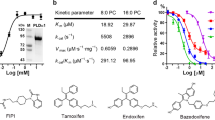
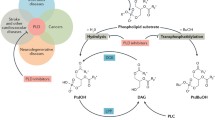
Abstract
Phospholipase D enzymes (PLDs) are ubiquitous phosphodiesterases that produce phosphatidic acid (PA), a key second messenger and biosynthetic building block. Although an orthologous bacterial Streptomyces sp. strain PMF PLD structure was solved two decades ago, the molecular basis underlying the functions of the human PLD enzymes (hPLD) remained unclear based on this structure due to the low homology between these sequences. Here, we describe the first crystal structures of hPLD1 and hPLD2 catalytic domains and identify novel structural elements and functional differences between the prokaryotic and eukaryotic enzymes. Furthermore, structure-based mutation studies and structures of inhibitor–hPLD complexes allowed us to elucidate the binding modes of dual and isoform-selective inhibitors, highlight key determinants of isoenzyme selectivity and provide a basis for further structure-based drug discovery and functional characterization of this therapeutically important superfamily of enzymes.





Similar content being viewed by others
Data availability
Structure data that support the findings of this study have been deposited in the RCSB Protein Data Bank (http://www.rcsb.org) under the following accession codes: PLD2-WO4, 6OHM; PLD2-apo, 6OHO; PLD2-1, 6OHP; PLD2-3, 6OHS; PLD2-4, 6OHQ; PLD1-5, 6OHR. The data that support the remaining findings of this study are available from the corresponding author upon reasonable request.
References
Peng, X. & Frohman, M. A. Mammalian phospholipase D physiological and pathological roles. Acta Physiol. (Oxf.) 204, 219–226 (2012).
Arhab, Y., Abousalham, A. & Noiriel, A. Plant phospholipase D mining unravels new conserved residues important for catalytic activity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864, 688–703 (2019).
Celma, L. et al. Structural basis for the substrate selectivity of Helicobacter pylori NucT nuclease activity. PLoS One 12, e0189049 (2017).
Bozatzi, P. & Sapkota, G. P. The FAM83 family of proteins: from pseudo-PLDs to anchors for CK1 isoforms. Biochem. Soc. Trans. 46, 761–771 (2018).
Sung, T. C. et al. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 16, 4519–4530 (1997).
Secundo, F., Carrea, G., D’Arrigo, P. & Servi, S. Evidence for an essential lysyl residue in phospholipase D from Streptomyces sp. by modification with diethyl pyrocarbonate and pyridoxal 5-phosphate. Biochemistry 35, 9631–9636 (1996).
Iwasaki, Y., Horiike, S., Matsushima, K. & Yamane, T. Location of the catalytic nucleophile of phospholipase D of Streptomyces antibioticus in the C-terminal half domain. Eur. J. Biochem. 264, 577–581 (1999).
Ponting, C. P. & Kerr, I. D. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 5, 914–922 (1996).
Koonin, E. V. A duplicated catalytic motif in a new superfamily of phosphohydrolases and phospholipid synthases that includes poxvirus envelope proteins. Trends Biochem. Sci. 21, 242–243 (1996).
Sung, T. C., Altshuller, Y. M., Morris, A. J. & Frohman, M. A. Molecular analysis of mammalian phospholipase D2. J. Biol. Chem. 274, 494–502 (1999).
Selvy, P. E., Lavieri, R. R., Lindsley, C. W., & Brown, H. A. Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 111, 6064–6119 (2011).
Leiros, I., McSweeney, S. & Hough, E. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J. Mol. Biol. 339, 805–820 (2004).
Leiros, I. et al. Crystallization and preliminary X-ray diffraction studies of phospholipase D from Streptomyces sp. Acta Crystallogr. D 56, 466–468 (2000).
Uesugi, Y. & Hatanaka, T. Phospholipase D mechanism using Streptomyces PLD. Biochim. Biophys. Acta 1791, 962–969 (2009).
Hammond, S. M. et al. Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-alpha. J. Biol. Chem. 272, 3860–3868 (1997).
Colley, W. C. et al. Cloning and expression analysis of murine phospholipase D1. Biochem J. 326, 745–753 (1997).
