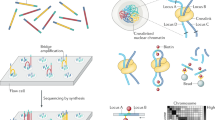
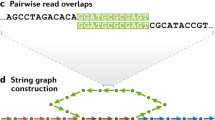
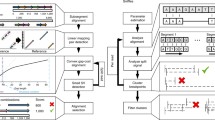
Abstract
As long-read sequencing technologies are becoming increasingly popular, a number of methods have been developed for the discovery and analysis of structural variants (SVs) from long reads. Long reads enable detection of SVs that could not be previously detected from short-read sequencing, but computational methods must adapt to the unique challenges and opportunities presented by long-read sequencing. Here, we summarize over 50 long-read-based methods for SV detection, genotyping and visualization, and discuss how new telomere-to-telomere genome assemblies and pangenome efforts can improve the accuracy and drive the development of SV callers in the future.



Similar content being viewed by others
References
Zook, J. M. et al. A robust benchmark for detection of germline large deletions and insertions. Nat. Biotechnol. 38, 1347–1355 (2020). This study represents a gold-standard SV benchmark for the HG002 genome, containing nearly 10,000 insertions and deletions validated by several orthogonal technologies.
Sudmant, P. H. et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015).
Cameron, D. L., Di Stefano, L. & Papenfuss, A. T. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat. Commun. 10, 3240 (2019).
Kosugi, S. et al. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 20, 117 (2019).
Mahmoud, M. et al. Structural variant calling: the long and the short of it. Genome Biol. 20, 246 (2019).
Bickhart, D. & Liu, G. The challenges and importance of structural variation detection in livestock. Front. Genet. 5, 37 (2014).
Conrad, D. F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010).
Kidd, J. M. et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143, 837–847 (2010).
Korbel, J. O. et al. Paired-end mapping reveals extensive structural variation in the human genome. Science 318, 420–426 (2007). An important study demonstrating extensive presence of SVs in human genomes using paired-end sequencing.
Sudmant, P. H. et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010).
Cortés-Ciriano, I. et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 52, 331–341 (2020).
Rees, E. & Kirov, G. Copy number variation and neuropsychiatric illness. Curr. Opin. Genet. Dev. 68, 57–63 (2021).
Stankiewicz, P. & Lupski, J. R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61, 437–455 (2010).
Feuk, L., Carson, A. R. & Scherer, S. W. Structural variation in the human genome. Nat. Rev. Genet. 7, 85–97 (2006).
Geiss, G. K. et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325 (2008).
Ye, K., Schulz, M. H., Long, Q., Apweiler, R. & Ning, Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 (2009).
Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012).
Layer, R. M., Chiang, C., Quinlan, A. R. & Hall, I. M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15, R84 (2014).
Chan, E. K. F. et al. Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 28, 726–738 (2018).
Kloosterman, W. P. & Cuppen, E. Chromothripsis in congenital disorders and cancer: similarities and differences. Curr. Opin. Cell Biol. 25, 341–348 (2013).
Dai, Y. et al. Single-molecule optical mapping enables quantitative measurement of D4Z4 repeats in facioscapulohumeral muscular dystrophy (FSHD). J. Med. Genet. 57, 109–120 (2020).
Alkan, C., Sajjadian, S. & Eichler, E. E. Limitations of next-generation genome sequence assembly. Nat. Methods 8, 61–65 (2011).
Sharp, A. J. et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77, 78–88 (2005).
Branton, D. et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 26, 1146–1153 (2008).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Marx, V. Method of the year: long-read sequencing. Nat. Methods 20, 6–11 (2023).
Wang, O. et al. Efficient and unique cobarcoding of second-generation sequencing reads from long DNA molecules enabling cost-effective and accurate sequencing, haplotyping, and de novo assembly. Genome Res. 29, 798–808 (2019).
Chen, Z. et al. Ultralow-input single-tube linked-read library method enables short-read second-generation sequencing systems to routinely generate highly accurate and economical long-range sequencing information. Genome Res. 30, 898–909 (2020).
Olson, N. D. et al. PrecisionFDA Truth Challenge V2: calling variants from short and long reads in difficult-to-map regions. Cell Genom. 2, 100129 (2022).
Mantere, T., Kersten, S. & Hoischen, A. Long-read sequencing emerging in medical genetics. Front. Genet. 10, 426 (2019).
