency between MOSAIC data and related trait diversity within clades. Users need to be mindful of the scope of the questions that they are setting out to answer and to be aware of the influence of sample sizes and coherency or heterogeneity of traits across different taxonomic levels.
Extensions and relevance
From databases to data networks
Broad aperture digitisation efforts (e.g., BIEN7,45 and TRY23) have helped resolve many answers to demographic questions. Examples include whether there are trait spectra and key trade-off patterns amongst functional traits and whether these are correlated with particular environments and life history strategies77,78,79, Trait-based ecology and Trait Driver Theory80 are indebted to such opportunity-driven research programmes. More generally, however, the trait-based ecology paradigm has failed to support clear answers to many research questions of central interest to demographers29,81. This limited reach of the functional trait programme coincides with a dearth of species-specific overlap across the range of functional traits that are collected by the functional trait databases.
The proliferation of databases and open data initiatives over the last two decades82 evidences an interest in improving both data access and data usability18,83,84. While existing databases standardize trait fields, collate records, and link associated metadata, existing databases often store data for simple, quantitative traits. Relatively few ecological trait databases store diverse data types (such as rate arrays, population time series, physiological rates at different structural levels, and habitat shape files) that may be associated with multidimensional, ecological study systems (but see CESTES17, GFBio85, DarwinCore86).
The digitisation and standardisation of existing data and their integration with complementary, new data presents a growing set of challenges and opportunities in ecology87. Efforts to gap-fill records can leverage the value of existing datasets while expediting the schedule for specific research outcomes. As trait datasets grow, the importance of targeted, gap-filling initiatives to address bias and to capitalize on existing data will also increase88. The value of existing records is further enhanced through improvements in the interoperability of datasets. Much of this work is done manually, at a high cost, and with little support from funding agencies87, and yet it has been effective at facilitating research and creating new value for old data. In recent years, initiatives have sought to improve the interoperability of datasets by guiding prospective data structure or retroactively harmonising existing datasets. These include programmes that develop universal standards to improve global interoperability (such as DarwinCore86 and Frictionless [https://frictionlessdata.io/]data standards) or that contain guidelines for data metastructure (such as the FAIR principles (findability, accessibility, interoperability, and reusability, sensu Wilkinson et al., 2016) and the OpenTraits framework18). These initiatives address emerging and scaling challenges of ecoinformatics, such as the protocols by which we share data, search data, and preserve provenance in data storage structures. These protocols will be essential in centralising datasets as diverse as government monitoring datasets (e.g., those stored in U.S. Data clearing houses [https://www.data.gov; https://www.dataone.org/]; National Biodiversity Atlas [https://nbnatlas.org/]); centralized monitoring and experimental networks (e.g., LTER and NEON), raw or reanalyzed remote sensing datasets (e.g., Landsat data, NASA EarthData datasets, ERA-5 data), and private datasets (https://www.natureserve.org/) that will demand versatile and navigationally efficient data structures.
Population ecology has benefitted from widespread open-access databases but requires further dataset integration to answer its central questions. Understanding whether and how some morphological or physiological traits predict demographic outcomes and why others fail to do so is of central interest to questions in physiological, population, and community ecology29,69,89,90. Population ecologists routinely use data that are distributed across a wide range of databases. Comparative and macroecological researchers use phylogenies91,92,93, adult bodymass61,94,95,96, and high-resolution, global climate information97,98,99 to answer relevant biological, evolutionary and ecological questions and to contextualize their findings. Population ecologists frequently examine a subset of physiological, morphological, and behavioural attributes associated with demographic outcomes (i.e., functional traits100). The trait-based research programme seeks to, among other aims6, identify the intrinsic and extrinsic regulators of vital rates and the causes of variation and constraints on possible trait values65,66. Not all traits predict demographic outcomes and functional traits may exercize influence on only a few demographic pathways68,101. The answers to these questions rely on the existence of vital rate and trait data, the overlap of which has been limited in the absence of targeted attention. For instance, of the hundreds of thousands of records available across thousands of plant species in TRY23 and over 345 plant species in COMPADRE46, Adler et al. (2014) report functional trait-vital rate relationships for only 222 plant species due to their limited data overlap.
