Abstract
The selectivity of neuronal responses arises from the architecture of excitatory and inhibitory connections. In the primary visual cortex, the selectivity of a neuron in layer 2/3 for stimulus orientation and direction is thought to arise from intracortical inputs that are similarly selective1,2,3,4,5,6,7,8. However, the excitatory inputs of a neuron can have diverse stimulus preferences1,2,3,4,6,7,9, and inhibitory inputs can be promiscuous10 and unselective11. Here we show that the excitatory and inhibitory intracortical connections to a layer 2/3 neuron accord with its selectivity by obeying precise spatial patterns. We used rabies tracing1,12 to label and functionally image the excitatory and inhibitory inputs to individual pyramidal neurons of layer 2/3 of the mouse visual cortex. Presynaptic excitatory neurons spanned layers 2/3 and 4 and were distributed coaxial to the preferred orientation of the postsynaptic neuron, favouring the region opposite to its preferred direction. By contrast, presynaptic inhibitory neurons resided within layer 2/3 and favoured locations near the postsynaptic neuron and ahead of its preferred direction. The direction selectivity of a postsynaptic neuron was unrelated to the selectivity of presynaptic neurons, but correlated with the spatial displacement between excitatory and inhibitory presynaptic ensembles. Similar asymmetric connectivity establishes direction selectivity in the retina13,14,15,16,17. This suggests that this circuit motif might be canonical in sensory processing.




Similar content being viewed by others
Data availability
Source data for the figures are deposited at https://github.com/lfedros/Rossi-et-al-2020. All other relevant data are available from the corresponding author upon reasonable request.
Code availability
Code used in this study is deposited at https://github.com/lfedros/Rossi-et-al-2020.
References
Wertz, A. et al. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science 349, 70–74 (2015).
Ko, H. et al. Functional specificity of local synaptic connections in neocortical networks. Nature 473, 87–91 (2011).
Iacaruso, M. F., Gasler, I. T. & Hofer, S. B. Synaptic organization of visual space in primary visual cortex. Nature 547, 449–452 (2017).
Lee, W. C. et al. Anatomy and function of an excitatory network in the visual cortex. Nature 532, 370–374 (2016).
Wilson, D. E., Whitney, D. E., Scholl, B. & Fitzpatrick, D. Orientation selectivity and the functional clustering of synaptic inputs in primary visual cortex. Nat. Neurosci. 19, 1003–1009 (2016).
Cossell, L. et al. Functional organization of excitatory synaptic strength in primary visual cortex. Nature 518, 399–403 (2015).
Wilson, D. E., Scholl, B. & Fitzpatrick, D. Differential tuning of excitation and inhibition shapes direction selectivity in ferret visual cortex. Nature 560, 97–101 (2018).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Jia, H., Rochefort, N. L., Chen, X. & Konnerth, A. Dendritic organization of sensory input to cortical neurons in vivo. Nature 464, 1307–1312 (2010).
Packer, A. M. & Yuste, R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J. Neurosci. 31, 13260–13271 (2011).
Hofer, S. B. et al. Differential connectivity and response dynamics of excitatory and inhibitory neurons in visual cortex. Nat. Neurosci. 14, 1045–1052 (2011).
Wickersham, I. R. et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647 (2007).
Barlow, H. B. & Levick, W. R. The mechanism of directionally selective units in rabbit’s retina. J. Physiol. (Lond.) 178, 477–504 (1965).
Fried, S. I., Münch, T. A. & Werblin, F. S. Mechanisms and circuitry underlying directional selectivity in the retina. Nature 420, 411–414 (2002).
Briggman, K. L., Helmstaedter, M. & Denk, W. Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–188 (2011).
Vaney, D. I., Sivyer, B. & Taylor, W. R. Direction selectivity in the retina: symmetry and asymmetry in structure and function. Nat. Rev. Neurosci. 13, 194–208 (2012).
Kim, J. S. et al. Space-time wiring specificity supports direction selectivity in the retina. Nature 509, 331–336 (2014).
Alonso, J. M. & Martinez, L. M. Functional connectivity between simple cells and complex cells in cat striate cortex. Nat. Neurosci. 1, 395–403 (1998).
