Abstract
Alternative cleavage and polyadenylation (APA) is a widespread mechanism to generate mRNA isoforms with alternative 3′ untranslated regions (UTRs). The expression of alternative 3′ UTR isoforms is highly cell type specific and is further controlled in a gene-specific manner by environmental cues. In this Review, we discuss how the dynamic, fine-grained regulation of APA is accomplished by several mechanisms, including cis-regulatory elements in RNA and DNA and factors that control transcription, pre-mRNA cleavage and post-transcriptional processes. Furthermore, signalling pathways modulate the activity of these factors and integrate APA into gene regulatory programmes. Dysregulation of APA can reprogramme the outcome of signalling pathways and thus can control cellular responses to environmental changes. In addition to the regulation of protein abundance, APA has emerged as a major regulator of mRNA localization and the spatial organization of protein synthesis. This role enables the regulation of protein function through the addition of post-translational modifications or the formation of protein–protein interactions. We further discuss recent transformative advances in single-cell RNA sequencing and CRISPR–Cas technologies, which enable the mapping and functional characterization of alternative 3′ UTRs in any biological context. Finally, we discuss new APA-based RNA therapeutics, including compounds that target APA in cancer and therapeutic genome editing of degenerative diseases.
Similar content being viewed by others
Introduction
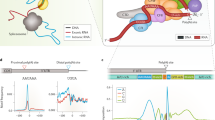
RNA polymerase II (Pol II) transcribes DNA and produces pre-mRNAs that are processed into mature mRNAs. The processing steps include capping, splicing, and cleavage and polyadenylation (CPA) at the 3′ end of the transcripts1,2. Alternative cleavage and polyadenylation (APA) in the last exon generates mRNA isoforms that encode the same protein, but with different 3′ untranslated regions (UTRs)3,4,5 (Fig. 1a). The use of intronic polyadenylation sites (PASs) generates truncated proteins, but also changes the 3′ UTR of the resulting mRNA6,7.
a | Alternative cleavage and polyadenylation (APA) generates mRNA isoforms that differ in their 3′ untranslated regions (UTRs). mRNA processing at intronic polyadenylation (IPA) sites generates mRNA isoforms (IPA isoforms) that encode proteins with alternative C-termini (light blue part of protein). mRNA processing at proximal or distal polyadenylation sites (PASs) in terminal exons generates mRNA isoforms with short 3′ UTRs (SU) or long 3′ UTRs (LU) that encode proteins with the same amino acid sequence. The grey boxes in the protein symbols represent protein domains. Introns are not drawn to scale. b | During transcription by RNA polymerase II (Pol II), the cleavage and polyadenylation (CPA) machinery binds the PAS hexamer motif AAUAAA and surrounding sequence elements in the nascent RNAs. Shown are CPA factors bound to the pre-mRNA (or nascent RNA) and to the C-terminal domain of Pol II while it is transcribing DNA. The different CPA protein complexes are colour-coded. Protein names or symbols (and gene symbols (in parentheses) when different) are given in the boxes. c | Sequence context of functional PASs. Endonucleolytic cleavage of the nascent RNA occurs ~20 nucleotides downstream of a PAS hexamer that is located in a suitable sequence context containing the UGUA and (G+U)-rich or U-rich sequence upstream and downstream, respectively. The colours of the RNA elements correspond to the colours of the complexes in part b that interact with them. CFI, cleavage factor I; CFII, cleavage factor II; CLP1, cleavage factor polyribonucleotide kinase subunit 1; CPSF1, CPA specificity factor subunit 1; CSTF, cleavage stimulation factor; FIP1, factor interacting with PAPOLA and CPSF1; nt, nucleotides; PAF1, RNA polymerase II-associated factor 1; PABPN1, poly(A)-binding protein 2; PAP, poly(A) polymerase; RBBP6, RB-binding protein 6; SCAF4, serine and arginine-related C-terminal domain-associated factor 4; TSS, transcription start site; WDR33, WD repeat domain 33.
The different 3′ UTRs contain a large number of regulatory elements that control mRNA and protein abundance, mRNA and protein localization, and protein complex assembly and therefore protein function8,9,10. In addition to determining 3′ UTR length, CPA is essential for the generation of mature mRNAs as incompletely processed transcripts are subject to degradation and cannot serve as the templates for protein synthesis. Moreover, PAS cleavage is an important determinant of mRNA expression levels11,12,13,14.
mRNA 3′-end formation occurs co-transcriptionally, and is accomplished by the CPA machinery, which comprises a multiprotein core complex and dozens of associated factors15,16,17,18,19 (Fig. 1b). CPA specificity factor (CPSF) recognizes the PAS hexamer motif AAUAAA (or variants thereof) in the nascent RNA. Additional upstream and downstream regulatory RNA sequence elements recruit further CPA factors that promote 3′-end processing. These elements include UGUA, the binding site of cleavage factor I (CFI), and the downstream U-rich or (G+U)-rich region, which is bound by cleavage stimulation factor (CSTF)2,14,20 (Fig. 1b,c). The additional elements are necessary to define functional PASs, as on average a polyadenylation signal hexamer occurs every 500 nucleotides in pre-mRNAs yet it is not used for CPA at these sites3.
Alternative 3′ UTR isoforms are expressed in a highly gene-specific and cell-type-specific manner3,5,21. In this Review, we discuss how expression of 3′ UTR isoforms is regulated. As the CPA machinery carries out APA, the concentration of CPA factors is an important determinant of APA. Depletion of these factors often shifts APA globally and changes 3′ UTR isoform expression of hundreds of genes22,23,24,25,26,27,28,29,30. However, environmental changes during development, stress and metabolic adaptation induce APA changes of only subsets of genes31,32,33,34,35.
We discuss how gene-specific 3′ UTR isoform changes are accomplished, including cell-type-specific abundance of regulatory factors and enzymes that post-translationally modify factors that control transcription elongation, transcription termination and CPA. Gene-specific sequence elements in promoters and enhancers also affect 3′ UTR isoform choice14,36,37,38. These layers of regulation allow cells to integrate environmental signals and respond with appropriate changes in gene and 3′ UTR isoform expression.
