Abstract
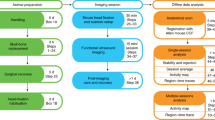
We extended the capabilities of functional ultrasound to whole-brain four-dimensional (4D) neuroimaging. Our multiplane-wave transmission scheme on matrix arrays at thousands of frames per second provides volumetric recordings of cerebral blood volume changes at high spatiotemporal resolution. We illustrated the approach in rats while providing multiple sensory stimuli, for 4D functional connectivity and during instantaneous tracking of epileptiform events.


Similar content being viewed by others
Data availability
All data supporting the findings of this study associated with figures are available upon request. Example data can be downloaded in the Supplementary Information.
Code availability
Example encoding and post-processing codes associated with figures are provided as Supplementary Software. Step-by-step instructions are available in a Readme pdf document to guide the user. A binary executable file for GPU-based 3D beamforming is included to process decoded sample radio frequency data. The low-level beamforming code is available in the framework of an official collaboration between academic institutions.
References
Friston, K. J. Science 326, 399–403 (2009).
Xia, J. & Wang, L. V. IEEE Trans. Biomed. Eng. 61, 1380–1389 (2014).
Razansky, D., Buehler, A. & Ntziachristos, V. Nat. Protoc. 6, 1121–1129 (2011).
Gottschalk, S., Felix Fehm, T., Luís Deán-Ben, X. & Razansky, D. J. Cereb. Blood Flow. Metab. 35, 531–535 (2015).
Macé, E. et al. Nat. Methods 8, 662–664 (2011).
Mace, E. et al. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 60, 492–506 (2013).
Deffieux, T., Demene, C., Pernot, M. & Tanter, M. Curr. Opin. Neurobiol. 50, 128–135 (2018).
Tanter, M. & Fink, M. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 61, 102–119 (2014).
Tiran, E. et al. Phys. Med. Biol. 60, 8549–8566 (2015).
Errico, C. et al. NeuroImage 124, 752–761 (2016).
Osmanski, B.-F., Pezet, S., Ricobaraza, A., Lenkei, Z. & Tanter, M. Nat. Commun. 5, 5023 (2014).
De Luca, M., Beckmann, C. F., De Stefano, N., Matthews, P. M. & Smith, S. M.NeuroImage 29, 1359–1367 (2006).
van den Heuvel, M. P. & Hulshoff Pol, H. E. Eur. Neuropsychopharmacol. 20, 519–534 (2010).
Ayata, C. & Lauritzen, M. Physiol. Rev. 95, 953–993 (2015).
Cain, S. M. et al. Proc. Natl Acad. Sci. USA 114, 2401–2406 (2017).
Dizeux, A. et al. Nat. Commun. 10, 1400 (2019).
Bourdeau, R. W. et al. Nature 553, 86–90 (2018).
Sieu, L.-A. et al. Nat. Methods 12, 831–834 (2015).
Demene, C. et al. Sci. Transl. Med. 9, eaah6756 (2017).
Provost, J. et al. Phys. Med. Biol. 59, L1 (2014).
Bercoff, J. et al. IEEE Trans. Ultrason Ferroelectr. Freq. Control 58, 134–147 (2011).
Demene, C. et al. IEEE Trans. Med. Imaging 34, 2271–2285 (2015).
Demené, C. et al. NeuroImage 127, 472–483 (2016).
Papp, E. A., Leergaard, T. B., Calabrese, E., Johnson, G. A. & Bjaalie, J. G. NeuroImage 97, 374–386 (2014).
Hernandez, V. et al. Front. Neuroinform. 5, 26 (2011).
Bowden, D. M., Song, E., Kosheleva, J. & Dubach, M. F. Neuroinform. 10, 97–114 (2012).
Acknowledgements
This work was supported by the European Research Council under the European Union’s Seventh Framework Program (no. FP7/2007-2013)/ERC Advanced grant agreement no. 339244-FUSIMAGINE) and by a funding from the Human Brain Project, Project no. FUSIMICE ANR-15-HBPR-0004. This work was also supported by the AXA Research Fund.
