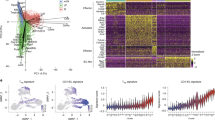
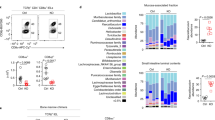
Abstract
Type 2 innate lymphoid cells (ILC2) contribute to immune homeostasis, protective immunity and tissue repair. Here we demonstrate that functional ILC2 cells can arise in the embryonic thymus from shared T cell precursors, preceding the emergence of CD4+CD8+ (double-positive) T cells. Thymic ILC2 cells migrated to mucosal tissues, with colonization of the intestinal lamina propria. Expression of the transcription factor RORα repressed T cell development while promoting ILC2 development in the thymus. From RNA-seq, assay for transposase-accessible chromatin sequencing (ATAC-seq) and chromatin immunoprecipitation followed by sequencing (ChIP-seq) data, we propose a revised transcriptional circuit to explain the co-development of T cells and ILC2 cells from common progenitors in the thymus. When Notch signaling is present, BCL11B dampens Nfil3 and Id2 expression, permitting E protein–directed T cell commitment. However, concomitant expression of RORα overrides the repression of Nfil3 and Id2 repression, allowing ID2 to repress E proteins and promote ILC2 differentiation. Thus, we demonstrate that RORα expression represents a critical checkpoint at the bifurcation of the T cell and ILC2 lineages in the embryonic thymus.







Similar content being viewed by others
Data availability
All high-throughput data in this study were deposited at the Gene Expression Omnibus (GEO) under accession number GSE146745. Source data are provided with this paper.
References
Walker, J. A. & McKenzie, A. N. Development and function of group 2 innate lymphoid cells. Curr. Opin. Immunol. 25, 148–155 (2013).
Vivier, E. et al. Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018).
Halim, T. Y. et al. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat. Immunol. 17, 57–64 (2016).
Oliphant, C. J. et al. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4+ T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity 41, 283–295 (2014).
Petrie, H. T. & Zuniga-Pflucker, J. C. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu. Rev. Immunol. 25, 649–679 (2007).
Porritt, H. E. et al. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20, 735–745 (2004).
Wong, S. H. et al. Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13, 229–236 (2012).
Lu, M. et al. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J. Immunol. 175, 5848–5856 (2005).
Kernfeld, E. M. et al. A single-cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immunity 48, 1258–1270 (2018).
Jones, R. et al. Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur. J. Immunol. 48, 1481–1491 (2018).
Schmitt, T. M., Ciofani, M., Petrie, H. T. & Zuniga-Pflucker, J. C. Maintenance of T cell specification and differentiation requires recurrent Notch receptor–ligand interactions. J. Exp. Med. 200, 469–479 (2004).
Hosokawa, H. & Rothenberg, E. V. Cytokines, transcription factors, and the initiation of T-cell development. Cold Spring Harb. Perspect. Biol. 10, a028621 (2018).
Koga, S. et al. Peripheral PDGFRα+gp38+ mesenchymal cells support the differentiation of fetal liver-derived ILC2. J. Exp. Med. 215, 1609–1626 (2018).
Miyazaki, M. et al. The E–Id protein axis specifies adaptive lymphoid cell identity and suppresses thymic innate lymphoid cell development. Immunity 46, 818–834 (2017).
Longabaugh, W. J. R. et al. BCL11B and combinatorial resolution of cell fate in the T-cell gene regulatory network. Proc. Natl Acad. Sci. USA 114, 5800–5807 (2017).
Hosokawa, H. et al. BCL11B sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat. Immunol. 19, 1427–1440 (2018).
Li, L., Leid, M. & Rothenberg, E. V. An early T cell lineage commitment checkpoint dependent on the transcription factor BCL11B. Science 329, 89–93 (2010).
Li, P. et al. Reprogramming of T cells to natural killer–like cells upon Bcl11b deletion. Science 329, 85–89 (2010).
Moro, K. et al. Innate production of TH2 cytokines by adipose tissue-associated c-KIT+SCA-1+ lymphoid cells. Nature 463, 540–544 (2010).
Seillet, C. et al. NFIL3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med. 211, 1733–1740 (2014).