Ecological data are complex and their structures will need consistent rules to link datasets together. It will be important for future datasets to adopt database designs that render large, thematically, and structurally diverse data to be readily locatable and usable without expert knowledge. Here, we show one such example in the scope of comparative research, using thematic groups and a strategy of achieving adequate record breadth before revisiting depth of records for specific species. The need for open access data, integrated workflows, and interoperable data systems is increasing with the scaling of data collection through use of robotics and technologies. The gaps in existing data systems, interoperability, and data acquisition can be filled strategically for specific applications, offering targeted and efficient dataset development. With data interoperability guiding the structure of new datasets, the modular development of area-specific datasets will enable more generalized use over time and help meet the aims of existing database initiatives.
Code availability
Convenience functions for navigating the MOSAIC database are included in the supplemental material (S5) and on the MOSAIC website https://mosaicdatabase.web.ox.ac.uk and associated GitHub repository https://github.com/mosaicdatabase/Rmosaic. All code is open access without restrictions on access.
References
Reichman, O. J., Jones, M. B. & Schildhauer, M. P. Challenges and opportunities of open data in ecology. Science (80-.). 331, 703–705 (2011).
Marx, V. The big challenges of big data. Nature 498, 255–260 (2013).
Farley, S. S., Dawson, A., Goring, S. J. & Williams, J. W. Situating ecology as a big-data science: Current advances, challenges, and solutions. Bioscience 68, 563–576 (2018).
Maldonado, C. et al. Estimating species diversity and distribution in the era of Big Data: To what extent can we trust public databases? Glob. Ecol. Biogeogr. 24, 973–984 (2015).
Troia, M. J. & McManamay, R. A. Filling in the GAPS: evaluating completeness and coverage of open-access biodiversity databases in the United States. Ecol. Evol. 6, 4654–4669 (2016).
Díaz, S. et al. The global spectrum of plant form and function. Nature 529, 167–171 (2016).
Maitner, B. S. et al. The bien r package: A tool to access the Botanical Information and Ecology Network (BIEN) database. Methods Ecol. Evol. 9, 373–379 (2018).
Enquist, B. J. et al. The commonness of rarity: Global and future distribution of rarity across land plants. Sci. Adv. 5, 1–14 (2019).
Davy, R. & Kusch, E. Reconciling high resolution climate datasets using KrigR. Environ. Res. Lett. (2021).
Hersbach, H. et al. The ERA5 global reanalysis. Q. J. R. Meteorol. Soc. 146, 1999–2049 (2020).
Rowcliffe, J. M., Jansen, P. A., Kays, R., Kranstauber, B. & Carbone, C. Wildlife speed cameras: measuring animal travel speed and day range using camera traps. Remote Sens. Ecol. Conserv. 2, 84–94 (2016).
Wilmoth, J. R., Andreev, K., Jdanov, D. & Glei, D. A. Methods Protocol for the Human Mortality. Database. Database 2007, 1–80 (2007).
Jasilioniene, A. et al. Methods Protocol for the Human Fertility Database. Max Planck Institute for Demographic Research (2015).
De Magalhᾶes, J. P. & Costa, J. A database of vertebrate longevity records and their relation to other life-history traits. J. Evol. Biol. 22, 1770–1774 (2009).
Dornelas, M. et al. BioTIME: A database of biodiversity time series for the Anthropocene. Glob. Ecol. Biogeogr. 27, 760–786 (2018).
Fortuna, M. A., Ortega, R. & Bascompote, J. The web of life. arXiv https://doi.org/10.4324/9780203410134 (2014).