One of the surprising findings in the past 10 years was the observation that gene expression and APA are largely independent, as genes that change their 3′ UTR isoforms often do not considerably change their overall expression level3,5,27,39,40,41. Moreover, genetic variants associated with gene expression changes rarely overlap with genetic variants that affect APA42,43,44. These data indicate that single-UTR genes, which contain one functional PAS at their 3′ ends, and multi-UTR genes, which have at least two functional PASs, differ substantially in their mode of regulation. We discuss the implications of such independent regulation and highlight the functions of alternative 3′ UTRs beyond the regulation of mRNA abundance35,45,46,47,48,49.
We feature new transformative tools that were developed through recent advances in single-cell RNA sequencing (scRNA-seq) and CRISPR–Cas technologies. These methods allow the reliable quantification of 3′ UTR isoforms in any cell type from publicly available datasets5,50,51,52, and facilitate manipulation of individual 3′ UTRs at endogenous gene loci to study their regulatory logic and function53,54,55. Together with small molecules that modulate PAS cleavage activity to control mRNA and protein expression, the insights gained by these novel tools will enable the development of RNA therapeutics that take advantage of isoform-specific and compartment-specific targeting.
Two classes of APA
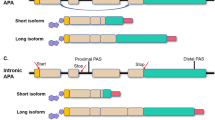
APA that occurs in the terminal exon generates tandem 3′ UTR isoforms (short and long isoforms) and is often called ‘3′ UTR APA’. This class of APA does not change the protein coding sequence. By contrast, APA can also occur in introns — termed ‘intronic APA’ — thereby generating truncated proteins.
3′ UTR APA
Sequences downstream of the coding region of genes contain many cis-regulatory elements. 3′ UTR length, and thus the regulatory potential, is determined by PAS choice. Multi-UTR genes generate mRNA isoforms with short 3′ UTRs or long 3′ UTRs3 (Fig. 1a). Although mRNA isoforms that differ only in their 3′ UTRs encode the same protein, the different 3′ UTRs can determine the half-life of mRNAs or proteins and their subcellular localization8,9. Differences in 3′ UTRs of gene homologues, which often encode proteins with highly similar amino acid sequences, allow differential regulation of each protein49,56. Both single-UTR and multi-UTR genes encode proteins with similar size distribution and a median coding region length of ~1,250 nucleotides. Sequencing methods that identify mRNA 3′-end cleavage sites transcriptome-wide and measure alternative 3′ UTR isoform expression are called 3′-end sequencing methods and are abbreviated to 3′-seq here. 3′-seq revealed that 3′ UTR length differs substantially between single-UTR genes and multi-UTR genes and resulted in an estimated median 3′ UTR length of ~600 and ~1,900 nucleotides (for the longest isoform), respectively6. The use of the coding-sequence proximal PAS in multi-UTR genes results in expression of short 3′ UTR isoforms with a median 3′ UTR length of ~300 nucleotides41. As APA often removes most 3′ UTR regulatory elements, 3′ UTR-mediated regulation may be particularly important for multi-UTR genes.
Intronic APA
Transcripts that are generated from intronic PASs are called ‘intronic polyadenylation (IPA) isoforms’ or ‘alternative last exon isoforms’. Many IPA isoforms have a physiological role and contribute to proteome diversification by generating proteins with alternative carboxy termini (Fig. 1a). IPA isoforms are often expressed in a cell-type-specific manner6,7,35,57,58,59,60. However, IPA can occur also in the context of premature CPA, in which no functional protein is produced33,34,59,61. This type of IPA arises following mutation of CPA factors in cancer, or it can be induced experimentally through manipulation of transcript elongation or splicing factors33,34,59,61. In B cell leukaemia, which is a disease with very few DNA mutations, inactivation of tumour-suppressor genes occurs mostly through the generation of truncated proteins by IPA instead of by DNA mutations59.
Premature CPA is also used by cells to downregulate the expression of full-length mRNAs. For example, IPA is upregulated following heat shock, which results in the downregulation of many genes not directly involved in the stress response62. IPA also determines the expression of the CPA factors PCF11 and CSTF3 through feedback autoregulation29,63,64. IPA is tightly controlled by co-transcriptional splicing and Pol II elongation rates. For example, binding of the splicing factor U1 small nuclear RNA strongly inhibits intronic PAS use, thereby allowing the generation of full-length mRNAs61. Conversely, a decrease in splicing factor availability or interference with splice site recognition increases IPA. As splicing factors and CPA-promoting factors physically interact with Pol II, they can compete for access to the elongating polymerase and tip the balance towards expression of IPA or full-length mRNAs33,34,65,66.
Regulation of alternative 3′ UTRs
APA is regulated both co-transcriptionally and post-transcriptionally (Fig. 2). As CPA occurs while Pol II is transcribing a gene, Pol II elongation dynamics have a major role in regulating APA. Pol II termination factors extensively interact with RNA-binding proteins such as splicing factors and CPA factors that recognize RNA sequence elements to control where and when a pre-mRNA is cleaved and polyadenylated2,14,20 (Fig. 2). RNA-binding proteins that are involved in the regulation of APA were reviewed previously1,2, and their effects are summarized in Table 1. All the factors involved can be regulated at the levels of abundance and activity, often in a cell-type-specific and condition-specific manner, thereby allowing APA at individual genes.
Alternative cleavage and polyadenylation (APA) is regulated both co-transcriptionally and post-transcriptionally. Cis-regulatory elements in the RNA and the DNA are necessary but not sufficient for cleavage and polyadenylation (CPA). As CPA occurs while RNA polymerase II (Pol II) transcribes a gene, Pol II elongation dynamics regulate APA. A selection of APA-regulating factors is given. Pol II termination factors show extensive crosstalk with RNA-binding proteins such as splicing factors and CPA factors that recognize RNA sequence elements and control where and when a pre-mRNA is cleaved and polyadenylated. Following processing of 3′ untranslated region isoforms, the expression level of individual APA isoforms is further regulated by post-transcriptional processes. Both the abundance and the activity of factors that control APA can be regulated co-transcriptionally and post-transcriptionally, often in a cell-type-specific and condition-specific manner, thereby allowing dynamic regulation of APA at individual genes. NEXT, nuclear exosome targeting (a trimeric protein complex); NXF1, nuclear RNA export factor 1; PAF1, RNA polymerase II-associated factor 1; PAXT, poly(A) tail exosome targeting (a trimeric protein complex); SCAF4, serine and arginine-related C-terminal domain-associated factor 4; SPT5, suppressor of Ty 5; SRSF, serine and arginine-rich splicing factor 3; THOC5, THO complex 5.