Nelson, R. K. & Frohman, M. A. Physiological and pathophysiological roles for phospholipase D. J. Lipid Res. 56, 2229–2237 (2015).
Frohman, M. A. The phospholipase D superfamily as therapeutic targets. Trends Pharm. Sci. 36, 137–144 (2015).
Gomez-Cambronero, J. & Ganesan, R. Targeting phospholipase D genetically and pharmacologically for studying leukocyte function. Methods Mol. Biol. 1835, 297–314 (2018).
Brown, H. A., Thomas, P. G. & Lindsley, C. W. Targeting phospholipase D in cancer, infection and neurodegenerative disorders. Nat. Rev. Drug Discov. 16, 351–367 (2017).
Waterson, A. G. et al. Isoform selective PLD inhibition by novel, chiral 2,8-diazaspiro[4.5]decan-1-one derivatives. Bioorg. Med. Chem. Lett. 28, 3670–3673 (2018).
Aloulou, A., Rahier, R., Arhab, Y., Noiriel, A. & Abousalham, A. Phospholipases: an overview. Methods Mol. Biol. 1835, 69–105 (2018).
Uesugi, Y., Arima, J., Iwabuchi, M. & Hatanaka, T. C-terminal loop of Streptomyces phospholipase D has multiple functional roles. Protein Sci. 16, 197–207 (2007).
Sung, T. C., Zhang, Y., Morris, A. J. & Frohman, M. A. Structural analysis of human phospholipase D1. J. Biol. Chem. 274, 3659–3666 (1999).
Stuckey, J. A. & Dixon, J. E. Crystal structure of a phospholipase D family member. Nat. Struct. Biol. 6, 278–284 (1999).
Morris, A. J., Engebrecht, J. & Frohman, M. A. Structure and regulation of phospholipase D. Trends Pharm. Sci. 17, 182–185 (1996).
Hammond, S. M. et al. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J. Biol. Chem. 270, 29640–29643 (1995).
Exton, J. H. Phospholipase D. Ann. N. Y. Acad. Sci. 905, 61–68 (2000).
Liu, M. Y., Gutowski, S. & Sternweis, P. C. The C terminus of mammalian phospholipase D is required for catalytic activity. J. Biol. Chem. 276, 5556–5562 (2001).
Sciorra, V. A. et al. Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 18, 5911–5921 (1999).
Du, G. et al. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 162, 305–315 (2003).
De Cuyper, H., van Praag, H. M. & Verstraeten, D. The clinical significance of halopemide, a dopamine-blocker related to the butyrophenones. Neuropsychobiology 12, 211–216 (1984).
Lavieri, R. R. et al. Design, synthesis, and biological evaluation of halogenated N-(2-(4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-8-yl)ethyl)benzamides: discovery of an isoform-selective small molecule phospholipase D2 inhibitor. J. Med. Chem. 53, 6706–6719 (2010).
Lewis, J. A. et al. Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: impact of alternative halogenated privileged structures for PLD1 specificity. Bioorg. Med. Chem. Lett. 19, 1916–1920 (2009).
Monovich, L. et al. Optimization of halopemide for phospholipase D2 inhibition. Bioorg. Med. Chem. Lett. 17, 2310–2311 (2007).
O’Reilly, M. C. et al. Development of dual PLD1/2 and PLD2 selective inhibitors from a common 1,3,8-triazaspiro[4.5]decane core: discovery of Ml298 and Ml299 that decrease invasive migration in U87-MG glioblastoma cells. J. Med. Chem. 56, 2695–2699 (2013).
Scott, S. A. et al. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 5, 108–117 (2009).
Scott, S. A. et al. Discovery of desketoraloxifene analogues as inhibitors of mammalian, Pseudomonas aeruginosa, and NAPE phospholipase D enzymes. ACS Chem. Biol. 10, 421–432 (2015).
Ganesan, R., Mahankali, M., Alter, G. & Gomez-Cambronero, J. Two sites of action for PLD2 inhibitors: the enzyme catalytic center and an allosteric, phosphoinositide biding pocket. Biochim. Biophys. Acta 1851, 261–272 (2015).
Kam, Y. & Exton, J. H. Dimerization of phospholipase D isozymes. Biochem. Biophys. Res. Commun. 290, 375–380 (2002).