Jeliazkov, A. et al. A global database for metacommunity ecology, integrating species, traits, environment and space. Sci. Data 7, 1–15 (2020).
Gallagher, R. V. et al. Open Science principles for accelerating trait-based science across the Tree of Life. Nat. Ecol. Evol. 4, 294–303 (2020).
Kissling, W. D. et al. Towards global data products of Essential Biodiversity Variables on species traits. Nat. Ecol. Evol. 2, 1531–1540 (2018).
Wilkinson, M. D. et al. Comment: The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 1–9 (2016).
Nadrowski, K. et al. Harmonizing, annotating and sharing data in biodiversity-ecosystem functioning research. Methods Ecol. Evol. 4, 201–205 (2013).
Culina, A. et al. Navigating the unfolding open data landscape in ecology and evolution. Nat. Ecol. Evol. 2, 420–426 (2018).
Kattge, J. et al. TRY plant trait database – enhanced coverage and open access. Glob. Chang. Biol. 26, 119–188 (2020).
Schneider, F. D. et al. Towards an ecological trait-data standard. Methods Ecol. Evol. 10, 2006–2019 (2019).
Edwards, J. L., Lane, M. A. & Nielsen, E. S. Interoperability of biodiversity databases: Biodiversity information on every desktop. Science (80-.). 289, 2312–2314 (2000).
Maurer, S. M., Firestone, R. B. & Scriver, C. R. Science’s neglected legacy: Large, sophisticated databases cannot be left to chance and improvisation. Nature 405, 117–120 (2000).
Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Vitousek, M. N., Johnson, M. A. & Husak, J. F. Illuminating endocrine evolution: The power and potential of large-scale comparative analyses. Integr. Comp. Biol. 58, 712–719 (2018).
Laughlin, D. C., Gremer, J. R., Adler, P. B., Mitchell, R. M. & Moore, M. M. The Net Effect of Functional Traits on Fitness. Trends Ecol. Evol. 35, 1037–1047 (2020).
Swenson, N. G. et al. A reframing of trait–demographic rate analyses for ecology and evolutionary biology. Int. J. Plant Sci. 181, 33–43 (2020).
Guerrero-Ramírez, N. R. et al. Global root traits (GRooT) database. Glob. Ecol. Biogeogr. 30, 25–37 (2021).
Froese, R. & Pauly, D. FishBase. (2010).
Madin, J. S. et al. The Coral Trait Database, a curated database of trait information for coral species from the global oceans. Sci. data 4, 170174 (2016).
Lintulaakso, K. MammalBase—database of recent mammals. (2013).
Oliveira, B. F., São-Pedro, V. A., Santos-Barrera, G., Penone, C. & Costa, G. C. AmphiBIO, a global database for amphibian ecological traits. Sci. Data 4, 1–7 (2017).
Ashman, T. L. et al. Tree of Sex: A database of sexual systems. Sci. Data 1, 1–8 (2014).
Boettiger, C. & Temple Lang, D. Treebase: An R package for discovery, access and manipulation of online phylogenies. Methods Ecol. Evol. 3, 1060–1066 (2012).
Borghetti, M., Gentilesca, T., Colangelo, M., Ripullone, F. & Rita, A. Xylem Functional Traits as Indicators of Health in Mediterranean Forests. Curr. For. Reports 6, 220–236 (2020).
Jetz, W., Thomas, G. H., Joy, J. B., Hartmann, K. & Mooers, A. O. The global diversity of birds in space and time. Nature 491, 444–448 (2012).
Santini, L., Isaac, N. J. B. & Ficetola, G. F. TetraDENSITY: A database of population density estimates in terrestrial vertebrates. Glob. Ecol. Biogeogr. 27, 787–791 (2018).
Monnet, A. C. et al. WOODIV, a database of occurrences, functional traits, and phylogenetic data for all Euro-Mediterranean trees. Sci. Data 8, 1–11 (2021).
