Abstract
Microtubules are regulated by post-translational modifications of tubulin. The ligation and cleavage of the carboxy-terminal tyrosine of α-tubulin impact microtubule functions during mitosis, cardiomyocyte contraction and neuronal processes. Tubulin tyrosination and detyrosination are mediated by tubulin tyrosine ligase and the recently discovered tubulin detyrosinases, vasohibin 1 and 2 (VASH1 and VASH2) bound to the small vasohibin-binding protein (SVBP). Here, we report the crystal structures of human VASH1–SVBP alone, in complex with a tyrosine-derived covalent inhibitor and bound to the natural product parthenolide. The structures and subsequent mutagenesis analyses explain the requirement for SVBP during tubulin detyrosination, and reveal the basis for the recognition of the C-terminal tyrosine and the acidic α-tubulin tail by VASH1. The VASH1–SVBP–parthenolide structure provides a framework for designing more effective chemical inhibitors of vasohibins, which can be valuable for dissecting their biological functions and may have therapeutic potential.





Similar content being viewed by others
Data availability
The coordinates and structure factors of the crystal structures of human VASH1–SVBP, VASH1–SVBP in complex with epoY and VASH1–SVBP bound to parthenolide have been deposited into the Protein Data Bank with the accession codes 6OCF, 6OCG and 6OCH, respectively. Source data for Figs. 2d, 4c, Supplementary Fig. 3d and Table 2 are available in Supplementary Dataset 2. All other data are available from the corresponding authors upon request.
References
Dogterom, M. & Koenderink, G. H. Actin-microtubule crosstalk in cell biology. Nat. Rev. Mol. Cell Biol. 20, 38–54 (2019).
Mitchison, T. & Kirschner, M. Dynamic instability of microtubule growth. Nature 312, 237–242 (1984).
Desai, A. & Mitchison, T. J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117 (1997).
Brouhard, G. J. & Rice, L. M. Microtubule dynamics: an interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol. 19, 451–463 (2018).
Song, Y. & Brady, S. T. Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol. 25, 125–136 (2015).
Gadadhar, S., Bodakuntla, S., Natarajan, K. & Janke, C. The tubulin code at a glance. J. Cell Sci. 130, 1347–1353 (2017).
Magiera, M. M., Singh, P., Gadadhar, S. & Janke, C. Tubulin posttranslational modifications and emerging links to human disease. Cell 173, 1323–1327 (2018).
Magiera, M. M., Singh, P. & Janke, C. SnapShot: functions of tubulin posttranslational modifications. Cell 173, 1552–1552.e1 (2018).
Nieuwenhuis, J. & Brummelkamp, T. R. The tubulin detyrosination cycle: function and enzymes. Trends Cell Biol. 29, 80–92 (2019).
Arce, C. A., Rodriguez, J. A., Barra, H. S. & Caputo, R. Incorporation of l-tyrosine, l-phenylalanine and l-3,4-dihydroxyphenylalanine as single units into rat brain tubulin. Eur. J. Biochem. 59, 145–149 (1975).
Raybin, D. & Flavin, M. An enzyme tyrosylating alpha-tubulin and its role in microtubule assembly. Biochem. Biophys. Res. Commun. 65, 1088–1095 (1975).
Hallak, M. E., Rodriguez, J. A., Barra, H. S. & Caputto, R. Release of tyrosine from tyrosinated tubulin. Some common factors that affect this process and the assembly of tubulin. FEBS Lett. 73, 147–150 (1977).
Ersfeld, K. et al. Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 120, 725–732 (1993).
Szyk, A., Deaconescu, A. M., Piszczek, G. & Roll-Mecak, A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 18, 1250–1258 (2011).
Arce, C. A. & Barra, H. S. Release of C-terminal tyrosine from tubulin and microtubules at steady state. Biochem. J. 226, 311–317 (1985).
Gundersen, G. G., Khawaja, S. & Bulinski, J. C. Postpolymerization detyrosination of alpha-tubulin: a mechanism for subcellular differentiation of microtubules. J. Cell Biol. 105, 251–264 (1987).
Steinmetz, M. O. & Akhmanova, A. Capturing protein tails by CAP-Gly domains. Trends Biochem. Sci. 33, 535–545 (2008).
Erck, C. et al. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc. Natl Acad. Sci. USA 102, 7853–7858 (2005).