O’Reilly, M. C. et al. Discovery of a highly selective PLD2 inhibitor (ML395): a new probe with improved physiochemical properties and broad-spectrum antiviral activity against influenza strains. ChemMedChem 9, 2633–2637 (2014).
O’Reilly, M. C., Scott, S. A., Brown, H. A. & Lindsley, C. W. Further evaluation of novel structural modifications to scaffolds that engender PLD isoform selective inhibition. Bioorg. Med. Chem. Lett. 24, 5553–5557 (2014).
Abergel, C., Walburger, A., Chenivesse, S. & Lazdunski, C. Crystallization and preliminary crystallographic study of the peptidoglycan-associated lipoprotein from Escherichia coli. Acta Crystallogr. D 57, 317–319 (2001).
Mahankali, M., Alter, G. & Gomez-Cambronero, J. Mechanism of enzymatic reaction and protein–protein interactions of PLD from a 3D structural model. Cell Signal 27, 69–81 (2015).
Frohman, M. A., Sung, T. C. & Morris, A. J. Mammalian phospholipase D structure and regulation. Biochim. Biophys. Acta 1439, 175–186 (1999).
Panda, A. et al. Functional analysis of mammalian phospholipase D enzymes. Biosci. Rep. 38, BSR20181690 (2018).
Auerbach, A. Activation of endplate nicotinic acetylcholine receptors by agonists. Biochem. Pharm. 97, 601–608 (2015).
Dvir, H., Silman, I., Harel, M., Rosenberry, T. L. & Sussman, J. L. Acetylcholinesterase: from 3D structure to function. Chem. Biol. Interact. 187, 10–22 (2010).
Pettitt, T. R., McDermott, M., Saqib, K. M., Shimwell, N. & Wakelam, M. J. Phospholipase D1b and D2a generate structurally identical phosphatidic acid species in mammalian cells. Biochem. J. 360, 707–715 (2001).
Lerchner, A., Mansfeld, J., Kuppe, K. & Ulbrich-Hofmann, R. Probing conserved amino acids in phospholipase D (Brassica oleracea var. capitata) for their importance in hydrolysis and transphosphatidylation activity. Protein Eng. Des. Sel. 19, 443–452 (2006).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Saerens, D. et al. Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies. J. Mol. Biol. 352, 597–607 (2005).
Hall, D. R. & Enyedy, I. J. Computational solvent mapping in structure-based drug design. Future Med. Chem. 7, 337–353 (2015).
Acknowledgements
We acknowledge the Paul Scherrer Institut, Villigen, Switzerland for provision of synchrotron radiation beamtime at beamlines PXI and PXII of the SLS and thank J. Diez at Expose GMBH for data collection at SLS and D. Esposito at Leidos (Frederick National Laboratory) for T. ni cells and small-scale expression and purification of hPLD1b.
Author information
Authors and Affiliations
Contributions
J.V.C. solved the structures of hPLD2 and C.M.M. solved the structure of hPLD1b. M.C. and P.R.K. expressed and purified hPLD proteins. I.J.E. performed modeling experiments and K.M. performed SPR experiments. E.A.P., T.L.M.-D. and T.C. contributed to compound design and synthesis. J.C.S., P.M. and K.A.S. performed biochemical assays. All authors contributed to data interpretation. J.V.C., C.M.M. and E.A.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Biogen funded these studies. C.M.M., E.A.P., J.C.S., I.J.E., P.M., T.C., K.M., M.C., K.A.S., P.R.K., T.L.M.-D. and J.V.C. are employees and shareholders of Biogen.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Figs. 1–4 and Note.
Rights and permissions
About this article
Cite this article
Metrick, C.M., Peterson, E.A., Santoro, J.C. et al. Human PLD structures enable drug design and characterization of isoenzyme selectivity. Nat Chem Biol 16, 391–399 (2020). https://doi.org/10.1038/s41589-019-0458-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0458-4
- Springer Nature America, Inc.
This article is cited by
-
Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C
Nature Communications (2023)
-
Inhibition of phospholipase D1 induces immunogenic cell death and potentiates cancer immunotherapy in colorectal cancer
Experimental & Molecular Medicine (2022)
-
Structural insights into PA3488-mediated inactivation of Pseudomonas aeruginosa PldA
Nature Communications (2022)
-
Illustrating human PLD
Nature Chemical Biology (2020)