Middleton, O., Svensson, H., Scharlemann, J. P. W., Faurby, S. & Sandom, C. CarniDIET 1.0: A database of terrestrial carnivorous mammal diets. Glob. Ecol. Biogeogr. 30, 1175–1182 (2021).
Myhrvold, N. P. et al. An amniote life-history database to perform comparative analyses with birds, mammals, and reptiles. Ecology 96, (2015).
Jones, K. E. et al. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648–2648 (2009).
Enquist, B. J., Condit, R., Peet, R. K., Schildhauer, M. & Thiers, B. The Botanical Information and Ecology Network (BIEN): Cyberinfrastructure for an integrated botanical information network to investigate the ecological impacts of global climate change on plant biodiversity. PeerJ Prepr. (2016).
Salguero-Gómez, R. et al. The compadre Plant Matrix Database: An open online repository for plant demography. J. Ecol. https://doi.org/10.1111/1365-2745.12334 (2015).
Salguero-Gómez, R. et al. COMADRE: A global data base of animal demography. J. Anim. Ecol. https://doi.org/10.1111/1365-2656.12482 (2016).
Levin, S. C. et al. Rpadrino: an R package to access and use PADRINO, an open access database of Integral Projection Models. bioRxiv (2022).
Caswell, H. Matrix population models. (Sinauer, 2001).
Morris, W. F. et al. Longevity can buffer plant and animal populations against chnaging climatic variability. Ecology 89, 19–25 (2008).
Smallegange, I. M., Deere, J. A. & Coulson, T. Correlative changes in life-history variables in response to environmental change in a model organism. Am. Nat. 183, 784–797 (2014).
Crouse, D. T., Crowder, L. B. & Caswell, H. A stage‐based population model for loggerhead sea turtles and implications for conservation. Ecology 68, 1412–1423 (1987).
Easterling, M. R., Ellner, S. P. & Dixon, P. M. Size-specific sensitivity: Applying a new structured population model. Ecology 81, 694–708 (2000).
Jongejans, E., Shea, K., Skarpaas, O., Kelly, D. & Ellner, S. P. Importance of individual and environmental variation for invasive species spread: A spatial integral projection model. Ecology 92, 86–97 (2011).
Ozgul, A. et al. Coupled dynamics of body mass and population growth in response to environmental change. Nature 466, 482–485 (2010).
Metcalf, C. J. E., Graham, A. L., Martinez-Bakker, M. & Childs, D. Z. Opportunities and challenges of Integral Projection Models for modelling host-parasite dynamics. J. Anim. Ecol. 85, 343–355 (2016).
Bernard, C. et al. MOSAIC trait database. figshare https://doi.org/10.6084/m9.figshare.21035857.v1 (2022).
Messier, J., McGill, B. J. & Lechowicz, M. J. How do traits vary across ecological scales? A case for trait-based ecology. Ecol. Lett. 13, 838–848 (2010).
Siefert, A. et al. A global meta-analysis of the relative extent of intraspecific trait variation in plant communities. Ecol. Lett. 18, 1406–1419 (2015).
Albert, C. H. Intraspecific trait variability matters. J. Veg. Sci. 26, 7–8 (2015).
Capdevila, P. et al. Longevity, body dimension and reproductive mode drive differences in aquatic versus terrestrial life-history strategies. Funct. Ecol. 34, 1613–1625 (2020).
Gaillard, J.-M., Festa-Bianchet, M. & Yoccoz, N. G. Population dynamics of large herbivores: variable recruitment with constant adult survival. Trends Ecol. Evol. 13, 249–251 (1998).
Csergő, A. M. et al. Less favourable climates constrain demographic strategies in plants. Ecol. Lett. 20, 969–980 (2017).
McGill, B. & Collins, C. A unified theory for macroecology based on spatial patterns of abundance. Evol. Ecol. Res. 5, 469–492 (2003).