Shi, L. et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat. Commun. 7, 12065 (2016).
Parikh, H. et al. svclassify: a method to establish benchmark structural variant calls. BMC Genomics 17, 64 (2016).
Pendleton, M. et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods 12, 780–786 (2015).
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018).
Ebert, P. et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 372, eabf7117 (2021). A study on SV detection from haplotype-resolved assemblies generated from long-reads and Strand-seq that identified three times as many SVs as short reads.
Merker, J. D. et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med. 20, 159–163 (2018).
Carneiro, M. O. et al. Pacific Biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics 13, 375 (2012).
Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162 (2019).
Menegon, M. et al. On site DNA barcoding by nanopore sequencing. PLoS ONE 12, e0184741 (2017).
Krishnakumar, R. et al. Systematic and stochastic influences on the performance of the MinION Nanopore sequencer across a range of nucleotide bias. Sci. Rep. 8, 3159 (2018).
Li, C. et al. INC-Seq: accurate single molecule reads using nanopore sequencing. GigaScience 5, 34 (2016).
Karst, S. M. et al. High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. Nat. Methods 18, 165–169 (2021).
Aganezov, S. et al. Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing. Genome Res. 30, 1258–1273 (2020).
Sone, J. et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet. 51, 1215–1221 (2019).
Miao, H. et al. Long-read sequencing identified a causal structural variant in an exome-negative case and enabled preimplantation genetic diagnosis. Hereditas 155, 32 (2018).
Zhou, A., Lin, T. & Xing, J. Evaluating nanopore sequencing data processing pipelines for structural variation identification. Genome Biol. 20, 237 (2019).
Luan, M.-W., Zhang, X.-M., Zhu, Z.-B., Chen, Y. & Xie, S.-Q. Evaluating structural variation detection tools for long-read sequencing datasets in Saccharomyces cerevisiae. Front. Genet. 11, 159 (2020).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022). The study describes the first complete human reference genome, T2T-CHM13, which allows SV detection in the centromeric region, the telomeric region and other complex regions.
Jiang, T. et al. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 21, 189 (2020).
Zhou, Y., Leung, A. W., Ahmed, S. S., Lam, T. W. & Luo, R. Duet: SNP-assisted structural variant calling and phasing using Oxford Nanopore sequencing. BMC Bioinformatics 23, 465 (2022).
Tham, C. Y. et al. NanoVar: accurate characterization of patients’ genomic structural variants using low-depth nanopore sequencing. Genome Biol. 21, 56 (2020).
Heller, D. & Vingron, M. SVIM: structural variant identification using mapped long reads. Bioinformatics 35, 2907–2915 (2019).
Gong, L. et al. Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat. Methods 15, 455–460 (2018).
Leung, H. C. M. et al. Detecting structural variations with precise breakpoints using low-depth WGS data from a single Oxford Nanopore MinION flowcell. Sci. Rep. 12, 4519 (2022).
Cretu Stancu, M. et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 8, 1326 (2017).
English, A. C., Salerno, W. J. & Reid, J. G. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics 15, 180 (2014).
Liu, Y. et al. SKSV: ultrafast structural variation detection from circular consensus sequencing reads. Bioinformatics 37, 3647–3649 (2021).
Chen, Y. et al. Deciphering the exact breakpoints of structural variations using long sequencing reads with DeBreak. Nat. Commun. 14, 283 (2023).
Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018). This study describes a highly accurate alignment-based long-read SV caller and its companion aligner, NGMLR. Sniffles is one of the earliest methods for long-read SV calling and is still widely used today.
Smolka, M. et al. Comprehensive structural variant detection: from mosaic to population-level. Preprint at bioRxiv https://doi.org/10.1101/2022.04.04.487055 (2022).
Kielbasa, S. M., Wan, R., Sato, K., Horton, P. & Frith, M. C. Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493 (2011).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Ahsan, M. U., Liu, Q., Fang, L. & Wang, K. NanoCaller for accurate detection of SNPs and indels in difficult-to-map regions from long-read sequencing by haplotype-aware deep neural networks. Genome Biol. 22, 261 (2021).
Luo, R. et al. Exploring the limit of using a deep neural network on pileup data for germline variant calling. Nat. Mach. Intell. 2, 220–227 (2020).
Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987 (2018).
