Abstract
Heparan sulfate (HS) proteoglycans are extended (-GlcAβ1,4GlcNAcα1,4-)n co-polymers containing decorations of sulfation and epimerization that are linked to cell surface and extracellular matrix proteins. In mammals, HS repeat units are extended by an obligate heterocomplex of two exostosin family members, EXT1 and EXT2, where each protein monomer contains distinct GT47 (GT-B fold) and GT64 (GT-A fold) glycosyltransferase domains. In this study, we generated human EXT1–EXT2 (EXT1–2) as a functional heterocomplex and determined its structure in the presence of bound donor and acceptor substrates. Structural data and enzyme activity of catalytic site mutants demonstrate that only two of the four glycosyltransferase domains are major contributors to co-polymer syntheses: the EXT1 GT-B fold β1,4GlcA transferase domain and the EXT2 GT-A fold α1,4GlcNAc transferase domain. The two catalytic sites are over 90 Å apart, indicating that HS is synthesized by a dissociative process that involves a single catalytic site on each monomer.







Similar content being viewed by others
Data availability
The cryo-EM 3D maps of human exostosin-1 and exostosin-2 heterodimer in the apo form (3.1 Å), bound to a 4-mer acceptor substrate (3.0 Å), bound to a 7-mer acceptor substrate (2.8 Å), complexed with UDP-GlcNAc (3.3 Å) or complexed with UDP-GlcA (3.0 Å), have been deposited in the Electron Microscopy Data Bank under accession codes EMD-25035, EMD-25036, EMD-25037, EMD-26701 and EMD-26702, respectively. The associated atomic models have been deposited in the Protein Data Bank (PDB) under accession codes 7SCH, 7SCJ, 7SCK, 7UQX and 7UQY, respectively. The crystal structure of the mouse EXTL2 (PDB accession code 1ON6) was used for comparison with human EXT1–2 structure. These data are available from the authors upon reasonable request. Source data are provided with this paper.
References
Carlsson, P. & Kjellen, L. in: Heparin—A Century of Progress. (eds Lever, R., Mulloy, B. & Page, C. P.) 23–40 (Springer, 2012).
Esko, J. D. & Selleck, S. B. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71, 435–471 (2002).
Sarrazin, S., Lamanna, W. C. & Esko, J. D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3, a004952 (2011).
Bishop, J. R., Schuksz, M. & Esko, J. D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 (2007).
Kim, S. H., Turnbull, J. & Guimond, S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 209, 139–151 (2011).
Busse-Wicher, M., Wicher, K. B. & Kusche-Gullberg, M. The exostosin family: proteins with many functions. Matrix Biol. 35, 25–33 (2014).
Sugahara, K. & Kitagawa, H. Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr. Opin. Struct. Biol. 10, 518–527 (2000).
Sugahara, K. & Kitagawa, H. Heparin and heparan sulfate biosynthesis. IUBMB Life 54, 163–175 (2002).
Annaval, T. et al. Heparan sulfate proteoglycans biosynthesis and post synthesis mechanisms combine few enzymes and few core proteins to generate extensive structural and functional diversity. Molecules 25, 4215 (2020).
McCormick, C., Duncan, G., Goutsos, K. T. & Tufaro, F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc. Natl Acad. Sci. USA 97, 668–673 (2000).
Kitagawa, H. & Nadanaka, S. in: Handbook of Glycosyltransferases and Related Genes (eds Taniguchi N. et al.) 905–923 (Springer Tokyo, 2014).
Busse, M. & Kusche-Gullberg, M. In vitro polymerization of heparan sulfate backbone by the EXT proteins. J. Biol. Chem. 278, 41333–41337 (2003).
Wei, G. et al. Location of the glucuronosyltransferase domain in the heparan sulfate copolymerase EXT1 by analysis of Chinese hamster ovary cell mutants. J. Biol. Chem. 275, 27733–27740 (2000).
Kim, B. T., Kitagawa, H., Tanaka, J., Tamura, J. & Sugahara, K. In vitro heparan sulfate polymerization: crucial roles of core protein moieties of primer substrates in addition to the EXT1–EXT2 interaction. J. Biol. Chem. 278, 41618–41623 (2003).