Salguero-Gómez, R., Violle, C., Gimenez, O. & Childs, D. Delivering the promises of trait-based approaches to the needs of demographic approaches, and vice versa. Funct. Ecol. 32, 1424–1435 (2018).
Buckley, Y. M. & Puy, J. The macroecology of plant populations from local to global scales. New Phytol. 233, 1038–1050 (2022).
Carmona, C. P., de Bello, F., Azcárate, F. M., Mason, N. W. H. & Peco, B. Trait hierarchies and intraspecific variability drive competitive interactions in Mediterranean annual plants. J. Ecol. 107, 2078–2089 (2019).
Pistón, N. et al. Multidimensional ecological analyses demonstrate how interactions between functional traits shape fitness and life history strategies. J. Ecol. 107, 2317–2328 (2019).
Adler, P. B. et al. Functional traits explain variation in plant life history strategies. Proc. Natl. Acad. Sci. USA 111, 10019 (2014).
Poorter, L. et al. Are functional traits good predictors of demographic rates? Evidence from five neotropical forests. Ecology 89, 1908–1920 (2008).
Violle, C. et al. The return of the variance: Intraspecific variability in community ecology. Trends Ecol. Evol. 27, 244–252 (2012).
Albert, C. H. et al. Intraspecific functional variability: Extent, structure and sources of variation. J. Ecol. 98, 604–613 (2010).
Bolnick, D. I. et al. Why intraspecific trait variation matters in community ecology. Trends Ecol. Evol. 26, 183–192 (2011).
McElreath, R. Statistical rethinking: A Bayesian course with examples in R and Stan. (Chapman and Hall/CRC, 2020).
Kendall, B. E. et al. Persistent problems in the construction of matrix population models. Ecol. Modell. 406, 33–43 (2019).
Römer, G., Dahlgren, J. P., Salguero-Gómez, R., Stott, I. M. & Jones, O. R. Plant demographic knowledge is biased towards short-term studies of temperate-region herbaceous perennials. bioRxiv 1–46 (2021).
Reich, P. B. et al. The evolution of plant functional variation: Traits, spectra, and strategies. Int. J. Plant Sci. 164, (2003).
Laughlin, D. C. Rugged fitness landscapes and Darwinian demons in trait-based ecology. New Phytol. 217, 501–503 (2018).
Wilkes, M. A. et al. Trait-based ecology at large scales: Assessing functional trait correlations, phylogenetic constraints and spatial variability using open data. Glob. Chang. Biol. 26, 7255–7267 (2020).
Enquist, B. J. et al. Scaling from traits to ecosystems: developing a general trait driver theory via integrating trait-based and metabolic scaling theories. in Advances in ecological research 249–318 (2015).
Bellier, E., Kéry, M. & Schaub, M. Relationships between vital rates and ecological traits in an avian community. J. Anim. Ecol. 87, 1172–1181 (2018).
Cheruvelil, K. S. & Soranno, P. A. Data-intensive ecological research is catalyzed by open science and team science. Bioscience 68, 813–822 (2018).
Whitlock, M. C. Data archiving in ecology and evolution: Best practices. Trends Ecol. Evol. 26, 61–65 (2011).
Michener, W. K. Meta-information concepts for ecological data management. Ecol. Inform. 1, 3–7 (2006).
Diepenbroek, M. et al. Towards an Integrated Biodiversity and Ecological Research Data Management and Archiving Platform: The…. Inform. 2014 – Big Data Komplexität meistern. GI-Edition Lect. Notes Informatics – Proc. 232 1711–1721 (2014).
Wieczorek, J. et al. Darwin core: An evolving community-developed biodiversity data standard. PLoS One 7, (2012).
Salguero-Gómez, R., Jackson, J. & Gascoigne, S. J. L. Four key challenges in the open-data revolution. J. Anim. Ecol. 90, 2000–2004 (2021).