Author information
Authors and Affiliations
Contributions
M.T. and T.D. conceived the study. C.R., V.F., M.P. and M.C. developed sequence acquisitions, C.R. and S.P. acquired data. C.R., T.D. and M.T. performed data processing. C.R., S.P., T.D. and M.T. interpreted the results. C.R. and M.T. wrote the first draft of the manuscript with substantial contribution from T.D., S.P. and M.P. All authors edited and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
T.D., M.P. and M.T are co-founders and shareholders of Iconeus company commercializing ultrasound neuroimaging scanners.
Additional information
Peer review information Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Subject-level correlation matrices of four animals estimated from 103 common cortical and subcortical regions of interest (ROI) under anesthesia.
Connectivity matrices display the pairwise correlation (Pearson’s product-moment) between all the 103 common ROIs signals. The neocortex regions are well intra-correlated in rats 1, 2 and 3 : [Rat 1: r(intra-neocortex)= 0.37; r(neocortex/others)=0,19], [Rat 2: r(intra-neocortex) = 0.37; r (neocortex/others)=0,09] [Rat 3: r(intra-neocortex) = 0.40; r(neocortex/others)=0,13] [Rat 4: r(intra-neocortex) = 0.24; r (neocortex/others)=0,21] as well as the endbrain regions in all rats: [Rat 1: r(intra-endbrain) = 0.29; r(endbrain/others)=0,21], [Rat 2: r(intra-endbrain) = 0.33; r(endbrain/others)=0,11] [Rat 3: r(intra- endbrain) = 0.28; r(endbrain /others)=0,11] [Rat 4: r(intra- endbrain) = 0.36; r(endbrain /others)=0,21]. Legends: r(intra-neocortex) = mean pairwise Pearson’s product-moment of regions belonging to the neocortex. r(neocortex/others) = mean pairwise Pearson’s product-moment between regions belonging to the neocortex and all the other regions of the brain. r(intra-endbrain) = mean pairwise Pearson’s product-moment of regions belonging to the endbrain. r(endbrain/others) = mean pairwise Pearson’s product-moment between regions belonging to the endbrain and all the other regions of the brain.
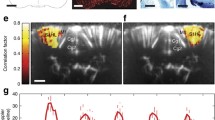
Supplementary Figure 2 Spatial mapping of the time of arrival of spreading depression activity.
Power Doppler (a) volume with overlaid volume color-coded map and (b-c) 2D planes with overlaid isochronous maps (one line every 7s) showing the propagation of a cortical depression wave during the seizure presented in Supplementary Video 2 (from time T=40 s to T=120 s) and Supplementary Video 3 (« Left Cortical Spreading Depression Wave »).
Supplementary Figure 3 Propagation of cortical spreading depressions both in ipsi and contralateral hemispheres.
The space-time representation of temporal CBV changes along segments A1-A2, B1-B2, C1-C2, D1-D2 highlights successive propagation patterns. Measurements of the slope in this space-time representation give access to the propagation speed centered around 3.0 +/- 0.3 mm/min.
Supplementary Figure 4 Hadamard decoding process on received Radio-Frequency (RF) data from a 80µm static nylon wire.
We show here ultrasonic raw data recorded on one set of 32 channels from the 32x32 matrix array probe. a) Reception of the eight echoes from the wire after the emission of eight successive tilted polarized plane waves. The polarizations of the wavefronts coming back from the wire are shown. The +1/-1 emitted polarizations correspond to the coefficients of the Hadamard matrix H8. b) Coherent summations (following the coefficient of H8) of the eight receptions. In the first combination, the contributions of the echoes from reception#2 to reception#8 disappear thanks to the multiplicative factors applied, whereas the contributions of the first echo are added ((+1)+(+1)+ +(+1)=8). Therefore, we obtain the same image as if only one individual transmit was sent with eight-fold amplitude. The time delay τn corresponding to the time before the emission of the wave #n is then compensated and the eight resulting volumes are coherently summed c) To obtain the final power volume, we square the imaging volume. d)-e) Axial and lateral resolution calculated with the point-spread function of the power image of the static nylon wire of 80µm. The spatial resolution is quantified as the full width at half maximum. We calculated Δz=180µm and Δx=240µm. The same experiment was performed with the wire rotated 90 degrees and a similar lateral resolution was found(Δy=240µm). f) On the edge of the volume, the axial resolution does not change but lateral resolution increases up to 320 µm.
Supplementary Figure 5 Virtual sources emitting a multiplane waves sequence.
Acquisitions are defined by virtual ultrasonic sources provided on a virtual array located far behind the probe (60m). For each individual source, delays are computed and tilted plane waves are obtained.