Priebe, N. J. Mechanisms of orientation selectivity in the primary visual cortex. Annu. Rev. Vis. Sci. 2, 85–107 (2016).
Isaacson, J. S. & Scanziani, M. How inhibition shapes cortical activity. Neuron 72, 231–243 (2011).
Liu, B. H. et al. Broad inhibition sharpens orientation selectivity by expanding input dynamic range in mouse simple cells. Neuron 71, 542–554 (2011).
Znamenskiy, P. et al. Functional selectivity and specific connectivity of inhibitory neurons in primary visual cortex. Preprint at https://doi.org/10.1101/294835 (2018).
Lee, S. H. et al. Activation of specific interneurons improves V1 feature selectivity and visual perception. Nature 488, 379–383 (2012).
Atallah, B. V., Bruns, W., Carandini, M. & Scanziani, M. Parvalbumin-expressing interneurons linearly transform cortical responses to visual stimuli. Neuron 73, 159–170 (2012).
Wilson, N. R., Runyan, C. A., Wang, F. L. & Sur, M. Division and subtraction by distinct cortical inhibitory networks in vivo. Nature 488, 343–348 (2012).
Ma, W. P. et al. Visual representations by cortical somatostatin inhibitory neurons–selective but with weak and delayed responses. J. Neurosci. 30, 14371–14379 (2010).
Monier, C., Chavane, F., Baudot, P., Graham, L. J. & Frégnac, Y. Orientation and direction selectivity of synaptic inputs in visual cortical neurons: a diversity of combinations produces spike tuning. Neuron 37, 663–680 (2003).
Livingstone, M. S. Mechanisms of direction selectivity in macaque V1. Neuron 20, 509–526 (1998).
Xu, X. et al. Primary visual cortex shows laminar-specific and balanced circuit organization of excitatory and inhibitory synaptic connectivity. J. Physiol. (Lond.) 594, 1891–1910 (2016).
Adesnik, H. & Scanziani, M. Lateral competition for cortical space by layer-specific horizontal circuits. Nature 464, 1155–1160 (2010).
Holmgren, C., Harkany, T., Svennenfors, B. & Zilberter, Y. Pyramidal cell communication within local networks in layer 2/3 of rat neocortex. J. Physiol. (Lond.) 551, 139–153 (2003).
Weiler, S. et al. Relationship between input connectivity, morphology and orientation tuning of layer 2/3 pyramidal cells in mouse visual cortex. Preprint at bioRxiv https://doi.org/10.1101/2020.06.03.127191 (2020).
Sun, W., Tan, Z., Mensh, B. D. & Ji, N. Thalamus provides layer 4 of primary visual cortex with orientation- and direction-tuned inputs. Nat. Neurosci. 19, 308–315 (2016).
Ringach, D. L. et al. Spatial clustering of tuning in mouse primary visual cortex. Nat. Commun. 7, 12270 (2016).
Marques, T., Nguyen, J., Fioreze, G. & Petreanu, L. The functional organization of cortical feedback inputs to primary visual cortex. Nat. Neurosci. 21, 757–764 (2018).
Bonin, V., Histed, M. H., Yurgenson, S. & Reid, R. C. Local diversity and fine-scale organization of receptive fields in mouse visual cortex. J. Neurosci. 31, 18506–18521 (2011).
Garrett, M. E., Nauhaus, I., Marshel, J. H. & Callaway, E. M. Topography and areal organization of mouse visual cortex. J. Neurosci. 34, 12587–12600 (2014).
Haider, B., Häusser, M. & Carandini, M. Inhibition dominates sensory responses in the awake cortex. Nature 493, 97–100 (2013).
Liu, B. H. et al. Intervening inhibition underlies simple-cell receptive field structure in visual cortex. Nat. Neurosci. 13, 89–96 (2010).
Li, Y. T., Liu, B. H., Chou, X. L., Zhang, L. I. & Tao, H. W. Strengthening of direction selectivity by broadly tuned and spatiotemporally slightly offset inhibition in mouse visual cortex. Cereb. Cortex 25, 2466–2477 (2015).