Kerr, J. P. et al. Detyrosinated microtubules modulate mechanotransduction in heart and skeletal muscle. Nat. Commun. 6, 8526 (2015).
Robison, P. et al. Detyrosinated microtubules buckle and bear load in contracting cardiomyocytes. Science 352, aaf0659 (2016).
Barisic, M. et al. Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science 348, 799–803 (2015).
Aillaud, C. et al. Vasohibins/SVBP are tubulin carboxypeptidases (TCPs) that regulate neuron differentiation. Science 358, 1448–1453 (2017).
Nieuwenhuis, J. et al. Vasohibins encode tubulin detyrosinating activity. Science 358, 1453–1456 (2017).
Iqbal, Z. et al. Loss of function of SVBP leads to autosomal recessiv e intellectual disability, microcephaly, ataxia, and hypotonia. Genet. Med. https://doi.org/10.1038/s41436-018-0415-8 (2019).
Fonrose, X. et al. Parthenolide inhibits tubulin carboxypeptidase activity. Cancer Res. 67, 3371–3378 (2007).
Chatterjee, D., Boyd, C. D., O’Toole, G. A. & Sondermann, H. Structural characterization of a conserved, calcium-dependent periplasmic protease from Legionella pneumophila. J. Bacteriol. 194, 4415–4425 (2012).
Sanchez-Pulido, L. & Ponting, C. P. Vasohibins: new transglutaminase-like cysteine proteases possessing a non-canonical Cys-His-Ser catalytic triad. Bioinformatics 32, 1441–1445 (2016).
Lopez-Franco, O. et al. Parthenolide modulates the NF-κB-mediated inflammatory responses in experimental atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 26, 1864–1870 (2006).
Guzman, M. L. et al. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood 105, 4163–4169 (2005).
Duan, D., Zhang, J., Yao, J., Liu, Y. & Fang, J. Targeting thioredoxin reductase by parthenolide contributes to inducing apoptosis of HeLa cells. J. Biol. Chem. 291, 10021–10031 (2016).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D 63, 32–41 (2007).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Ti, S. C. et al. Mutations in human tubulin proximal to the kinesin-binding site alter dynamic instability at microtubule plus- and minus-ends. Dev. Cell 37, 72–84 (2016).
Acknowledgements
We thank Z. Chen and D. Tomchick for assistance with X-ray diffraction data collection and X. Ye and L. Rice for providing the recombinant human tubulin and for helpful discussions. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This study is supported by grants from the National Institutes of Health (grant no. GM107415, to X.L.), Cancer Prevention and Research Institute of Texas (grant nos. RP160255, to X.L., and RP160667-P2, to H.Y.) and the Welch Foundation (grant nos. I-1932, to X.L., and I-1441, to H.Y.). H.Y. is an Investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
F.L. performed protein purification, crystallization, structure determination and functional studies in vitro and in human cells and wrote the initial draft of the paper. Y.H. and S.Q. performed the immunofluorescence assays in human cells. X.L. assisted in structure refinement, analyzed data and supervised the research. H.Y. analyzed data, supervised the research and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Structural comparison between VASH1–SVBP and LapG.
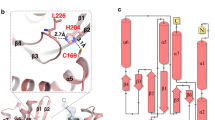
a, Ribbon diagram of the transglutaminase-like cysteine protease LapG (PDB ID: 4FGP), with the catalytic triad shown as sticks. The N- and C-lobes of LapG are colored gray and beige, respectively. b, c, Overlay of the ribbon diagrams of VASH1–SVBP and LapG structures in two different orientations, with their catalytic triad shown as sticks. The color scheme of LapG is the same as in a. The NTE, N-lobe, and C-lobe of VASH1 are colored light blue, blue, and cyan, respectively. SVBP is colored orange. d, Close-up view of the overlay of the active site of VASH1 and LapG, with the catalytic triad of LapG shown as gray sticks and that of VASH1 shown as blue and cyan sticks.