Pol II elongation dynamics
DNA sequence and chromatin structure, together with transcript elongation factors, determine Pol II dynamics and the functional state of the polymerase. Pol II elongation and termination factors cooperate or compete with CPA factors to control pre-mRNA CPA.
DNA sequence and topology
The speed of transcript elongation is decreased by Pol II pausing or backtracking67,68. Increased Pol II pausing downstream of a functional PAS promotes CPA at this site and contributes to APA regulation67,69,70 (Fig. 3a). In addition to promoter-proximal Pol II pausing, which is not discussed here, Pol II pausing in gene bodies is caused by certain DNA sequences, termed ‘pause sites’, or by structures in RNA and DNA, such as G-quadruplexes67,70.
a | A roadblock in the DNA can affect RNA polymerase II (Pol II) translocation and use of alternative polyadenylation sites (PASs). The roadblock can be a DNA sequence element, such as a pause site, a structural element, such as a G-quadruplex, or a large protein that impedes Pol II elongation, thereby increasing the use of a proximal PAS. b | Cyclin-dependent kinase 12 (CDK12) is required for transcript elongation and suppression of premature cleavage and polyadenylation (CPA) of long genes. CDK12 promotes transcript elongation by phosphorylating Pol II. Loss of CDK12 predominantly affects DNA damage repair genes because of their extensive lengths and their lower U1 small nuclear RNA to PAS ratio. The lower processivity of Pol II results in premature CPA, thereby reducing expression of full-length mRNAs and protein output. c | Discovery of diverse alternative cleavage and polyadenylation (APA) regulators in a single genetic screen in Caenorhabditis elegans. unc-44 encodes the cytoskeleton protein ankyrin. Use of an intronic PAS in unc-44 generates intronic polyadenylation (IPA) mRNA isoforms, which encode ankyrin that is ubiquitously expressed, including in immature neurons. Giant ankyrin is expressed only in mature neurons, where it is generated from the full-length (FL) mRNA isoform through suppression of the intronic PAS. Worms that lack casein kinase 1δ (CK1δ; encoded by kin-20) have impaired movement owing to a block in neuron maturation that is caused by the lack of giant ankyrin expression. A screen for suppressor mutations identified 13 genes that, when knocked out, were able to restore the expression of giant ankyrin and the generation of mature neurons in kin-20-KO worms. The screen identified mutations that disrupt the intronic PAS sequence of the ankyrin gene, and mutations in several factors that control transcript elongation, transcription termination and CPA and in enzymes that regulate their activity. 3′-seq, 3′-end sequencing; CTD, C-terminal domain; KO, knockout; LU, long 3′ untranslated region isoform; SU, short 3′ untranslated region isoform.
Increased Pol II pausing can also be accomplished by a change in the chromatin environment71,72. For example, DNA methylation in gene bodies is important to prevent binding of the insulator protein CTCF. CTCF binding recruits the cohesin complex and induces APA changes across a few hundred genes71. Mechanistically, CTCF and cohesin may act as roadblock for Pol II. In addition, CTCF and cohesin binding gave rise to new chromatin loops and altered the phosphorylation status of the Pol II C-terminal domain (CTD) (see Pol II-associated elongation and termination factors). As the CTD is a crucial regulator of Pol II-mediated regulation of mRNA processing at different stages of transcription, the changes in Pol II CTD modifications may contribute to the increased use of proximal PASs through reduced DNA methylation71.
Pol II-associated elongation and termination factors
The speed of Pol II largely depends on CTD-associated elongation factors and on the phosphorylation status of the CTD65,66,71,73,74,75, which functionally orients Pol II with respect to its location within a gene74. The CTD consists of 52 heptad repeats of the consensus sequence YSPTSPS. Several of these residues are phosphorylated by cyclin-dependent kinase 9 (CDK9), CDK12 and CDK13 and dephosphorylated by the phosphatases SSU72, FCP1 (also known as CTD phosphatase 1) and PP2A33,34,74,76,77,78,79. The association between CTD phosphorylation and APA was discovered in the context of cancer, where CDK12 loss-of-function mutations were associated with increased expression of IPA isoforms of DNA damage repair genes33,34. The increased IPA expression caused lower expression of full-length proteins, which inactivated the DNA damage response33,34,80. Loss of CDK12 impairs transcript elongation rates at many genes, but DNA repair genes are predominantly affected because of their substantial gene lengths and lower U1 small nuclear RNA to PAS ratios, which makes them more susceptible to intronic PAS use34 (Fig. 3b).
Transcription termination involves the dissociation of Pol II from the DNA template several kilobases downstream of the PAS66,81. CPA and transcription termination are separate events, but they are functionally linked. Pol II transition through a functional PAS is required for transcription termination, and slowdown of Pol II is required for proper CPA75,81. The phosphorylation status of the transcription elongation factor SPT5 is crucial for elongation and termination, and is regulated by CDK9 and the phosphatase PNUTS–PP1 (ref.75). PAS use was altered by depletion of CDK9 or SPT5 in a reporter assay14. Impaired activity of enzymes that regulate elongation and termination causes premature transcription termination, which leads to degradation of the nascent transcript before the distal PAS is transcribed, resulting in an APA pattern that may appear as increased IPA isoform or short 3′ UTR isoform expression and reduced full-length mRNA expression14,33,34.