Drula, E. et al. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577 (2022).
Senay, C. et al. The EXT1/EXT2 tumor suppressors: catalytic activities and role in heparan sulfate biosynthesis. EMBO Rep. 1, 282–286 (2000).
Moremen, K. W. et al. Expression system for structural and functional studies of human glycosylation enzymes. Nat. Chem. Biol. 14, 156–162 (2018).
Pedersen, L. C. et al. Crystal structure of an α1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis. J. Biol. Chem. 278, 14420–14428 (2003).
Lairson, L. L., Henrissat, B., Davies, G. J. & Withers, S. G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008).
Moremen, K. W. & Haltiwanger, R. S. Emerging structural insights into glycosyltransferase-mediated synthesis of glycans. Nat. Chem. Biol. 15, 853–864 (2019).
Keys, T. G. et al. Engineering the product profile of a polysialyltransferase. Nat. Chem. Biol. 10, 437–442 (2014).
Urbanowicz, B. R., Pena, M. J., Moniz, H. A., Moremen, K. W. & York, W. S. Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J. 80, 197–206 (2014).
Chang, A., Singh, S., Phillips, G. N. Jr. & Thorson, J. S. Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr. Opin. Biotechnol. 22, 800–808 (2011).
Hecht, J. T. et al. Hereditary multiple exostoses (EXT): mutational studies of familial EXT1 cases and EXT-associated malignancies. Am. J. Hum. Genet. 60, 80–86 (1997).
Farhan, S. M. et al. Old gene, new phenotype: mutations in heparan sulfate synthesis enzyme, EXT2 leads to seizure and developmental disorder, no exostoses. J. Med. Genet. 52, 666–675 (2015).
Fokkema, I. et al. The LOVD3 platform: efficient genome-wide sharing of genetic variants. Eur. J. Hum. Genet. 29, 1796–1803 (2021).
Bukowska-Olech, E. et al. Hereditary multiple exostoses—a review of the molecular background, diagnostics, and potential therapeutic strategies. Front. Genet. 12, 759129 (2021).
Clement, N. & Porter, D. Hereditary multiple exostoses: anatomical distribution and burden of exostoses is dependent upon genotype and gender. Scott. Med. J. 59, 35–44 (2014).
Cook, A. et al. Genetic heterogeneity in families with hereditary multiple exostoses. Am. J. Hum. Genet. 53, 71–79 (1993).
Stenson, P. D. et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 136, 665–677 (2017).
Bovee, J. V. et al. EXT-mutation analysis and loss of heterozygosity in sporadic and hereditary osteochondromas and secondary chondrosarcomas. Am. J. Hum. Genet. 65, 689–698 (1999).
Raskind, W. H. et al. Evaluation of locus heterogeneity and EXT1 mutations in 34 families with hereditary multiple exostoses. Hum. Mutat. 11, 231–239 (1998).
Wuyts, W. et al. Mutations in the EXT1 and EXT2 genes in hereditary multiple exostoses. Am. J. Hum. Genet. 62, 346–354 (1998).
Philippe, C. et al. Mutation screening of the EXT1 and EXT2 genes in patients with hereditary multiple exostoses. Am. J. Hum. Genet. 61, 520–528 (1997).
Morimoto, K. et al. Transgenic expression of the EXT2 gene in developing chondrocytes enhances the synthesis of heparan sulfate and bone formation in mice. Biochem. Biophys. Res. Commun. 292, 999–1009 (2002).
Li, Z. et al. Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat. Chem. Biol. 13, 757–763 (2017).
Lira-Navarrete, E. & Hurtado-Guerrero, R. A perspective on structural and mechanistic aspects of protein O-fucosylation. Acta Crystallogr F Struct. Biol. Commun. 74, 443–450 (2018).
Lira-Navarrete, E. et al. Structural insights into the mechanism of protein O-fucosylation. PLoS ONE 6, e25365 (2011).
Urbanowicz, B. R. et al. Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism. Plant J. 91, 931–949 (2017).