Supplementary Figure 2 Sequence alignment of VASH1 and SVBP proteins.
a, Sequence alignment of the protease domain of the VASH1 proteins from different species, with the secondary structure elements of human VASH1 shown at the top. The conserved residues are highlighted with the software Jalview2.8.1 (http://www.jalview.org/). All non-conserved residues are shaded white. For the conserved residues, the color scheme is as follows: hydrophobic aliphatic residues, blue; positively charged residues, red; negatively charged residues, magenta; polar residues, green; cysteines, pink; glycines, orange; prolines, yellow; aromatic residues, cyan. The catalytic triad is indicated with red dots. Residues critical for substrate recognition are indicated with blue dots. Residues important for SVBP binding are indicated with green dots. b, Sequence alignment of the SVBP proteins from different species, with the secondary structure elements of human VASH1 shown at the top. Residues involved in VASH1 binding are indicated with orange dots.
Supplementary Figure 3 Inhibition of VASH1–SVBP by epoY.
a, Inhibition of VASH1–SVBP-dependent microtubule detyrosination by epoY in vitro. In this assay, VASH1–SVBP was incubated with the indicated amounts of epoY at room temperature for 10 min. The treated enzyme was then mixed with polymerized microtubules as substrates and incubated at 37˚C for 15 min. The reaction mixtures were blotted with the indicated antibodies. Uncropped blot images are included in Supplementary Data Set 1. b, Surface diagram of VASH1–SVBP, with epoY shown as sticks. c, Interactions between epoY and the active site residues of VASH1. Hydrogen bonds are indicated by dashed lines with the distances labeled. The figure is generated with the program LIGPLOT. d, Quantification of the relative detyrosination levels of α tubulin in Fig. 3f (mean ± SD, n = 3 independent experiments). Source data for d are available in Supplementary Data Set 2.
Supplementary Figure 4 Requirement for basic residues lining the substrate-binding cleft in tubulin detyrosination.
a, Sequence alignment of the C-terminal tails of human α-tubulin isoforms. b,c, Tubulin detyrosination assays of VASH1–SVBP in human cells. Lysates of HeLa Tet-On cells transfected with the indicated plasmids were blotted with the indicated antibodies. WT, wild-type; deY-tubulin, detyrosinated α tubulin. Uncropped blot images are included in Supplementary Data Set 1.
Supplementary Figure 5 Analysis of tubulin detyrosination by VASH2–SVBP.
a, Sequence alignment of the protease domain of human VASH1 and VASH2. The conserved residues are highlighted using the software Jalview2.8.1 (http://www.jalview.org/) with the same color scheme as in Supplementary Fig. 2. The catalytic triad and residues important for substrate recognition in VASH1 are indicated with red dots. b, Tubulin detyrosination assays of VASH2–SVBP in human cells. Lysates of HeLa Tet-On cells transfected with the indicated plasmids were blotted with the indicated antibodies. WT, wild-type; deY-tubulin, detyrosinated α tubulin. Uncropped blot images are included in Supplementary Data Set 1.
Supplementary Figure 6 Binding of parthenolide does not induce large conformational changes of VASH1.
Superimposition of the structures of apo-VASH1–SVBP (blue and cyan ribbons) and parthenolide-bound VASH1–SVBP (gray ribbons).
Supplementary information
Supplementary Information
Supplementary Figs. 1–6, Supplementary Table 1
Supplementary Dataset 1
Uncropped gel and blotted images for Figs. 1, 2, 3, 5, Supplementary Figs. 3, 4 and 5
Supplementary Dataset 2
Source data for Figs. 2, 4, Supplementary Fig. 3 and Table 2
Rights and permissions
About this article
Cite this article
Li, F., Hu, Y., Qi, S. et al. Structural basis of tubulin detyrosination by vasohibins. Nat Struct Mol Biol 26, 583–591 (2019). https://doi.org/10.1038/s41594-019-0242-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0242-x
- Springer Nature America, Inc.
This article is cited by
-
Tubulin engineering by semi-synthesis reveals that polyglutamylation directs detyrosination
Nature Chemistry (2023)
-
Tension of plus-end tracking protein Clip170 confers directionality and aggressiveness during breast cancer migration
Cell Death & Disease (2022)
-
MARK4 controls ischaemic heart failure through microtubule detyrosination
Nature (2021)
-
Tubulin carboxypeptidase activity of vasohibin-1 inhibits angiogenesis by interfering with endocytosis and trafficking of pro-angiogenic factor receptors
Angiogenesis (2021)
-
The tubulin code and its role in controlling microtubule properties and functions
Nature Reviews Molecular Cell Biology (2020)