Factors that bridge Pol II and the CPA complex
The strong effect of CDK12 on APA is also caused by its ability to recruit the PAF1 complex to Pol II (refs.65,82,83). The PAF1 complex bridges Pol II and the CPA machinery, and thus integrates the regulation of transcription elongation and termination and CPA. It connects elongating Pol II with the CPA machinery through its interaction with the CPSF and CSTF complexes82. Loss of the PAF1 complex and of PAF1-recruited CPSF3 from elongating Pol II leads to a transcription termination defect and is important for the regulation of APA and IPA14,35,83,84.
Additional connections between Pol II and the CPA complex are formed by RNA-binding proteins with Pol II CTD-interacting domains, including SR-related and CTD-associated factor 4 (SCAF4), SCAF8 and PCF11 (refs.29,66). PCF11 is a subunit of cleavage factor II (CFII) and regulates expression or APA of ~50% of human genes. PCF11 enhances transcription termination and stimulates CPA at proximal PASs, thereby resulting in 3′ UTR shortening, but regulation of expression or regulation of APA occurs at different genes29. SCAF4 and SCAF8 interact with Pol II, the PAF1 complex and CPA factors66. In human cells, SCAF4 and SCAF8 suppress transcription termination at intronic PASs to allow expression of full-length transcripts. Absence of both SCAF4 and SCAF8 upregulates more than 1,300 IPA isoforms in a sequence-specific manner, as most of these genes contain SCAF4- and SCAF8-binding sites upstream of their PASs66.
Identification of all classes of APA regulators in a single genetic screen
The importance of Pol II dynamics for APA regulation was revealed by a single genetic suppressor screen performed in Caenorhabditis elegans35 (Fig. 3c). This screen identified Pol II-associated factors, enzymes that regulate their activity and factors that bridge Pol II and the CPA machinery. APA at unc-44, which encodes the cytoskeleton protein ankyrin, was shown to be essential for neuron maturation in C. elegans. Immature neurons mostly express unc-44 mRNA isoforms generated by use of an intronic PAS, which encode ankyrin (Fig. 3c). By contrast, mature neurons produce longer mRNA isoforms that include additional exons, which encode giant ankyrin. Casein kinase 1δ (CK1δ; encoded by kin-20) was identified as a regulator of the switch from IPA to full-length ankyrin expression35. CK1δ-deficient worms do not express giant ankyrin, which results in impaired maturation of motor neurons and largely immobile worms. In a subsequent screen for suppressor mutations, 13 genes were identified that, when knocked out, were able to restore the generation of mature neurons in kin-20-knockout worms35.
All the identified mutations enable expression of giant ankyrin and were either involved in CPA or in transcription elongation and termination35,85 (Fig. 3c). They include mutations that disrupt the intronic PAS of the ankyrin gene, and mutations in SSU72, a direct target of CK1δ that acts as a Pol II CTD phosphatase with a crucial role in the transition of Pol II from elongation to termination77,86. Moreover, the screen identified PIN1, which is a prolyl cis–trans isomerase that enhances the phosphatase activity of SSU72, and genes that encode components of Pol II, CPA factors and two subunits of the PAF1 complex65,82,83,87. That study represents a paradigm for context-dependent regulation of individual mRNA isoforms at specific time points during development by enzymes that control ubiquitous processes, such as transcription elongation and termination. Although CK1δ appears to regulate intronic PAS readthrough only at a single gene, the mechanism by which this enzyme acts in a gene-specific manner is currently unclear.
Gene-specific APA regulation
All elongation, termination and CPA factors involved in APA can be regulated at the levels of abundance and activity. In this section we explain how cell states or cell types induce changes in mRNA processing of individual genes. In some examples, APA changes can reinforce the cell state.
Early studies detected CPA factors at promoters88,89,90,91,92. In yeast, promoters are known to regulate many post-transcriptional processes, including mRNA stability, cytoplasmic localization and translation93,94,95,96,97. Two major conceptual models by which promoters regulate processes at mRNA 3′ ends have been proposed and are currently difficult to disentangle. In the promoter loading model, factors that are recruited to promoters travel along the gene with Pol II, thus making them available at downstream sites98. This model is supported by a cell-type-specific binding pattern of RNA-binding proteins at human promoters and enhancers99. As RNA-binding proteins are hubs for protein–protein interactions100,101,102,103, RNA-binding proteins at active promoters have the potential to be major regulators of APA. In the gene looping model, the transcription start site and the PAS physically interact through looping of the chromatin between them, thus allowing promoters to directly regulate 3′-end formation104,105,106. Gene loops between the promoter and either of the alternative PASs in CYC1 in yeast were detected; interestingly, changes in gene loop structure were observed upon alteration of growth conditions, which correlated with alternative PAS use106. Both models are supported by the findings we discuss later herein.
Alternative transcription start sites
Regulation of 3′ UTR isoforms of individual genes by transcription factors can be explained by the existence of alternative transcription start sites composed of different cis-regulatory elements, which allow the binding of factors that determine 3′-end formation. Evidence for this hypothesis was found in fly neurons, where the presence of the RNA-binding protein Elav at gene promoters was associated with the production of mRNA isoforms with long 3′ UTRs36. Accordingly, swapping of promoter sequences between genes changed 3′ UTR length (Fig. 4a). Combined evidence from RNA-seq and long-read sequencing further revealed significant co-occurrence between specific transcription start sites and specific alternative 3′ UTR isoforms in human cells, organoids and rat hippocampal neurons37,38. In C. elegans, promoters were found to determine different modes of transcription termination107.
a | Differential binding of factors at alternative transcription start sites can mediate gene-specific regulation of alternative polyadenylation site (PAS) use. In fly neurons, the RNA-binding protein embryonic lethal abnormal visual system (Elav) binds to specific promoters and transcription start sites (TSS) and thus controls the expression of mRNAs with longer 3′ untranslated regions (UTRs). b | Transcription factors that bind to specific enhancers regulate APA in a gene-specific manner. Signalling-induced activation of nuclear factor-κB (NF-κB) induces its binding to a specific enhancer of the phosphatase and tensin homologue gene (PTEN) in human cell lines. NF-κB activation does not change the production or stability of the PTEN mRNA, but induces a change in 3′ UTR isoform expression. Deletion of the enhancer or silencing of NF-κB impairs this signalling-induced APA change. c | Signalling-induced phosphorylation of cleavage and polyadenylation specificity factor subunit 6 (Cpsf6) increases its activity and promotes autophagy. In flies, inactivation of the mTOR pathway allows expression of two kinases (not shown) that phosphorylate Cpsf6 in the cytoplasm, thereby promoting its translocation to the nucleus and RNA-binding activity. Cpsf6 changes the APA pattern of two master regulators of autophagy, the autophagy-related protein 1 gene (Atg1) and Atg8a. Increased expression of their long 3′ UTR (LU) isoforms supports high-level protein expression, thereby inducing autophagy upon mTOR inhibition to allow intracellular nutrient uptake. 3′-seq, 3′-end sequencing; SU, short 3′ UTR.