Walker, J. A. et al. BCL11B is essential for group 2 innate lymphoid cell development. J. Exp. Med. 212, 875–882 (2015).
Hosokawa, H. et al. Cell type-specific actions of BCL11B in early T-lineage and group 2 innate lymphoid cells. J. Exp. Med. 217, e20190972 (2019).
Walker, J. A. et al. Polychromic reporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity 51, 104–118 (2019).
Sagar et al. Deciphering the regulatory landscape of fetal and adult γδ T-cell development at single-cell resolution. EMBO J. 39, e104159 (2020).
Spidale, N. A. et al. Interleukin-17-producing γδ T cells originate from SOX13+ progenitors that are independent of γδTCR signaling. Immunity 49, 857–872 (2018).
Fiorini, E. et al. Cutting edge: thymic crosstalk regulates Delta-like 4 expression on cortical epithelial cells. J. Immunol. 181, 8199–8203 (2008).
Neill, D. R. et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010).
Halim, T. Y. et al. Retinoic-acid-receptor-related orphan nuclear receptor α is required for natural helper cell development and allergic inflammation. Immunity 37, 463–474 (2012).
Schmitt, T. M. & Zuniga-Pflucker, J. C. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity 17, 749–756 (2002).
Yagi, R. et al. The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity 40, 378–388 (2014).
Zhu, J. T helper 2 (TH2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine 75, 14–24 (2015).
Li, L. et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood 122, 902–911 (2013).
Gascoyne, D. M. et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10, 1118–1124 (2009).
Tang, Y. et al. Emergence of NK-cell progenitors and functionally competent NK-cell lineage subsets in the early mouse embryo. Blood 120, 63–75 (2012).
Stenstad, H. et al. Gut-associated lymphoid tissue–primed CD4+ T cells display CCR9-dependent and -independent homing to the small intestine. Blood 107, 3447–3454 (2006).
von Moltke, J., Ji, M., Liang, H. E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016).
Ikawa, T. et al. An essential developmental checkpoint for production of the T cell lineage. Science 329, 93–96 (2010).
Blackburn, C. C. & Manley, N. R. Developing a new paradigm for thymus organogenesis. Nat. Rev. Immunol. 4, 278–289 (2004).
Masuda, K. et al. Notch activation in thymic epithelial cells induces development of thymic microenvironments. Mol. Immunol. 46, 1756–1767 (2009).
White, A. J. et al. A type 2 cytokine axis for thymus emigration. J. Exp. Med. 214, 2205–2216 (2017).
Zhang, J. A., Mortazavi, A., Williams, B. A., Wold, B. J. & Rothenberg, E. V. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149, 467–482 (2012).
Barlow, J. L. et al. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J. Allergy Clin. Immunol. 129, 191–198 (2012).
Fallon, P. G. et al. IL-4 induces characteristic TH2 responses even in the combined absence of IL-5, IL-9, and IL-13. Immunity 17, 7–17 (2002).
Monticelli, L. A. et al. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat. Immunol. 17, 656–665 (2016).
Taghon, T., Yui, M. A. & Rothenberg, E. V. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat. Immunol. 8, 845–855 (2007).
Xu, W. et al. E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood 121, 1534–1542 (2013).
Schwartz, R., Engel, I., Fallahi-Sichani, M., Petrie, H. T. & Murre, C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc. Natl Acad. Sci. USA 103, 9976–9981 (2006).
Longabaugh, W. J., Davidson, E. H. & Bolouri, H. Computational representation of developmental genetic regulatory networks. Dev. Biol. 283, 1–16 (2005).
Schlenner, S. M. et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32, 426–436 (2010).
Hardman, C. S., Panova, V. & McKenzie, A. N. IL-33 citrine reporter mice reveal the temporal and spatial expression of IL-33 during allergic lung inflammation. Eur. J. Immunol. 43, 488–498 (2013).
Chang, H. C. et al. Dissection of the human CD2 intracellular domain. Identification of a segment required for signal transduction and interleukin 2 production. J. Exp. Med. 169, 2073–2083 (1989).
Baddeley, A., Rubak, E. & Turner, R. Spatial Point Patterns: Methodology and Applications with R. (CRC Press, Taylor & Francis Group, 2016).