Lien, A. D. & Scanziani, M. Tuned thalamic excitation is amplified by visual cortical circuits. Nat. Neurosci. 16, 1315–1323 (2013).
Li, Y. T., Ibrahim, L. A., Liu, B. H., Zhang, L. I. & Tao, H. W. Linear transformation of thalamocortical input by intracortical excitation. Nat. Neurosci. 16, 1324–1330 (2013).
Hillier, D. et al. Causal evidence for retina-dependent and -independent visual motion computations in mouse cortex. Nat. Neurosci. 20, 960–968 (2017).
Cruz-Martín, A. et al. A dedicated circuit links direction-selective retinal ganglion cells to the primary visual cortex. Nature 507, 358–361 (2014).
Lien, A. D. & Scanziani, M. Cortical direction selectivity emerges at convergence of thalamic synapses. Nature 558, 80–86 (2018).
Thiele, A., Distler, C., Korbmacher, H. & Hoffmann, K. P. Contribution of inhibitory mechanisms to direction selectivity and response normalization in macaque middle temporal area. Proc. Natl Acad. Sci. USA 101, 9810–9815 (2004).
Madisen, L. et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015).
Wekselblatt, J. B., Flister, E. D., Piscopo, D. M. & Niell, C. M. Large-scale imaging of cortical dynamics during sensory perception and behavior. J. Neurophysiol. 115, 2852–2866 (2016).
Gorski, J. A. et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002).
Mayford, M. et al. Control of memory formation through regulated expression of a CaMKII transgene. Science 274, 1678–1683 (1996).
Peron, S. P., Freeman, J., Iyer, V., Guo, C. & Svoboda, K. A cellular resolution map of barrel cortex activity during tactile behavior. Neuron 86, 783–799 (2015).
Wickersham, I. R., Finke, S., Conzelmann, K. K. & Callaway, E. M. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 4, 47–49 (2007).
Marshel, J. H., Mori, T., Nielsen, K. J. & Callaway, E. M. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron 67, 562–574 (2010).
Rancz, E. A. et al. Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nat. Neurosci. 14, 527–532 (2011).
Osakada, F. et al. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron 71, 617–631 (2011).
Goldey, G. J. et al. Removable cranial windows for long-term imaging in awake mice. Nat. Protocols 9, 2515–2538 (2014).
Judkewitz, B., Rizzi, M., Kitamura, K. & Häusser, M. Targeted single-cell electroporation of mammalian neurons in vivo. Nat. Protocols 4, 862–869 (2009).
Kitamura, K., Judkewitz, B., Kano, M., Denk, W. & Häusser, M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat. Methods 5, 61–67 (2008).
Pologruto, T. A., Sabatini, B. L. & Svoboda, K. ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2, 13 (2003).
Kim, E. J., Jacobs, M. W., Ito-Cole, T. & Callaway, E. M. Improved monosynaptic neural circuit tracing using engineered rabies virus glycoproteins. Cell Rep. 15, 692–699 (2016).
Drobizhev, M., Makarov, N. S., Tillo, S. E., Hughes, T. E. & Rebane, A. Two-photon absorption properties of fluorescent proteins. Nat. Methods 8, 393–399 (2011).
Brondi, M., Sato, S. S., Rossi, L. F., Ferrara, S. & Ratto, G. M. Finding a needle in a haystack: identification of EGFP tagged neurons during calcium imaging by means of two-photon spectral separation. Front. Mol. Neurosci. 5, 96 (2012).
Kleiner, M. et al. What’s new in psychtoolbox-3. Perception 36, 1–16 (2007).
Burgess, C. P. et al. High-yield methods for accurate two-alternative visual psychophysics in head-fixed mice. Cell Rep. 20, 2513–2524 (2017).
Pachitariu, M. et al. Suite2p beyond 10,000 neurons with standard two-photon microscopy. Preprint at https://doi.org/10.1101/061507 (2016).
Dipoppa, M. et al. Vision and locomotion shape the interactions between neuron types in mouse visual cortex. Neuron 98, 602–615 (2018).
Berens, P. CircStat: a MATLAB toolbox for circular statistics. J. Stat. Softw. 31, 1–21 (2009).