Negishi, M., Dong, J., Darden, T. A., Pedersen, L. G. & Pedersen, L. C. Glucosaminylglycan biosynthesis: what we can learn from the X-ray crystal structures of glycosyltransferases GlcAT1 and EXTL2. Biochem. Biophys. Res. Commun. 303, 393–398 (2003).
Albesa-Jove, D., Sainz-Polo, M. A., Marina, A. & Guerin, M. E. Structural snapshots of α-1,3-galactosyltransferase with native substrates: insight into the catalytic mechanism of retaining glycosyltransferases. Angew. Chem. Int. Ed. Engl. 56, 14853–14857 (2017).
Yu, H. et al. Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat. Chem. Biol. 11, 847–854 (2015).
Kreuger, J. & Kjellen, L. Heparan sulfate biosynthesis: regulation and variability. J. Histochem. Cytochem. 60, 898–907 (2012).
Kitagawa, H., Shimakawa, H. & Sugahara, K. The tumor suppressor EXT-like gene EXTL2 encodes an α1, 4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of heparan sulfate. J. Biol. Chem. 274, 13933–13937 (1999).
Wilson, L. F. L. et al. The structure of EXTL3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis. Nat. Commun. 13, 3314 (2022).
Kim, B. T. et al. Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode α1,4- N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/ heparin biosynthesis. Proc. Natl Acad. Sci. USA 98, 7176–7181 (2001).
Osawa, T. et al. Crystal structure of chondroitin polymerase from Escherichia coli K4. Biochem. Biophys. Res. Commun. 378, 10–14 (2009).
Katz, M. & Diskin, R. The homodimeric structure of the LARGE1 dual glycosyltransferase. Preprint at https://www.biorxiv.org/content/10.1101/2022.05.11.491581v1.full (2022).
Joseph, S. et al. Structure and mechanism of LARGE1 matriglycan polymerase. Preprint at https://www.biorxiv.org/content/10.1101/2022.05.12.491222v1 (2022).
Maloney, F. P. et al. Structure, substrate recognition and initiation of hyaluronan synthase. Nature 604, 195–201 (2022).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Buchan, D. W. A. & Jones, D. T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 47, W402–W407 (2019).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Miteva, M. A., Guyon, F. & Tuffery, P. Frog2: efficient 3D conformation ensemble generator for small compounds. Nucleic Acids Res. 38, W622–W627 (2010).
Acknowledgements
Cryo-EM images were collected in the David Van Andel Advanced Cryo-Electron Microscopy Suite at the Van Andel Institute. We thank G. Zhao and X. Meng for facilitating data collection, R. Weiss for helpful discussions on HS biology and S. Archer-Hartman for LC–MS analysis of oligosaccharide substrates (supported by National Institutes of Health (NIH) grant R24GM137782 to P. Azadi). This work was supported by NIH grants R01GM130915 and P41GM103390 (to K.W.M.) and R01CA231466 (to Huilin Li) and the Van Andel Institute (to Huilin Li). R.A.A was supported by funding from the US Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences and Biosciences Division, under award DE-SC0015662. Materials generated in this study are freely available upon sending a request to the corresponding authors.
Author information
Authors and Affiliations
Contributions
K.W.M. and Huilin Li conceived and designed the experiments. D.C. expressed and purified proteins and performed enzymatic and functional assays on wild-type and mutant proteins. A.R. generated all mammalian expression constructs and mutants. R.A.A. and D.C. performed analysis of enzymatic products by mass spectrometry. Hua Li performed cryo-EM, image analysis, 3D reconstruction, atomic model building and molecular docking. Hua Li, D.C., K.W.M. and Huilin Li analyzed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Kamil Godula and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
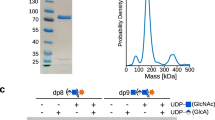
Extended Data Fig. 1 Expression and purification of the EXT1-2 complex for structural studies.