Conde, D. A. et al. Data gaps and opportunities for comparative and conservation biology. Proc. Natl. Acad. Sci. USA 116, 9658–9664 (2019).
Chalmandrier, L. et al. Linking functional traits and demography to model species-rich communities. Nat. Commun. 12, (2021).
Yang, J., Cao, M. & Swenson, N. G. Why Functional Traits Do Not Predict Tree Demographic Rates. Trends Ecol. Evol. 33, 326–336 (2018).
Levin, S. C., Crandall, R. M., Pokoski, T., Stein, C. & Knight, T. M. Phylogenetic and functional distinctiveness explain alien plant population responses to competition: Phylogeny and traits explain dominance. Proc. R. Soc. B Biol. Sci. 287, (2020).
Daskalova, G. N., Myers-Smith, I. H. & Godlee, J. L. Rare and common vertebrates span a wide spectrum of population trends. Nat. Commun. 11, 1–13 (2020).
Roper, M., Capdevila, P. & Salguero-Gómez, R. Senescence: Why and where selection gradients might not decline with age. Proc. R. Soc. B Biol. Sci. 288, (2021).
Healy, K. et al. Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc. R. Soc. B Biol. Sci. 281, (2014).
Terry, J. C. D., O’Sullivan, J. D. & Rossberg, A. G. No pervasive relationship between species size and local abundance trends. Nat. Ecol. Evol. 6, 140–144 (2022).
Williams, N. F., McRae, L., Freeman, R., Capdevila, P. & Clements, C. F. Scaling the extinction vortex: Body size as a predictor of population dynamics close to extinction events. Ecol. Evol. 11, 7069–7079 (2021).
Daskalova, G. N., Bowler, D., Myers-Smith, I. H. & Dornelas, M. Representation of global change drivers across biodiversity datasets. EcoEvoRxiv 1–36 (2021).
Paniw, M., Maag, N., Cozzi, G., Clutton-Brock, T. & Ozgul, A. Life history responses of meerkats to seasonal changes in extreme environments. Science (80-.). 363, 631–635 (2019).
Stenseth, N. C. & Mysterud, A. Climate, changing phenology, and other life history traits: Nonlinearity and match-mismatch to the environment. Proc. Natl. Acad. Sci. USA 99, 13379–13381 (2002).
Violle, C. et al. Let the concept of trait be functional! Oikos 116, 882–892 (2007).
Salguero-Gómez, R. & Laughlin, D. C. Not all traits are functional: the Panglossian paradigm. Authorea, https://doi.org/10.22541/au.163940711.10447233/v1 (2021).
Acknowledgements
We thank Mark Roper, Sara Middleton, Etienne Cousin, and Thomas Marrien for their contributions to discussion at the beginning of the project, Jessica Hass and Lauren Hinchcliffe for collecting data for the project, John Park for his contributions to the conception of citMOSAIC, and Sam Levin for valuable comments on the Rmosaic package. This work was supported by a NERC IRF (NE/M018458/1) to RS-G. RCRC acknowledges postdoctoral support from the Regional Valencian Government and the European Social Fund (APOSTD/2020/090).
Author information
Authors and Affiliations
Contributions
C.B. and R.S.G. conceived the ideas; C.B., R.S.G., G.S.S., J.D. and P.C. designed the methodology; C.B., G.S.S., J.D., P.C. and S.J.L.G. integrated databases; C.B., G.S.S., and J.D. collected primary records data; E.K. led the climate data acquisition and analysis of geospatial data; and C.B. led the writing of the manuscript with assistance from J.J. All authors contributed critically to the drafts and gave final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bernard, C., Santos, G.S., Deere, J.A. et al. MOSAIC - A Unified Trait Database to Complement Structured Population Models. Sci Data 10, 335 (2023). https://doi.org/10.1038/s41597-023-02070-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02070-w
- Springer Nature Limited
This article is cited by
-
DEBBIES Dataset to study Life Histories across Ectotherms
Scientific Data (2024)