Acknowledgements
We thank E. Callaway for sharing viruses via the Salk Institute Vector Core; M. Rizzi for advice on electroporation; M. Krumin for support with microscopy; C. Reddy for help with plasmid production; M. Drobizhev for providing the two-photon cross-section of fluorescent proteins; S. Bugeon for help with histology and anatomical registration; L. Mariotti for help with anatomical reconstructions and cell counting; M. Scanziani for suggestions on the manuscript; and K. Balint, S. Trenholm, B. Roska and M. Tripodi for advice and for genetic materials that we used in pilot studies. This work was supported by the Wellcome Trust (grant 099692 to L.F.R., and grants 205093 and 108726 to K.D.H. and M.C.). M.C. holds the GlaxoSmithKline/Fight for Sight Chair in Visual Neuroscience.
Author information
Authors and Affiliations
Contributions
L.F.R., K.D.H. and M.C. conceived the experiments. L.F.R. refined the techniques, performed the experiments and analysed the data. L.F.R., K.D.H. and M.C. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Hillel Adesnik and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Targeted single-neuron electroporation with survival control in vivo.
a, Experimental pipeline: electroporation of the postsynaptic neuron, targeted by shadow-imaging and expression of GCaMP6; imaging of the postsynaptic neuron, labelled by dsRed; injection of the modified rabies virus; imaging and tracing of the presynaptic neurons marked by dsRed. b–d, Schematic of the electroporation technique, performed under a two-photon microscope on a transgenic mouse expressing GCaMP6 in cortical excitatory neurons. b, A pipette filled with DNA plasmids and Alexa Fluor 594 is targeted to a craniotomy; an 820-nm laser (red) excites Alexa Fluor 594 fluorescence (magenta) and GCaMP6 fluorescence (cyan). The latter is insensitive to neural activity because 820 nm is an isosbestic wavelength, at which calcium-bound and calcium-free isoforms fluoresce approximately equally. c, Upon electroporation, DNA plasmids and Alexa Fluor 594 are transferred into a neuron expressing GCaMP6. d, A healthy neuron maintains its GCaMP6 concentration and the resulting calcium-insensitive fluorescence (top), whereas a neuron with a damaged membrane bleeds indicator, gradually darkening and disappearing against the surrounding neuropil (bottom). e, Time lapse of an electroporation in layer 2/3 of mouse V1, using Alexa Fluor 594 negative contrast and calcium-insensitive GCaMP6 fluorescence imaging: approach (left), electroporation (middle) and pipette withdrawal (right). f, The same neuron as in e, imaged the next 1–3 days at 920 nm, expressing the electroporated genes for dsRed (red) and maintaining healthy GCaMP6 expression (green). g, Average fluorescence (mean ± s.e.m.) of somatic Alexa Fluor 594 (magenta) and GCaMP6 (cyan), relative to neuropil background (dashed line represents unity) before and after electroporation (arrow, n = 10 neurons). h, i, As in e, f, but with a slower time lapse of a neuron that survived the procedure. Images in h are 30-s-long averages acquired 0–1 min (left), 2–3 min (centre) and 5–6 min (right) after electroporation. j, k, As in h, i, for a neuron that did not recover from the electroporation. l,As in g, for GCaMP6 somatic fluorescence (mean ± s.e.m.) in neurons that did (cyan, n = 18) or did not (grey, n = 10) survive the procedure. Scale bar, 15 μm (for all fluorescence images).
Extended Data Fig. 2 Photo-ablation of supernumerary postsynaptic neurons before rabies injection.