a, Single and co-expression of EXT1 and EXT2 as small N-terminal fusion proteins (pGEn1 vector) and larger N-terminal fusion proteins (pGEn2 vector). SDS-PAGE of the expressed and purified construct combinations is shown indicating only co-expression of EXT1 and EXT2 in either fusion vector format leads to appreciable secretion of the respective fusion proteins. b, The purified EXT1-2 complex prior to TEV cleavage was characterized by size exclusion-multiangle light scattering (SEC-MALS). A280 is shown by the green line, refractive index in blue, light scattering in red and calculated molar mass in black. The molecular mass derived from SEC-MALS analysis (~223 kDa) is in close agreement with the predicted size of the heterodimeric EXT1-2 complex with the respective fusion tags (EXT1: 88 kDa + EXT2: 109 kDa + 4 N-glycans). b, The EXT1-pGEn1 and EXT2-pGEn2 expression constructs were co-transfected into HEK293F cells and the secreted heterocomplex in the media (Crude media) was purified by Ni2+-NTA purification (IMAC run-through, Wash 1, Wash 2) to yield a highly-enriched enzyme preparation (IMAC1 elution). The enzyme was concentrated (IMAC1 elution conc) and cleaved with TEV protease to remove the fusion tag sequences (+TEV). Ni2+-NTA chromatography separated the unbound EXT1-2 complex from the bound tag sequences and TEV protease (IMAC2 runthru), as the latter were all His-tagged. Panels a and b are each representative of data from two biological replicate experiments. Original uncropped images are provided in the Source Data.
Extended Data Fig. 2 Structure of EXT1-2.
a, Superimposition of GT-B fold of EXT1 and EXT2. The intervening loop in red dash cycle. b, Superimposition of GT-A fold from EXT1, EXT2, and mEXTL2 (PDB ID: 1ON6). c, Top view of GT-B fold in EXT1-2, suggesting a pseudo-twofold symmetry. d, Superimposition of GT-A fold from EXT1-2 and mEXTL2 (PDB ID: 1ON6) in top view, suggesting a pseudo-twofold symmetry for GT-A fold in EXT1-2. EXT1-2 is colored the same as Fig. 1, and mEXTL2 in cyan.
Extended Data Fig. 3 Comparison of 4-mer complex with EXT1-2.
a, Superimposition of EXT1-2 and 4-mer complex. b, Superimposition of EXT1 GT-B fold from EXT1-2 and 4-mer complex. 4-mer complex colored the same as in Fig. 1, and EXT1-2 in light gray. Residues involved in active pocket are shown as sticks. The disordered loop H300-C302 in EXT1-2 marked in red dashed oval. The density of NAG at Asn330 is shown as 50% transparency.
Extended Data Fig. 4 Comparison of 7-mer complex with EXT1-2, 4-mer complex, and EXT1-2 complexed with UDP-GlcA.
a, Superimposition of EXT1-2, 4-mer and 7-mer complex. b, Orthogonal view highlighting the GT-A fold comparison. 7-mer complex colored the same as in Fig. 1, EXT1-2 in light gray, and 4-mer complex in wheat. c, Superimposition of EXT1 GT-B fold from EXT1-2 complexed with UDP-GlcA and 7-mer complex. 7-mer complex are colored the same as in Fig. 1, and EXT1-2 complexed with UDP-GlcA in cyan. Residues involved in active pocket are shown as sticks.
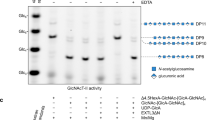
Extended Data Fig. 5 Development of an HS co-polymerase assay for EXT1-2.