Baddeley, A. & Turner, R. spatstat: an R package for analyzing spatial point patterns. J. Stat. Softw. https://doi.org/10.18637/jss.v012.i06 (2005).
Anderson, G. & Jenkinson, E. J. Fetal thymus organ culture. CSH Protoc. 2007, pdb.prot4808 (2007).
Rana, B. M. J. et al. A stromal cell niche sustains ILC2-mediated type-2 conditioning in adipose tissue. J. Exp. Med. 216, 1999–2009 (2019).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
McCarthy, D. J., Campbell, K. R., Lun, A. T. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5, 2122 (2016).
Schmidl, C., Rendeiro, A. F., Sheffield, N. C. & Bock, C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Sultana, T. et al. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol. Cell 74, 555–570 (2019).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Acknowledgements
We are grateful to the Ares staff, genotyping facility and flow cytometry core for their technical assistance. This study was supported by grants from the UK Medical Research Council (U105178805 to J.L.B., M.W.D.H., M.G., H.E.J., M.D., R.B., A.C.) and the Wellcome Trust (100963/Z/13/Z to A.C.F.F., J.A.W., P.A.C., S.K., A.L.). A.C.H.S. was supported by a Croucher Cambridge International Scholarship.
Author information
Authors and Affiliations
Contributions
A.C.F.F. designed and performed experiments and wrote the paper. J.A.W., P.A.C., A.C., J.L.B., M.W.D.H., S.K., A.L., M.G., A.C.H.S., R.B., M.D. and H.E.J. performed experiments, provided advice on experimental design and interpretation and commented on the manuscript. A.N.J.M. supervised the project, designed the experiments and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Peer reviewer reports are available. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of DN1 embryonic thymus populations using 5xpolychromILC mice.
(a) Flow-cytometry analysis of the indicated cell surface markers or transcription factors in the E13.5, E14.5, E15.5, E17.5 and E19.5 embryonic thymus of 5xpolychromILC mice. (b) Flow-cytometric analysis of cKit and CD44 expression in DN1-Id2–Bcl11b–, DN1-Id2+Bcl11b+, DN2 and DN3 cells in the E15.5 embryonic thymus of 5xpolychromILC mice. (c) Principal component analysis (PCA) of RNA-seq data from indicated thymic cell populations (Fig. 2a) at E19.5 (n = 3). (d) Heatmap of genes from bulk RNA-seq analysis selected from single-cell gene expression analysis data (Fig. 1c). (e) Relative gene expression (from bulk RNA-seq analysis) of Zbtb16 and Pdcd1, in different embryonic thymus populations at E15.5 and E19.5 (n=3). Mean ± s.e.m; two-way ANOVA with Tukey post-hoc test. (f) Flow-cytometry analysis of RORγt-Katushka, Bcl11b-tdtomato, GATA-3-hCD2, RORα-Teal and Id2-BFP expression in the double negative (DN) and double positive (DP) cell subsets from E17.5 embryonic thymus. (g) Flow-cytometry analysis of RORγt-Katushka, Bcl11b-tdtomato, GATA-3-hCD2, RORα-Teal and Id2-BFP expression in ETP and ILC2p (lin-ST2+IL7Ra+) from E17.5 embryonic thymus.
Extended Data Fig. 2 Thymic ILC2p express ST2 and IL-7Rα and are present in IL-33-deficient mice.
(a) Flow-cytometry analysis of Bcl11b-tdtomato, GATA-3-hCD2, RORα-Teal and Id2-BFP in IL-7Rα+ST2+ ILC2p during embryonic thymus development. (b) Flow-cytometry analysis of ST2 and NK1.1 expression in the DN1 population in E16.6 embryonic thymus from wildtype (WT), Il33+/- or Il33-/- mice.
Extended Data Fig. 3 In vitro differentiation of ETP can be modulated by RORα expression.