a, Cartoon of the protocol. The target neuron expresses both GCaMP6 (green) and dsRed (red), whereas surrounding neurons express only GCaMP6. The day after electroporation (day 1), the neuron is targeted with steady two-photon illumination at 820 nm, focused with intensity of approximately 150 mW, for 10–20 s (top). The target neuron is ablated and by the next day (day 2) it has disappeared (bottom). b, Time-lapse imaging during the photo-ablation of two neighbouring neurons, lasting approximately 10 min. Imaging at 920 nm shows calcium-sensitive fluorescence of GCaMP6 (top) and fluorescence of dsRed (bottom). Each of the neurons was targeted with two photo-ablation pulses (red triangles). Photo-ablation pulses lasted 20 s, and each neuron was imaged for 30 s afterwards. Each pulse increases cellular damage: localized photo-bleaching after the first pulse; elevated intracellular calcium and cell swelling after the second pulse. Neighbouring cells, not expressing dsRed, resist the photo-damage. c, The successful elimination of the target neurons (red, see insets) is confirmed the day following the photo-ablation procedure: by day 2, the target neuron has either disappeared (yellow arrow, top neuron) or gone in apoptosis (yellow arrow, bottom neuron). The surrounding tissue is unaffected, as shown by the normal activity detected in neighbouring neurons using Suite2p (blue-green ROIs and traces; scale bar, 30 s and 10 s.d.). d, Effectiveness of photo-ablation as a function of cortical distance from the photo-ablation target neuron (n = 9 attempts, mean ± s.e.m.).
Extended Data Fig. 3 Time course of rabies tracing and recordings.
a, Viability of postsynaptic neurons as a function of day after the rabies virus injection, based on n = 17 injections. b, Count of observed presynaptic neurons traced over the same period from n = 17 postsynaptic neurons (mean ± s.e.m.). c, Fraction of viable presynaptic neurons (red) over the total traced (grey), and worst-case-scenario population mortality of presynaptic neurons (dashed), estimated assuming the viability of each newly labelled presynaptic neuron degrades at the same rate as the viability of the postsynaptic neurons measured in a. d, Average distribution across mice (n = 17, mean ± s.e.m.) of the imaging sessions used to record the responses of presynaptic neurons, split by cortical layer. The time of imaging did not systematically change across layer (black triangle, median, first and third quartiles), and most of the data were acquired before presynaptic neurons suffered from the toxicity of the rabies virus. e, Distribution of responsivity of the presynaptic neurons across days (red violin plot with black median). Responsivity was measured as the maximum average stimulus-triggered response. To compare across sessions, presynaptic responsivity was normalized to the median responsivity of the local population (shaded line). f, Longitudinal imaging of presynaptic neurons identified before (left, black ROI) and 5–12 days after (right, red ROI) rabies virus infection. Scale bar, 25 μm. g, Average responses to drifting gratings of the same presynaptic neurons before (black) and after (red) the rabies virus infection. Responses (4-s long) were z-scored with the respect to blank trials. Scale bar, 1 s.d. h, Preferred orientation of presynaptic neurons recorded before and after the rabies virus infection. At the time of recording used in this study, the preferred orientation of presynaptic neurons is unperturbed by the rabies virus infection (n = 51 neurons from n = 4 mice, circular correlation 0.76, Pr = 1.6 × 10−6, Z test).
Extended Data Fig. 4 Classification of excitatory and inhibitory presynaptic neurons.
a, Three example excitatory presynaptic neurons showing similar expression of dsRed (left) and decreasing expression of GCaMP6 (middle). The somatic masks obtained from dsRed and the GCaMP6 signal were used to compute a map of phase correlation in the 5-μm-radius annulus around the somatic centre (right). In the first two examples, the stronger GCaMP6 expression in the soma compared to the surrounding neuropil results in positive peaks of the phase correlation. b, As in a, for three example inhibitory neurons. The lack of GCaMP6 expression in the soma results in negative peaks of the phase correlation. c, For each presynaptic neuron, the central peak of phase correlation is plotted against the s.d. of the phase correlation within the 5-μm annulus centred on the soma (top). Darker dots indicate neurons the identity of which was confirmed by immunostaining. Open dots represent example neurons shown in a, b. A classification boundary was used to split the clusters of excitatory (red) and inhibitory neurons (blue). This boundary was identified independently for each presynaptic ensemble with a bilinear fit. The average fit across datasets is shown (black line). The histogram (bottom) summarizes the classification across experiments, with the average classification boundary. d, Average expression of dsRed (left), GCaMP6 (middle) and map of phase correlation (right) for presynaptic neurons classified as excitatory neurons (top, n = 584) or inhibitory neurons (bottom, n = 426). e, Number of presynaptic neurons classified as inhibitory versus excitatory in experiments in CaMK2a–GCaMP6 mice (red upward triangles, n = 13) and GAD2–NLS–mCherry mice (red downward triangles, n = 4). A linear fit (blue, r = 0.6, Pr = 2.6 × 10−3, linear correlation, F-test) shows that the fraction of traced presynaptic neurons that are inhibitory tends towards 30% as the yield of tracing increases; yet, owing to the positive intercept, the fraction of inhibitory neurons may appear as high as 60% in experiments with low tracing yield. In control experiments in which the red marker tdTomato was expressed only in excitatory neurons (AAV-CaMK2-tomato in CaMK2a–GCaMP6 mice, grey circles), the fraction of neurons incorrectly classified as inhibitory was below 5%.