Shafin, K. et al. Haplotype-aware variant calling with PEPPER-Margin-DeepVariant enables high accuracy in nanopore long-reads. Nat. Methods 18, 1322–1332 (2021).
Luo, J. et al. BreakNet: detecting deletions using long reads and a deep learning approach. BMC Bioinformatics 22, 577 (2021).
Ding, H. & Luo, J. MAMnet: detecting and genotyping deletions and insertions based on long reads and a deep learning approach. Brief. Bioinform. 23, bbac195 (2022).
Lin, J. et al. SVision: a deep learning approach to resolve complex structural variants. Nat. Methods 19, 1230–1233 (2022). An innovative deep learning-based inference model for complex SV detection. It converts read alignment into an image that is analyzed by CNNs.
Popic, V. et al. Cue: a deep-learning framework for structural variant discovery and genotyping. Nat. Methods 20, 559–568 (2023).
Fang, L., Hu, J., Wang, D. & Wang, K. NextSV: a meta-caller for structural variants from low-coverage long-read sequencing data. BMC Bioinformatics 19, 180 (2018).
Sović, I. et al. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat. Commun. 7, 11307 (2016).
Jeffares, D. C. et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 8, 14061 (2017).
Dierckxsens, N., Li, T., Vermeesch, J. R. & Xie, Z. A benchmark of structural variation detection by long reads through a realistic simulated model. Genome Biol. 22, 342 (2021).
Pacific Biosciences. pbsv - PacBio structural variant (SV) calling and analysis tools. GitHub https://github.com/PacificBiosciences/pbsv (2018).
Fu, Y., Mahmoud, M., Muraliraman, V. V., Sedlazeck, F. J. & Treangen, T. J. Vulcan: improved long-read mapping and structural variant calling via dual-mode alignment. GigaScience 10, giab063 (2021).
Heller, D. & Vingron, M. SVIM-asm: structural variant detection from haploid and diploid genome assemblies. Bioinformatics 36, 5519–5521 (2020).
Nattestad, M. & Schatz, M. C. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics 32, 3021–3023 (2016).
Goel, M., Sun, H., Jiao, W.-B. & Schneeberger, K. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20, 277 (2019).
Shafin, K. et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol. 38, 1044–1053 (2020). This study describes the Shasta toolkit for fast de novo assembly from Oxford Nanopore sequencing, which allows a 6-h runtime for assembly.
Marx, V. Long road to long-read assembly. Nat. Methods 18, 125–129 (2021).
Garg, S. et al. Chromosome-scale, haplotype-resolved assembly of human genomes. Nat. Biotechnol. 39, 309–312 (2021). This study describes an accurate assembly tool for PB HiFi reads that can generate chromosome-scale and haplotype-resolved assemblies using trio or Hi-C data.
Lin, J., Jia, P., Wang, S., Kosters, W. & Ye, K. Comparison and benchmark of structural variants detected from long read and long-read assembly. Brief. Bioinform. https://doi.org/10.1093/bib/bbad188 (2023).
Zhao, X. et al. Expectations and blind spots for structural variation detection from long-read assemblies and short-read genome sequencing technologies. Am. J. Hum. Genet. 108, 919–928 (2021).
De Coster, W., Weissensteiner, M. H. & Sedlazeck, F. J. Towards population-scale long-read sequencing. Nat. Rev. Genet. 22, 572–587 (2021).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Kronenberg, Z. N. et al. High-resolution comparative analysis of great ape genomes. Science 360, eaar6343 (2018).
Chin, C. S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016).
Cheng, H. et al. Haplotype-resolved assembly of diploid genomes without parental data. Nat. Biotechnol. 40, 1332–1335 (2022).
Chaisson, M. J. P. et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 10, 1784 (2019). A key study that demonstrated a sixfold increase in SV detection from local assembly-based SV calling compared to short-read sequencing.
Rodriguez, O. L., Ritz, A., Sharp, A. J. & Bashir, A. MsPAC: a tool for haplotype-phased structural variant detection. Bioinformatics 36, 922–924 (2020).
Denti, L., Khorsand, P., Bonizzoni, P., Hormozdiari, F. & Chikhi, R. SVDSS: structural variation discovery in hard-to-call genomic regions using sample-specific strings from accurate long reads. Nat. Methods 20, 550–558 (2022).