Supplementary Figure 6 Comparison between Conventional Compounding and Multiplane Transmit Compounding.
Typical example of two coronal imaging planes in a trepanned rat, both acquired with a)-c): the conventional compounding method using successive transmission/receptions of tilted plane waves or b)-d): the multiplane wave transmit method using simultaneous transmission of multiple tilted plane waves. To be comparable, data are normalized and displayed in logarithmic scale (−40dB to 0dB). Those two coronal planes are representative of all imaging planes in the imaged volume.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Tables 1 and 2.
Supplementary Video 1
4D functional ultrasound imaging of whole-brain activity in rat brain. Time, 1–>10 s. Moving view of a cerebral vascular doppler volume of a whole rat brain embedded in a brain atlas. The Doppler volume is acquired in 350 ms thanks to the emission of successive plane waves with the matrix array probe that allows the insonification of the whole volume at each shot. From time = 11 s: functional activity of the rat brain following ON–OFF stimulations. Baseline signal (when no stimulation) is indicated in gray, and all voxels with an increase of the CBV above 15% of their baseline over time are overlaid in orange. Whiskers were stimulated with a cotton swab, followed by a visual stimulation.
Supplementary Video 2
Different views of the propagation of cortical depression waves in a single rat during an ictal event Right: four static views. Left, the camera moves around the brain. Cortical depression waves after injection of 1 µl of 4-aminopyridine (4AP) (15 mmol l−1) at T = 7 s.
Supplementary Video 3
3D time-of-arrival map showing a power Doppler cerebral volume with overlaid color-coded 3D map Time of arrival color-coded map of the first and third cortical depression waves presented in Supplementary Video 1. The location of the onset of the spreading depression is shown in in dark blue.
Supplementary Software and Code
Hadamard coding (Generate multiplane wave coded emissions that can be used in a transmit sequence in a 3D research scanner) and decoding (decode the received RF buffer using the multiplane wave scheme.). Beamforming of decoded RF buffer data to obtain a beamformed IQ data volume. Clutter filter to remove signals coming from tissue and compute the 3D power Doppler volume. Computation of the normalized correlation between one Doppler movie and one stimulus signal (whisker stimulation) Computation of the connectivity matrix from one Doppler movie and associated 3D mask of regions of interest In vivo data provided from functional imaging experiments on a trepanned rat. Step-by step instructions are provided in the Readme pdf document.
Dataset 1
Coded_emissions.mat and Encoded_RF_buffer.mat : input data for Script 1_Encode_Emissions.m - RF_decoded.mat: input data for Script3_Beamform_decoded_RF.m -stim_tot: : input data for Script5_Create_Activation_maps.m -ROI_3D.mat: input data for Script6_Create_connectivity_matrix.m
Dataset 2a
IQ_part1.mat: first part of the input data for Script4_Apply_Clutter_Filter3D.m.
Dataset 2b
IQ_part2.mat: second part of the input data for Script4_Apply_Clutter_Filter3D.m.
Dataset 2c
IQ_part3.mat: third part of the input data for Script4_Apply_Clutter_Filter3D.m.
Dataset 3
Doppler_acq_stim.mat: input data for Script5_Create_Activation_maps.m.
Dataset 4a
PW_connectivity_part1.mat: first part of the input data for Script6_Create_connectivity_matrix.m.
Dataset 4b
PW_connectivity_part2.mat: second part of the input data for Script6_Create_connectivity_matrix.m.
Rights and permissions
About this article
Cite this article
Rabut, C., Correia, M., Finel, V. et al. 4D functional ultrasound imaging of whole-brain activity in rodents. Nat Methods 16, 994–997 (2019). https://doi.org/10.1038/s41592-019-0572-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0572-y
- Springer Nature America, Inc.
This article is cited by
-
Decoding motor plans using a closed-loop ultrasonic brain–machine interface
Nature Neuroscience (2024)
-
Waxholm Space atlas of the rat brain: a 3D atlas supporting data analysis and integration
Nature Methods (2023)
-
Imaging and computing with disorder
Nature Physics (2022)
-
Functional ultrasound localization microscopy reveals brain-wide neurovascular activity on a microscopic scale
Nature Methods (2022)
-
Global dissociation of the posterior amygdala from the rest of the brain during REM sleep
Communications Biology (2022)