Extended Data Fig. 5 Immunohistochemical verification of presynaptic neuron classification method.
a, Coronal maximum projection from a z-stack acquired in vivo, showing a section through a presynaptic network labelled with RV-dsRed (red) in a CaMK2a–GCaMP6s transgenic mouse (green). Scale bar, 50 μm. b, Coronal brain slice matching the section in a. We could match n = 94 out of the n = 105 neurons traced in vivo. c, Inset from b (dashed) showing the immunostaining for the inhibitory neurons marker GAD (blue). d, ROC curve illustrating the sensitivity and specificity of the neuron classification method based on somatic GCaMP6 fluorescence (Extended Data Fig. 4) against the ground-truth measurements obtained by immunostaining against GAD, for a range of classification boundaries. The fitted classification boundary was optimal (blue dot), yielding a specificity of 94% and sensitivity of 91%. e, Two example presynaptic neurons classified as excitatory (n = 49) from the z-stack in a. f, Slice immunostaining of the two neurons in e. Neurons were confirmed as excitatory if they expressed GCaMP6 and were GAD-negative (n = 45). Scale bar, 40 μm. g, h, As in e, f, for two example presynaptic neurons classified as inhibitory. Neurons were confirmed as inhibitory if they did not express GCaMP6 and were GAD-positive (n = 42).
Extended Data Fig. 6 Spectral unmixing of dsRed and mCherry fluorescence.
a, Emission spectra of GCaMP6, dsRed and mCherry. Shaded areas indicate the emission band captured by the green (G) and red (R) channels of the microscope. The G channel collects mostly GCaMP6 fluorescence, and the R channel captures a mixture of mCherry, dsRed and GCaMP6 fluorescence. Emission spectra were normalized to their peak. b, Two-photon action cross-section of dsRed (top, red) and mCherry (bottom, purple). c, Ratio between the two-photon action cross-sections of dsRed and mCherry (red) and its inverse (purple). Shaded bands in b, c indicate the excitation wavelengths used for imaging: 780, 890, 970 and 1,020 nm. The four wavelengths were chosen to maximize the SNR of each fluorophore while ensuring the ratio between the two signals was maximal. d, Example field of view imaged at the four excitation wavelengths in the G channel. GCaMP6 was expressed in all neurons with an AAV2.1-Syn-GCaMP6s in a GAD–NLS–mCherry mouse; a subset of presynaptic neurons was traced with a dsRed rabies virus. e, As in d, for the R channel. f, The fluorescence in the R channel (Rλ) plotted against the fluorescence in the G channel (Gλ) for each pixel, and for each excitation wavelength λ. Because the GCaMP6 labelling was dense and both the dsRed and the mCherry signals were sparse, and because the contribution of dsRed and mCherry to Gλ was minimal, the GCaMP6 signal contributing to Rλ could be recovered by piecewise robust linear regression (αGλ + β). g, The image representing the linear mix of dsRed or mCherry signals, Mλ, was recovered by subtracting the scaled Gλ from the Rλ. h, An iterative algorithm was used to linearly unmix the two source images. Each unmixing iteration was constrained to minimize the quadratic reconstruction error over the data and return maximally uncorrelated sources RdsRed and RmCherry. i, j, The two source images RdsRed and RmCherry for the example field of view in d, e. The unmixing procedure correctly recovers the nuclear localization of mCherry without any prior. Scale bars, 50 μm. Similar results were obtained for all GAD–NLS–mCherry mice (n = 4).