Alternative enhancers
Enhancer-mediated recruitment of factors to promoters was recently suggested to regulate APA in human cells14. Although it is thought that transcriptional enhancers regulate transcript production, CRISPR–Cas-mediated deletion of a promoter-proximal enhancer did not change the production or stability of the PTEN mRNA, but instead it changed APA14. APA was regulated by the transcription factor NF-κB, which binds to the PTEN enhancer. Signalling-induced activation of NF-κB resulted in 3′ UTR shortening, whereas silencing of NF-κB impaired the signalling-induced APA change14 (Fig. 4b). Moreover, cell-type-specific enhancers were found to be significantly associated with 3′ UTR shortening during differentiation14.
The transcription factor BMAL1 controls expression of individual 3′ UTR isoforms
Circadian gene expression is regulated by transcriptional and translational negative feedback loops, initiated by the transcription factor CLOCK–BMAL1, which binds to promoters of genes expressed in a circadian manner108. As reported in a recent preprint, 3′-seq analyses performed in a period of 24 h identified hundreds of APA changes that correlated with the circadian rhythm109. When comparing circadian changes in gene expression and in APA, the study authors identified nearly 500 genes with a circadian expression pattern, whose alternative 3′ UTRs did not oscillate. Conversely, in a similar number of genes that were not rhythmically expressed, 3′ UTR isoforms with circadian oscillation were observed109. This recent study provides one of the most surprising findings in APA research, as it detected independent expression regulation of individual 3′ UTR isoforms. By contrast, in the past it was thought that alternative 3′ UTRs generated from a gene are linked, and it was predicted that upregulation of one isoform will downregulate the other isoform when gene expression remains constant3,27,39,40.
To identify how expression of individual 3′ UTR isoforms is regulated, the aforementioned study analysed APA changes in Bmal1-knockout mice (Bmal1 is also known as Arntl), and found that the majority of dysregulated APA events involved the circadian expression of individual 3′ UTR isoforms, while expression of the non-regulated isoforms remained fairly constant109. This result suggests a widespread lack of compensatory regulation, and instead indicates that BMAL1-bound promoters largely confer independent expression control of alternative 3′ UTR isoforms.
Regulation of APA factors by post-translational modifications
Gene-specific regulation of APA in response to environmental signals is further accomplished by kinases and ubiquitin ligases that post-translationally modify CPA factors and other RNA-binding proteins. Starvation inhibits mTOR and activates autophagy, a mechanism that provides nutrients from intracellular sources31. In flies, autophagy is stimulated chiefly by upregulation of autophagy-related protein 1 (Atg1) and Atg8a, through expression of their long 3′ UTR isoforms, which produce more protein than their short isoforms31 (Fig. 4c). This 3′ UTR lengthening was controlled by two kinases, which are expressed in cells with inactive mTOR and phosphorylate Cpsf6, a subunit of CFI. Phosphorylation of CPSF6 in human cells and of Cpsf6 in flies increases its activity by promoting its nuclear localization and RNA-binding capacity31,110 (Fig. 4c).
Ubiquitin ligases are recruited to specific substrates by adapter proteins. As adapter proteins are often expressed in a pathway-specific or cell-type-specific manner, this regulatory mode allows context-dependent regulation of widely expressed ubiquitin ligases111,112. For example, expression of the ubiquitin adapter melanoma-associated antigen 11 (MAGEA11) is restricted to germ cells, but it is activated in cancer, where it recruits the ubiquitin ligase HUWE1 to the substrate PCF11. PCF11 ubiquitylation changes the composition of the CPA machinery by expelling NUDT21 (also known as CPSF5), which results in predominant 3′ UTR shortening111. MAGEA11 upregulation in cancer may contribute to the observed 3′ UTR shortening in solid cancers, and could provide a new avenue for therapeutic intervention112,113,114.
These examples illustrate how APA is dynamically regulated and responds to diverse external signals. These signals include cold exposure, nutrient availability, immune pathway activation and developmental cues5,14,31,35,48,109,110,115. Environmental changes often affect only a subset of multi-UTR genes and can be accomplished by pathway-specific enzymes. The gene-specific APA response to environmental and developmental stimuli makes APA an integral component of all gene regulation circuits.
Post-transcriptional regulation
The production of 3′ UTR isoforms is controlled co-transcriptionally, but their expression can be further altered by several mechanisms of post-transcriptional regulation. It is estimated that 10–30% of multi-UTR genes are subject to this type of regulation3,27,39,40,41,109,116. The mechanisms include sequential CPA, nuclear RNA degradation, regulation of mRNA export and control of mRNA stability113,115,117,118,119,120,121 (Fig. 2). Differential nuclear export can shape 3′ UTR isoform ratios, as long 3′ UTRs rely much more on the presence of export receptors than do shorter 3′ UTRs117,118,119.
Most techniques that assess 3′ UTR isoforms cannot distinguish between transcriptional regulation and post-transcriptional regulation. However, 3′-seq performed on nuclear and cytoplasmic fractions revealed differential expression of 3′ UTRs for approximately 450 genes. In most of these genes, the longer 3′ UTRs were more abundant in the nucleus than in the cytoplasm116. Moreover, some genes undergo sequential CPA in the nucleus121. The distal PAS, which is often stronger, is processed first, but owing to incomplete mRNA processing at upstream elements such as introns, the mRNA is retained in the nucleus121. Subsequent processing at a proximal PAS can then release the mRNA for export to the cytoplasm. That study nicely revealed that the processing efficiency of specific 3′ UTRs does not always correlate with their cytoplasmic expression121.