(a) Representative flow-cytometry gating strategy for the purification of ETP cells. (b) Representative flow-cytometry analysis of GFP, CD44 and ICOS expression by cells generated in vitro after co-culture of ETPs, transduced with empty or RORα overexpressing vector, with OP9-DL1 stromal cells in the presence of growth factors (IL-7 and Flt3-L). GFP+ cells represent the positively transduced cells. (c) Flow-cytometric analysis of Bcl11b-Tom, Id2-BFP, RORα-TEAL and GATA-3-hCD2 expression in pro-T cells (ICOS-CD44-) and ILC2p (ICOS+CD44+) generated in vitro after co-culture of ETPs purified from 5xpolychromILC mice with OP9-DL1 stromal cells in the presence of growth factors (IL-7 and Flt3-L). (d) Western blot analysis from HEK cells transiently transfected with overexpressing constructs (pcDNA3) for RORα-FLAG-T2A (62 kD), RORα-FLAG (58 kD) or GATA-3-HA-T2A (48 kD), immunoprecipitated with anti-T2A or anti-FLAG antibody and detected using anti-FLAG or anti-T2A antibody, respectively. Data are representative of 2 independent experiments.
Extended Data Fig. 4 RORα binds to circadian rhythm associated genes in ILC2.
(a) Representative ATAC-seq tracks for thymic ETP, DN2, DN3, NKp and ILC2p, and binding profiles of RORα-T2A and GATA-3-T2A in ILC2 purified from lymph nodes of Rorateal/teal, Gata3hCD2TR/+ or wild type mice, and expanded in vitro with IL-7 and IL-33, around the Arntl and Clock loci. Tracks shown are representative of three independent experiments. (b) Representative ATAC-seq tracks for thymic ETP, DN2, DN3, NKp and ILC2p, and binding profiles of RORα-T2A and GATA-3-T2A in ILC2 (as in Fig. S4a) around the Il1rl1, Il2ra and Rora loci.
Extended Data Fig. 5 Genes associated with ILC2 function are among RORα target genes.
a) Gene expression (RPKM from bulk RNA-seq analysis) (top panel) and tSNE plots (log2 expression from single cell analysis) (lower panel) showing Arg1, Il13, Icos and Il1rl1, in different embryonic thymus populations. Data represent mean ± s.e.m. (n=3 biologically independent samples). (b) Representative ATAC-seq tracks for thymic ETP, DN2, DN3, NKp and ILC2p, and binding profiles of RORα-T2A and GATA-3-T2A in ILC2 purified from lymph nodes of Rorateal/teal, Gata3hCD2TR/+ or wild type mice, and expanded in vitro with IL-7 and IL-33, around the type-2 cytokine locus. Tracks shown are representative of three independent experiments.
Extended Data Fig. 6 RORα binds Id2 and Nfil3 regulatory elements.
(a) Representative ATAC-seq tracks for thymic ETP, DN2, DN3, NKp and ILC2p around the Bcl11b locus. Tracks shown are representative of three independent experiments. (b) Representative ATAC-seq tracks for thymic ETP, DN2, DN3, NKp and ILC2p and binding profiles of RORα-T2A and GATA-3-T2A in ILC2 in ILC2 purified from lymph nodes of Rorateal/teal, Gata3hCD2TR/+ or wild type mice, and expanded in vitro with IL-7 and IL-33 around the Id2 and Nfil3 locus. Tracks shown are representative of three independent experiments. (c) A luciferase assay shows the activity (relative to empty vector) of a DNA fragment containing the RORα-binding site from the Id2-associated -143 kb regulatory region, in the presence of increasing concentrations (as indicated) of RORα, GATA-3 or both. Data are representative of three independent experiments; mean ± s.e.m.; two-way ANOVA with Tukey post-hoc test.
Supplementary information
Source data
Source Data Extended Data Fig. 3
Unprocessed immunoblots
Rights and permissions
About this article
Cite this article
Ferreira, A.C.F., Szeto, A.C.H., Heycock, M.W.D. et al. RORα is a critical checkpoint for T cell and ILC2 commitment in the embryonic thymus. Nat Immunol 22, 166–178 (2021). https://doi.org/10.1038/s41590-020-00833-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-00833-w
- Springer Nature America, Inc.
This article is cited by
-
Nr4a1 marks a distinctive ILC2 activation subset in the mouse inflammatory lung
BMC Biology (2023)
-
Multiscale 3D genome organization underlies ILC2 ontogenesis and allergic airway inflammation
Nature Immunology (2023)
-
T cells in health and disease
Signal Transduction and Targeted Therapy (2023)