Extended Data Fig. 7 Distributions of individual excitatory and inhibitory presynaptic ensembles.
a, Density of excitatory presynaptic neurons around the postsynaptic neuron (black triangle) as a function of cortical depth and radial distance from the postsynaptic neuron. Density was normalized to its maximum for display purposes. Depth marginals are shown on the left of each density map. b, As in a, for inhibitory presynaptic neurons. c, Overlay of excitatory and inhibitory densities shown in a, b. Hue indicates the relative proportion of excitatory (red) and inhibitory (blue) inputs, and saturation indicates maximum normalized neuronal density. Data are shown for the 13 experiments that included substantial recordings from L4.
Extended Data Fig. 8 L2/3 neurons receive presynaptic excitation preferentially tuned to their orientation preference.
a, Average tuning across the postsynaptic neurons responding to drifting gratings (n = 16, mean ± s.e.m.), after alignment of their preferred stimulus direction. b, Distribution of preferred direction relative to the postsynaptic preferred direction, for excitatory presynaptic ensembles across all layers (n = 15, median ± m.a.d.). c, As in b, for excitatory presynaptic ensembles within L2/3 (top) and within L4 (bottom). d, As in b, for inhibitory presynaptic ensembles. e, Average orientation tuning across the postsynaptic neurons responding to drifting gratings (n = 16, mean ± s.e.m.), after alignment of their preferred stimulus orientation. f, Average distribution of preferred orientation relative to the postsynaptic preferred orientation, for excitatory presynaptic neurons pooled across all layers (n = 15, median ± m.a.d.). Presynaptic ensembles tuning for orientation (PKW = 5 × 10−8, two-sided one-way Kruskal–Wallis test) was significantly stronger than expected from random samples of the surrounding population (grey, median ± m.a.d.). g, As in f, for presynaptic ensembles within L2/3 (top, n = 15, PKW = 10−7) and within L4 (bottom, n = 13, PKW = 2 × 10−5). h, As in f, for inhibitory presynaptic ensembles (n = 4). i, The tuning of the distributions of preferred orientation of excitatory presynaptic ensembles across layers plotted against the preferred orientation of their postsynaptic neuron. The co-tuning for orientation (r = 0.75, circular correlation, Pr = 4 × 10−3, Z-test; PV = 9 × 10−8 circular V-test) was stronger than expected from random samples of the local population (Pr_rand < 10−4 for circular correlation, PV_rand < 10−4 for V statistic). Upwards triangles represent experiments in CaMK2a–GCaMP6 mice (n = 11); downward triangles indicate experiments in GAD2–NLS–mCherry mice (n = 4). j, As in i, for excitatory presynaptic ensembles within L2/3 (top, r = 0.92, Pr = 5 × 10−3, PV = 10−5, Pr_rand = 2 × 10−4, PV_rand < 10−4, n = 15) and within L4 (bottom, r = 0.76, Pr = 2 × 10−2, PV = 3 × 10−3, Pr_rand = 0.18, PV_rand = 3 × 10−3, n = 13). k, As in i, for inhibitory presynaptic ensembles. Inhibitory presynaptic ensembles were weakly biased to the orientation preference of the postsynaptic neuron (PV = 0.05, circular V-test, n = 4).
Extended Data Fig. 9 Mapping retinotopy using individual neurons versus wide-field signals.