Following successful nuclear export, mRNAs with longer 3′ UTRs were observed at intracellular membranes122. As reported in a recent preprint, compartment-specific 3′-seq further identified post-transcriptional regulation as the predominant regulatory mode of ~400 circadian 3′ UTR isoforms whose corresponding genes were not expressed in a circadian manner109.
Global 3′ UTR changes and phenotypes
Single-UTR genes and multi-UTR genes differ substantially in their expression patterns. Conceptually, chromatin regulators and CPA factors can fulfil similar roles in shaping the cellular transcriptome. Whereas chromatin regulators shape the gene expression landscape, CPA factors that induce global shifts in APA may set up 3′ UTR isoform landscapes that then accommodate further 3′ UTR isoform changes.
Genetic variants in 3′ UTRs have many phenotypes
To evaluate the functional relevance of 3′ UTRs, several genome-wide association studies assessed the contribution of 3′ UTR genetic variants to phenotypic traits42,43,44. Non-coding regions harbour most genetic variants123. In addition to genetic variants that correlate with gene expression level, genetic variants were identified that are associated with changes in APA, termed ‘APA quantitative trait loci’ (apaQTL) or ‘3′ aQTL’. Correlation of apaQTL with phenotypes revealed that 15–19% of apaQTL are associated with heritability of known human traits or diseases42,43,44. This association indicates that a large fraction of phenotypic variation observed across individuals can be tied to changes in APA, which most often alters the PASs or binding sites of RNA-binding proteins located in 3′ UTRs44. Interestingly, these studies found that the genetic variants associated with APA rarely overlap with genetic variants associated with gene expression changes, suggesting that the two regulatory mechanisms are independent43,44. These results suggest that most apaQTL contribute to human traits and diseases in a way that does not involve changes in mRNA abundance.
Control of single-UTR genes versus multi-UTR genes
Comparison of changes in gene expression and in 3′ UTR isoform use across cell types revealed that only in a minority (10–15%) of multi-UTR genes both parameters change simultaneously, suggesting that the two gene regulatory mechanisms are independent3,27,39,40,41. This finding was recently confirmed by a large-scale study, available as a preprint, that compared changes in the expression of gene and 3′ UTR isoforms across 120 mouse cell types5. The analysis showed that during differentiation, a gene either changes its expression or changes its APA, except for a small minority of genes that altered both. Remarkably, the number of genes with 3′ UTR isoform changes between cell types was fairly similar to the number of genes with expression changes.
Moreover, the mode of regulation of cell-type-specific expression differs substantially between single-UTR genes and multi-UTR genes (Fig. 5a,b). Gene expression of most (~80%) single-UTR genes is regulated developmentally3,5. Chromatin accessibility and transcription factor expression determine whether these genes are transcribed or not transcribed in specific cell types (Fig. 5a). The remaining ~20% of single-UTR genes are classified as ‘housekeeping genes’, whose transcription is switched on in nearly all cell types3,5.
a | Chromatin accessibility and transcription factor (TF) binding determine cell-type-specific transcription of genes whose mRNAs contain a single 3′ untranslated region (UTR) isoform (single-UTR genes). b | Multi-UTR genes are widely transcribed (like single-UTR housekeeping genes) yet change their 3′ UTR isoforms in a cell-type-specific manner. The cell-type-specific 3′ UTR landscape is determined largely by cell-type-specific expression of regulators of alternative cleavage and polyadenylation, such as cleavage and polyadenylation specificity factor subunit 5 (CPSF5; encoded by NUDT21). c | Cooperativity between a cell-type-specific 3′ UTR landscape and activation of signalling pathways or transcription factors determines phenotypic outcomes. 3′-seq, 3′-end sequencing; CPA, cleavage and polyadenylation; iPSC, induced pluripotent stem cell; LU, long 3′ UTR; OKSM, OCT4, KLF4, SOX2 and MYC; PAS, polyadenylation site; SU, short 3′ UTR.
At the transcriptional level, multi-UTR genes appear similar to housekeeping genes, as they are expressed in most cell types3,5 (Fig. 5b). Importantly, however, these genes express their alternative 3′ UTR isoforms in a cell-type-specific manner (Fig. 5b). This APA pattern is largely determined by cell-type-specific abundance of APA regulators, including CPSF5 or CPSF6, two subunits of CFI (refs.24,26,27,30,124) (Fig. 1). Many proteins with regulatory functions are encoded by multi-UTR genes (including transcription and chromatin regulators, ubiquitin enzymes, kinases and factors involved in mRNA or protein transport), suggesting that many of them are regulated by APA3.
APA regulators influence phenotypes by changing the 3′ UTR landscape
APA is globally dysregulated in cancer, predominantly exhibiting 3′ UTR shortening in solid tumours113,114. One of the APA regulators that is most relevant to 3′ UTR shortening is NUDT21 (CPSF5), whose downregulation by small hairpin RNAs results in 3′ UTR shortening of nearly 1,500 genes24,26,27,30. NUDT21 mRNA levels are significantly reduced in several solid tumours, and low NUDT21 expression levels correlate with a poor prognosis in several cancers26,125,126,127,128. Although global 3′ UTR shortening is widespread in cancer, it has been largely unclear what the shift towards expression of shorter 3′ UTRs means for cancer-specific phenotypes.
Notably, loss of NUDT21 results in 3′ UTR shortening of genes that are enriched in the RAS signalling pathway127. Depletion of NUDT21 enhances RAS-activation phenotypes, such as increased proliferation and migration of glioblastoma cell lines, and induces a RAS-activation multivulva phenotype in C. elegans127,129. Loss of NUDT21 strongly increased the oncogenic phenotype of human colon carcinoma cells with oncogenic RAS gene mutations129. These data suggest that a global APA regulator such as NUDT21 sets up a certain 3′ UTR isoform landscape that may alter the responsiveness of specific genes to additional signals (Fig. 5c). Cooperativity between the underlying 3′ UTR landscape and specific oncogenic pathways likely contributes to the susceptibility of certain cell types to particular oncogenes.