Lee, C., Grasso, C. & Sharlow, M. F. Multiple sequence alignment using partial order graphs. Bioinformatics 18, 452–464 (2002).
Stephens, Z., Wang, C., Iyer, R. K. & Kocher, J. P. Detection and visualization of complex structural variants from long reads. BMC Bioinformatics 19, 508 (2018).
Meng, G. et al. TSD: a computational tool to study the complex structural variants using PacBio targeted sequencing data. G3 9, 1371–1376 (2019).
Jiang, T., Fu, Y., Liu, B. & Wang, Y. Long-read based novel sequence insertion detection with rCANID. IEEE Trans. Nanobioscience 18, 343–352 (2019).
Jiang, T., Liu, B., Li, J. & Wang, Y. rMETL: sensitive mobile element insertion detection with long read realignment. Bioinformatics 35, 3484–3486 (2019).
Shao, H. et al. npInv: accurate detection and genotyping of inversions using long read sub-alignment. BMC Bioinformatics 19, 261 (2018).
Paulson, H. Repeat expansion diseases. Handb. Clin. Neurol. 147, 105–123 (2018).
Bates, G. P. et al. Huntington disease. Nat. Rev. Dis. Primers 1, 15005 (2015).
Liu, Q., Zhang, P., Wang, D., Gu, W. & Wang, K. Interrogating the ‘unsequenceable’ genomic trinucleotide repeat disorders by long-read sequencing. Genome Med. 9, 65 (2017).
Ummat, A. & Bashir, A. Resolving complex tandem repeats with long reads. Bioinformatics 30, 3491–3498 (2014).
Bakhtiari, M., Shleizer-Burko, S., Gymrek, M., Bansal, V. & Bafna, V. Targeted genotyping of variable number tandem repeats with adVNTR. Genome Res. 28, 1709–1719 (2018).
Fang, L. et al. Haplotyping SNPs for allele-specific gene editing of the expanded huntingtin allele using long-read sequencing. HGG Adv. 4, 100146 (2023).
Mitsuhashi, S. et al. Tandem-genotypes: robust detection of tandem repeat expansions from long DNA reads. Genome Biol. 20, 58 (2019).
Chiu, R., Rajan-Babu, I. S., Friedman, J. M. & Birol, I. Straglr: discovering and genotyping tandem repeat expansions using whole genome long-read sequences. Genome Biol. 22, 224 (2021).
Guo, R. et al. RepLong: de novo repeat identification using long read sequencing data. Bioinformatics 34, 1099–1107 (2018).
Giesselmann, P. et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat. Biotechnol. 37, 1478–1481 (2019).
De Roeck, A. et al. NanoSatellite: accurate characterization of expanded tandem repeat length and sequence through whole genome long-read sequencing on PromethION. Genome Biol. 20, 239 (2019).
Fang, L. et al. DeepRepeat: direct quantification of short tandem repeats on signal data from nanopore sequencing. Genome Biol. 23, 108 (2022).
Rheinbay, E. et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 578, 102–111 (2020).
Campbell, P. J. et al. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Sakamoto, Y., Zaha, S., Suzuki, Y., Seki, M. & Suzuki, A. Application of long-read sequencing to the detection of structural variants in human cancer genomes. Comput. Struct. Biotechnol. J. 19, 4207–4216 (2021).
Euskirchen, P. et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 134, 691–703 (2017).
Shiraishi, Y. et al. Precise characterization of somatic structural variations and mobile element insertions from paired long-read sequencing data with nanomonsv. Preprint at bioRxiv https://doi.org/10.1101/2020.07.22.214262 (2021).
Valle-Inclan, J. E. et al. Optimizing Nanopore sequencing-based detection of structural variants enables individualized circulating tumor DNA-based disease monitoring in cancer patients. Genome Med. 13, 86 (2021).
Fujimoto, A. et al. Whole-genome sequencing with long reads reveals complex structure and origin of structural variation in human genetic variations and somatic mutations in cancer. Genome Med. 13, 65 (2021).
Beyter, D. et al. Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nat. Genet. 53, 779–786 (2021). A pioneering study on SV genotyping and merging of large-scale SV callsets from a long-read dataset of a large cohort of the Icelandic population.
Belyeu, J. R. et al. Samplot: a platform for structural variant visual validation and automated filtering. Genome Biol. 22, 161 (2021).