a, The stimulus used for retinotopic mapping was a sparse random pattern of white and black squares on a grey background (top). The fluorescence time-course from the entire field of view was used to compute a global stimulus-triggered average response elicited by changes in luminance at each position. The centre of mass of this global receptive field (RF) was used to constrain the fits of wide field and neuronal receptive field to the appropriate retinotopic region (dashed rectangle). b, Maximal projection from an example field of view. In this example, the field of view was subsampled in a grid of 9 × 9 regions of interest (ROIs, red squares) to compute wide-field receptive fields. Scale bar, 100 μm. c, The wide-field receptive fields calculated for the ROIs in b, normalized to their maximum. The wide-field receptive field centres from the grid of ROIs were interpolated to estimate a retinotopic map, assigning a wide-field receptive field to each cortical location, whether it contained a responsive neuron, an unresponsive neuron or neuropil. d, Estimation of neuronal receptive fields. ON (red) and OFF (blue) receptive fields were estimated by regularized smooth pseudoinverse regression using either streams of white (ON) or black stimuli (OFF) as predictors, and assuming a common response kernel across neurons. ON and OFF subfields were then combined to estimate the receptive field centre (green dot). Receptive fields were considered significant if the cross validated correlation coefficient between predicted (red trace) and actual response (black trace) was greater 0.2. e, Azimuth of neuron receptive field centre versus wide-field receptive field centre for all excitatory presynaptic neurons (black dots, n = 113, rpre = 0.89, Pr_pre = 2.8 × 10−39, linear correlation, F-test) and surrounding excitatory neurons (red density, n = 25,677, rall = 0.88, Pr_all < 10−308, linear correlation F-test) across experiments. f, As in e for elevation (rpre = 0.80, Pr_pre = 9 × 10−27; rall = 0.85, Pr_all < 10−308). g, h, As in e, f, for inhibitory presynaptic neurons (black dots, n = 37, rpre = 0.92, Pr_pre = 1.3 × 10−15, for azimuth; rpre = 0.71, Pr_pre = 9.7 × 10−7, for elevation) and all inhibitory neurons (n = 1,963, rall = 0.95, Pr_all < 10−308, for azimuth; rall = 0.74, Pr_all < 10−308, for elevation).
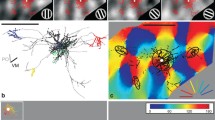
Extended Data Fig. 10 Spatial connectivity accords with direction selectivity in cortex and across layers.
a, Distribution of excitatory and inhibitory presynaptic neurons in cortex, pooled across experiments (n = 17), and polar tuning curves for each postsynaptic neuron (top). The colour hue indicates the average fraction of local excitatory (red) or inhibitory (blue) presynaptic neurons; the colour saturation indicates the maximum normalized input density, averaged across experiments. b, c, As in a, plotting the excitatory and inhibitory presynaptic neurons separately. d, As in a, after rotating each presynaptic cortical distribution to align the postsynaptic preferred direction (n = 16). The cortical angle of rotation, corresponding to the postsynaptic preferred direction, was calculated from the local retinotopic gradient at the postsynaptic location. After the alignment, the postsynaptic preferred orientation approximately maps to a line at the postsynaptic location (dashed line). e, As in d, for excitatory presynaptic neurons. f, As in d, for inhibitory presynaptic neurons. g, As in d, for the distribution of L1 and L2/3 excitatory and inhibitory presynaptic neurons in visual space, pooled across experiments after alignment to the preferred direction across postsynaptic neurons (n = 16). h, i, As in g, distinguishing between excitatory and inhibitory presynaptic neurons. j–l, As in g–i, for presynaptic neurons in L4 and in superficial L5. In all panels, upwards triangles and circles represent CaMK2a–GCaMP6 datasets; downward triangles and squares indicate GAD2–NLS–mCherry datasets.
Supplementary information
Rights and permissions
About this article
Cite this article
Rossi, L.F., Harris, K.D. & Carandini, M. Spatial connectivity matches direction selectivity in visual cortex. Nature 588, 648–652 (2020). https://doi.org/10.1038/s41586-020-2894-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2894-4
- Springer Nature Limited
This article is cited by
-
The logic of recurrent circuits in the primary visual cortex
Nature Neuroscience (2024)
-
Panoramic visual statistics shape retina-wide organization of receptive fields
Nature Neuroscience (2023)
-
Sampling-based Bayesian inference in recurrent circuits of stochastic spiking neurons
Nature Communications (2023)
-
A Novel Mouse Model for Polysynaptic Retrograde Tracing and Rabies Pathological Research
Cellular and Molecular Neurobiology (2023)
-
Ascertaining cells’ synaptic connections and RNA expression simultaneously with barcoded rabies virus libraries
Nature Communications (2022)