A co-polymerase assay was developed using the HS proteoglycan, glypican-1 (a and b), or heparosan primers (4-mer-pNP or 5-mer-pNP) (c) as acceptors and UDP-GlcA and UDP-GlcNAc (200 µM each) as donors. Individual assay components were tested in the reaction mixture and HS polymerization was detected using the UDP-Glo assay format (a and c). Recombinant human glypican-1 was expressed in HEK293 cells and purified for use as an acceptor substrate for HS extension of the proteoglycan. a, Low, but detectable activity was revealed when single UDP sugars were added to the reaction mix reflecting single sugar additions to the HS chains. When both UDP-GlcA and UDP-GlcNAc were added to the reaction, the enzyme activity increased ~40 fold indicating an iterative use of the sugar donors during the HS extension reaction. b, Reaction products were resolved by SDS-PAGE and Coomassie staining. Extended HS polymer product on glypican-1 appearing as a high molecular weight smear (red brackets) was detected only when enzyme and both sugar nucleotide donors were present in the co-polymerase reaction. The data are representative of n = 2 biologically independent samples. Original uncropped images are provided in the Source Data. c, UDP-Glo reactions using 4-mer-pNP and 5-mer-pNP as acceptors. Extension of the 4-mer primer with single sugar nucleotide donors only occurred when UDP-GlcA, but not UDP-GlcNAc, was used as donor as expected for an acceptor containing a non-reducing terminal GlcNAc residue. In contrast, extension of the 5-mer primer with single sugar nucleotide donors only occurred with UDP-GlcNAc, but not UDP-GlcA, was used as donor consistent with the presence of a non-reducing terminal GlcA residue on the acceptor. Addition of both donors in extension reactions with either the 4-mer or 5-mer as acceptor led to enhanced activity indicating an iterative use of the sugar donors during the HS extension reaction. Plots show the mean values (bar) ± s.d. (error bars) for n = 2 technical replicates (red circles).
Extended Data Fig. 6 Expression and purification of EXT1 and EXT2 mutants.
a. Diagrammatic representation of the positions of the EXT1 and EXT2 active site mutations that were generated. The respective GT-B and GT-A domains are indicated relative to the N-terminal GFP encoded by the pGEn2 vector. Residues for mutagenesis were chosen based on proximity to the respective GT-A or GT-B domain active sites. Truncation to form expression constructs encoding the respective single domains are also indicated. Termination codons were introduced in the linker regions between the GT-A and GT-B domains of EXT1 and EXT2 to generate the respective single GT-B domains (EXT1-GT-B and EXT2 GT-B). Each of the GT-A domains were isolated by PCR and transferred into the pGEn2 vector (EXT1 GT-A and EXT2 GT-A) to generate the respective single GT-A domains. Each of the mutants in the pGEn2 vector were co-expressed along with wild type versions of the corresponding partner EXT (in pGEn1 vector) to generate a secreted heterocomplex (for example EXT1 K269A mutant co-expressed with wild type EXT2) that was subsequently purified by Ni2+-NTA chromatography and resolved by SDS PAGE c, Initial expression level of the proteins in the culture media was monitored by GFP fluorescence b, since the mutant EXT form harbored a GFP fusion as a result of expression in the pGEn2 vector. The upper band on the SDS-PAGE gel (~115 kDa) corresponds to the mutant EXT form expressed in the pGEn2 vector, while the lower band (~90 kDa) corresponds to the wild-type EXT form (pGEn1 vector) that was co-expressed. Individual EXT1 and EXT2 isoforms and individual domains were also expressed singly or in combination. Of these latter expression tests, only EXT1 alone or the individual EXT2 GT-A or GT-B domains resulted in any appreciable secreted products. Each co-transfection experiment was performed once and two different SDS PAGE gels of the samples were generated. Data presented are representative of the respective experiments. Original uncropped images are provided in the Source Data.
Extended Data Fig. 7 Enzymatic extension of 4-mer and 5-mer acceptor primers with single sugar residues.
To complement the UDP-Glo assays performed in Fig. 4 and Extended Data Fig. 5, we analyzed the reaction products for glycan extension by MALDI-MS. Individual spectra were obtained for the indicated enzyme, sugar nucleotide donors (a and b), and the 4-mer-pNP (c) and 5-mer-pNP (e) acceptors. Electrospray mass spectra of each of the synthetic acceptors indicated a predicted parent mass of 897.75 and 1073.87 for the 4-mer-pNP and 5-mer-pNP acceptors, respectively (c and f). However, MALDI-MS of the compounds produced a broad pair of mass peaks where the smaller, higher mass peak matched the predicted mass (singly charged masses of 898 and 1074 for the 4-mer-pNP and 5-mer-pNP acceptor, respectively, insets for panels c and f at the top). For each compound, the more abundant second species was 15 mass units smaller than the predicted mass, consistent with an in-source rearrangement resulting in neutral mass loss during MALDI analysis. The cause of this 15 mass unit neutral loss is not clear at the present time, but it did not impact the ability of the 4-mer and 5-mer to act as acceptors for extension by EXT1-2. Reactions containing the 4-mer acceptor, which harbored a reducing terminal GlcNAc residue, could be extended by the mass of a GlcA unit in the presence of the UDP-GlcA donor (e), while reactions containing the UDP-GlcNAc donor led to no extension (d). By comparison, reactions containing the 5-mer acceptor (harboring a reducing terminal GlcA residue) could be extended by the mass of a GlcNAc residue in the presence of a UDP-GlcNAc donor (g), but reactions containing UDP-GlcA as donor led to no extension (h). Data presented are representative of n > 3 independent reactions.