Suppression of NUDT21 also showed a cooperative effect with transcription factors during reprogramming of induced pluripotent stem cells, as suppression of NUDT21 increased reprogramming efficiency by more than tenfold27. Depletion of NUDT21 in mouse embryonic fibroblasts resulted in 3′ UTR shortening of more than 1,500 genes. This 3′ UTR landscape makes cells receptive to the activity of reprogramming transcription factors, and simultaneously increases their resistance to differentiation signals (Fig. 5c).
Such a regulatory pattern suggests that NUDT21 influences the 3′ UTR landscape in a manner similar to the shaping of gene expression by chromatin regulators. Whereas chromatin regulators establish a distinct epigenetic state to influence cell-specific gene expression130, we propose that APA regulators set up a distinct 3′ UTRome, which makes cells resistant or receptive to additional gene or 3′ UTR isoform changes. Cooperativity between the gene expression programme, the underlying APA landscape and the gene-specific changes in the expression of 3′ UTR isoforms determines the resulting phenotype26,27,129 (Fig. 5b,c).
Functions of alternative 3′ UTRs
In this section, we discuss functions of alternative 3′ UTRs, including the regulation of protein output, and translation in specific subcellular environments that enable post-translational modifications or allow the formation of specific protein complexes.
Alternative 3′ UTRs can regulate the expression levels of their encoded proteins through several mechanisms, including modulation of mRNA export, mRNA stability and mRNA translation rates. RNA-binding proteins and microRNAs can differentially bind and regulate transcripts with long and short 3′ UTRs. In cancer cells, some oncogenes can escape suppression by microRNAs through upregulation of short 3′ UTRs lacking the microRNA-binding sites113. Analyses of mRNA stability regulation through APA revealed that for more than two-thirds of genes the short and long 3′ UTR isoforms have a similar mRNA stability41. Of the genes with different stability of the alternative 3′ UTR isoforms, in ~90% of them the long isoform was less stable41. However, APA can have a different effect on mRNA stability and translation in different genes, and several cases were reported where long 3′ UTR isoforms generate substantially more protein.
Regulation of protein abundance
Uncoupling protein 1 (UCP1) is a mitochondrial proton transporter that induces a proton leak across the mitochondrial membrane to shift energy production towards heat production. Cold exposure activates adrenergic signalling and increases UCP1 expression. Northern blot analysis revealed that mouse Ucp1 generates two mRNA isoforms with alternative 3′ UTRs48. It was observed that the long 3′ UTR isoform of Ucp1 contributes only 5–10% of the total Ucp1 mRNA level, but its deletion reduced UCP1 expression by 50–60%. To fully delete the long 3′ UTR isoform of Ucp1, CRISPR–Cas9 was used to eliminate the 3′ UTR downstream of the proximal PAS in mice48. This experiment showed that UCP1 levels are predominantly regulated by translation of the long 3′ UTR isoform, which contains binding sites for the translation regulator cytoplasmic polyadenylation element-binding protein 2 (CPEB2). CPEB2 was required for polysome association of the long 3′ UTR isoform of Ucp1, for low level translation in steady-state conditions and for signalling-induced upregulation of translation in cold conditions or the presence of adrenaline48 (Fig. 6a).
a | 3′ untranslated region (UTR)-dependent increase in protein synthesis. Cytoplasmic polyadenylation element-binding protein 2 (CPEB2) binds to the long 3′ UTR (LU) isoform of the uncoupling protein 1 gene (Ucp1), which increases translation efficiency and protein abundance of UCP1. b | 3′ UTR-dependent mRNA localization facilitates synthesis of proteins at their final destination. Shown is a general example of local protein synthesis at the synapse of neurons. c | 3′ UTR-dependent local translation in condensates enables protein complex formation. (A+U)-rich elements represent the binding sites for the RNA-binding protein TIS11B. mRNAs that contain these elements in their 3′ UTRs localize to TIS granules, whereas lack of these elements results in localization to the endoplasmic reticulum (ER) or to the cytoplasm. Local protein synthesis in TIS granules allows the formation of protein complexes that cannot be established upon translation outside this cytoplasmic condensate. Shown is the compartment-dependent assembly of the complex between CD47 and SET, which traffics CD47 more efficiently to the plasma membrane, where it represses phagocytosis. d | 3′ UTR-dependent protein complex assembly determines protein function. A change in 3′ UTR isoform expression of the gene encoding the ubiquitin ligase BIRC3 leads to assembly of a protein complex, which changes BIRC3 function in human cells. 3′-seq, 3′-end sequencing; LU, long 3′ untranslated region; SU, short 3′ untranslated region.
polo mRNA is another example in which loss of long 3′ UTR isoform expression (through deletion of the distal PAS) results in decreased protein abundance45. polo encodes a kinase involved in cell cycle progression in flies. Insufficient Polo expression impairs cell proliferation and causes defects in abdominal wall development, resulting in embryonic lethality45. Interestingly, the same phenotype was observed when the transcript elongation rate of Pol II was reduced45. The slower elongation rate was caused by a mutation in the largest subunit of Pol II and changed APA of polo towards an increase in short 3′ UTR isoform expression at the expense of long 3′ UTR isoform expression. This example illustrates how APA of polo is regulated by changes in transcription elongation rate.
Local translation in neurons
Local protein synthesis in neurons often depends on specific 3′ UTRs, because they contain elements that are necessary for mRNA localization58,60,131,132,133,134,135,136,137,138,139,140. For example, steady-state and signalling-induced local synthesis of mTOR in axons depends on the presence of its 3′ UTR137 (Fig. 6b). Axon injury increases the local production of mTOR and activates mTOR through phosphorylation. Active mTOR then induces local protein synthesis from hundreds of mRNAs that often contain mTOR-regulated elements. Absence of the Mtor 3′ UTR prevents stress-dependent upregulation of local protein synthesis, which leads to neuronal cell death following injury137.