Spies, N., Zook, J. M., Salit, M. & Sidow, A. svviz: a read viewer for validating structural variants. Bioinformatics 31, 3994–3996 (2015).
Lecompte, L., Peterlongo, P., Lavenier, D. & Lemaitre, C. SVJedi: genotyping structural variations with long reads. Bioinformatics 36, 4568–4575 (2020).
Zhao, X., Weber, A. M. & Mills, R. E. A recurrence-based approach for validating structural variation using long-read sequencing technology. GigaScience 6, 1–9 (2017).
Yang, J. & Chaisson, M. J. P. TT-Mars: structural variants assessment based on haplotype-resolved assemblies. Genome Biol. 23, 110 (2022).
Duan, X., Pan, M. & Fan, S. Comprehensive evaluation of structural variant genotyping methods based on long-read sequencing data. BMC Genomics 23, 324 (2022).
Robinson, J. T. et al. Integrative Genomics Viewer. Nat. Biotechnol. 29, 24–26 (2011).
Ahdesmaki, M. J. et al. Prioritisation of structural variant calls in cancer genomes. PeerJ 5, e3166 (2017).
Nattestad, M., Aboukhalil, R., Chin, C. S. & Schatz, M. C. Ribbon: intuitive visualization for complex genomic variation. Bioinformatics 37, 413–415 (2021).
English, A. C., Menon, V. K., Gibbs, R. A., Metcalf, G. A. & Sedlazeck, F. J. Truvari: refined structural variant comparison preserves allelic diversity. Genome Biol. 23, 271 (2022).
Sentieon. Hap-Eval - a VCF comparison engine for structual variant benchmarking. GitHub https://github.com/Sentieon/hap-eval (2022).
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Schneider, V. A. et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 27, 849–864 (2017).
Sherman, R. M. & Salzberg, S. L. Pan-genomics in the human genome era. Nat. Rev. Genet. 21, 243–254 (2020).
Eichler, E. E., Clark, R. A. & She, X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat. Rev. Genet. 5, 345–354 (2004).
Huddleston, J. et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 27, 677–685 (2017).
Vollger, M. R. et al. Segmental duplications and their variation in a complete human genome. Science 376, eabj6965 (2022).
Miga, K. H. & Wang, T. The need for a human pangenome reference sequence. Annu. Rev. Genomics Hum. Genet. 22, 81–102 (2021).
Wang, T. et al. The Human Pangenome Project: a global resource to map genomic diversity. Nature 604, 437–446 (2022). This study describes the development of a human pangenome reference from haplotype-resolved assemblies to accurately represent human genomic diversity by facilitating SV discovery.
Collins, R. L. et al. A structural variation reference for medical and population genetics. Nature 581, 444–451 (2020).
Byrska-Bishop, M. et al. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell 185, 3426–3440 (2022).
Li, H. et al. A synthetic-diploid benchmark for accurate variant-calling evaluation. Nat. Methods 15, 595–597 (2018).
Cao, S., Jiang, T., Liu, Y., Liu, S. & Wang, Y. Re-genotyping structural variants through an accurate force-calling method. Preprint at bioRxiv https://doi.org/10.1101/2022.08.29.505534 (2022).
Acknowledgements
We thank several readers who worked on SVs to provide valuable feedback on our preprint, including A. Gouru, Y. Zhou and Y. Hu. We thank the developers of the various software tools described in the text for making their tools available with detailed documentation. The survey is in part supported by NIH grant GM132713 and the CHOP Research Institute.
Author information
Authors and Affiliations
Contributions
Q.L. and K.W. conceived the study. M.U.A., Q.L. and K.W. wrote the initial version. J.P. and L.F. wrote several sections in the text and prepared figures. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Lin Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ahsan, M.U., Liu, Q., Perdomo, J.E. et al. A survey of algorithms for the detection of genomic structural variants from long-read sequencing data. Nat Methods 20, 1143–1158 (2023). https://doi.org/10.1038/s41592-023-01932-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-023-01932-w
- Springer Nature America, Inc.
This article is cited by
-
A sequence-aware merger of genomic structural variations at population scale
Nature Communications (2024)
-
Comparative evaluation of SNVs, indels, and structural variations detected with short- and long-read sequencing data
Human Genome Variation (2024)
-
Long-read sequencing and optical mapping generates near T2T assemblies that resolves a centromeric translocation
Scientific Reports (2024)