Extended Data Fig. 8 Time course of co-polymer extension of 4-mer and 5-mer primers.
While single sugar residue extensions were observed on 4-mer and 5-mer primers in the presence of UDP-GlcA and UDP-GlcNAc, respectively, a time course of co-polymer extension was examined by the addition of both donors to reactions containing the 4-mer or 5-mer primers. Blank reactions containing boiled enzyme (red asterisk) and both sugar nucleotides (a), 4-mer-pNP (b) or 5-mer-pNP (f) were examined for comparison to reactions containing 4-mer-pNP, enzyme and both donors (c-e) or 5-mer-pNP, enzyme and both donors (g-i) in a time course extension. Both reactions showed a similar set of extension products with the 4-mer being extended by the addition of a GlcA residue to form a 5-mer product followed by a GlcNAc addition and subsequent extension of a disaccharide unit terminating in a GlcNAc sugar residue indicating that the GlcA transferase activity was rate limiting (c-e). A similar extension of the 5-mer-pNP was observed with the extension to an 18-mer over the course of the 1 h reaction (g-i). Data presented are representative of n > 3 independent reactions.
Extended Data Fig. 9 Comparison of the catalytic pockets of GT-A domains and GT-B domains explain why some of these domains are inactive.
a, b, Electrostatic potential of GT-B cleft in EXT1 and EXT2. Positive charged surface colored in blue, and negative charged surface in red. Superimposition of GT-B fold of EXT1 and EXT2. c, d, Sequence alignment and close-up view of UDP binding pocket of EXT1 and EXT2 GT-B. The key residues are marked and shown in sticks. e, Superimposition of active pocket in hEXT1 GT-A, hEXT2 GT-A, and mEXTL2 (PDB ID: 1ON6). The residues involved in substrate interaction shown in stick, and residues in hEXT2 are labeled, except for V487, P489, and K684 those not conserved in hEXT1. hEXT1 and hEXT2 colored the same as in Fig. 1, mEXTL2 colored in cyan. f, Sequence alignment of hEXT1, hEXT2, and mEXTL2. The residues involved in substrate interaction marked in star, the same one in hEXT1 and hEXT2 in magenta, while different one in black. The first (loop487-495) and last (loop-C) loop are marked in olive dashed frame.
Extended Data Fig. 10 Mapping of disease related missense mutations onto the structure of EXT1 and EXT2.
Mutations in the EXT1 and EXT2 coding regions that lead to disease were identified in the Human Gene Mutation Database30 and are shown on the linear representations of the respective protein sequences as colored circles (c and d) and as cyan spheres on the structural representation of EXT1 (a) and EXT2 (b). EXT1-2 are colored the same as Fig. 1. Disease characteristics shown in the legend are based on the annotations for the respective mutations in the Human Gene Mutation Database.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Supplementary Figs. 1–7
Source data
Source Data Fig. 1
Unprocessed gel
Source Data Fig. 1
Unprocessed gel
Source Data Fig. 5
Unprocessed gel
Source Data Fig. 6
Unprocessed gel
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, H., Chapla, D., Amos, R.A. et al. Structural basis for heparan sulfate co-polymerase action by the EXT1–2 complex. Nat Chem Biol 19, 565–574 (2023). https://doi.org/10.1038/s41589-022-01220-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01220-2
- Springer Nature America, Inc.
This article is cited by
-
Structural and mechanistic characterization of bifunctional heparan sulfate N-deacetylase-N-sulfotransferase 1
Nature Communications (2024)
-
Molecular mechanism of decision-making in glycosaminoglycan biosynthesis
Nature Communications (2023)