Extracellular signals can promote local protein synthesis of a transcription factor at the synapse, followed by retrograde trafficking to the nucleus of the neuron and activation of a signalling-induced transcriptional programme135,136,141. For example, the 3′ UTR of the gene encoding the mouse chromatin remodeller high mobility group nucleosome-binding domain-containing protein 5 (HMGN5) directs its translation to growth cones of neurons136. Only HMGN5 that was locally synthesized in growth cones was phosphorylated and was able to mediate transcription activation after being trafficked to the nucleus136. In this example, 3′ UTRs are being used to allow translation in a special subcellular environment, such as the growth cone, the properties of which can be controlled by environmental stimuli.
Environmental signals often affect only particular sets of neurons141. This can be accomplished by differential expression of surface receptors or through alternative 3′ UTRs of transcription factors or other regulatory proteins. In the case of cyclic AMP-responsive element-binding protein (CREB), mTOR and HMGN5, only specific 3′ UTR isoforms enable local synthesis of their proteins, which can then respond to environmental signals. As only the neurons that express these 3′ UTR isoforms are able to respond, alternative 3′ UTR isoform expression can establish a selective response to the signals60,135,136,137.
Local translation in condensates
APA also allows mRNA isoforms with specific 3′ UTRs to be translated at membranes, or in biomolecular condensates such as TIS granules10,47,142,143. CD47, which encodes a plasma membrane protein that acts as a phagocytosis-suppression signal, generates mRNA isoforms with alternative 3′ UTRs46. CD47 is translated at the endoplasmic reticulum, but CD47 mRNA with a long 3′ UTR isoform is translated in endoplasmic reticulum-associated TIS granules, a new type of cytoplasmic membraneless organelle that is generated through assembly of the RNA-binding protein TIS11B (also known as ZFP36L1) together with the mRNAs that it binds47. Translation in TIS granules was required for assembly of a protein complex comprising CD47 and the trafficking factor SET, which promotes the localization of CD47 to the plasma membrane, thereby inhibiting phagocytosis of the cell46,47. mRNA localization to TIS granules, followed by local translation, requires (A+U)-rich elements in the long 3′ UTR of CD47 mRNA, which function as TIS11B-binding sites (Fig. 6c). As the short 3′ UTR isoform lacks these motifs, it is not preferentially translated in TIS granules and is unable to interact with SET, leading to diminished protection of the cells from phagocytosis46,47. This example shows that 3′ UTRs control protein complex assembly, which affects protein functions.
Formation of protein complexes
The membraneless processing bodies (P-bodies) are cytoplasmic condensates that concentrate RNA-binding proteins and RNAs. In yeast, P-bodies are nucleated through interactions between 40S ribosomal protein S28-B (Rps28b) and enhancer of mRNA-decapping protein 3 (Edc3)144. Intriguingly, 3′ UTR-dependent protein complex formation enhances assembly of P-bodies. Complex assembly between Rps28B and Edc3 is strongly promoted by the RPS28B 3′ UTR, which binds and recruits Edc3 to the site of Rps28b translation. In the absence of the RPS28B 3′ UTR, the formation of the protein–protein interaction is very inefficient. Importantly, the RPS28B 3′ UTR does not regulate the abundance of Rps28b144.
A similar scenario was observed for the human ubiquitin ligase BIRC3, which is best known as a regulator of cell death49,145. Cells highly expressing BIRC3 are better protected from apoptosis, indicating that this function is regulated by BIRC3 abundance and is mediated through interaction with caspases and factors of the NF-κB pathway49,145. BIRC3 also regulates cell migration through regulation of the abundance of C-X-C chemokine receptor type 4 (CXCR4) at the cell surface. However, only BIRC3 encoded by the mRNA with the long BIRC3 3′ UTR is able to control migration49. Mechanistically, only BIRC3 that was encoded by BIRC3 mRNA with the long 3′ UTR is incorporated in a 3′ UTR-dependent manner into a complex that regulates CXCR4 surface expression (Fig. 6d). Interestingly, BIRC3 abundance was not the determining factor for the control of migration, as leukaemia cells with very low expression of BIRC3, generated by the long 3′ UTR mRNA isoform, were still able to migrate in a long BIRC3 3′ UTR-dependent manner49. These examples illustrate how alternative 3′ UTRs can facilitate the assembly of alternative protein complexes, indicating that 3′ UTRs are integral regulators of alternative protein function.
Novel technologies for studying APA
New sequencing approaches provide unprecedented spatial and temporal resolution of APA isoform expression. Furthermore, research into the functional significance of APA has been catalysed by CRISPR–Cas technologies, which facilitate the manipulation of 3′ UTRs in their endogenous genomic contexts.
APA quantification by scRNA-seq
In the past, only a few laboratories analysed APA, as specialized 3′-end sequencing protocols were required for faithful 3′ UTR isoform quantification3,4,21,40,146. The recent development of 3′-tagged scRNA-seq protocols, together with several new computational pipelines, will change this situation as now publicly available datasets can be used to quantify gene and 3′ UTR isoform expression from basically any cell type5,50,51,52,147. Comparison of the technical variation of APA between previously used bulk 3′-end sequencing protocols and 10x Genomics scRNA-seq datasets found that the new method is substantially more reproducible5. This technological advance has enabled APA analysis at all stages of cell differentiation and revealed global shortening of 3′ UTRs during haematopoiesis and spermatogenesis5,51,52,148,149. These results have challenged the prior notion that differentiation processes generally induce expression of longer 3′ UTRs. Interestingly, APA analysis using single-nucleus RNA-seq of post-mortem brain tissue revealed predominantly longer 3′ UTRs in people with autism in comparison with healthy individuals150,151. These analyses demonstrate how technical improvement may soon enable even more sophisticated studies of APA-related disease phenotypes.
Manipulation of alternative 3′ UTRs
Functional analyses of 3′ UTRs used to rely mainly on reporter gene assays and transient overexpression of plasmid-encoded constructs113,115. Although luciferase or GFP reporters are sensitive tools for measuring expression differences, they are not suitable for evaluating other 3′ UTR-mediated functions8. This bias in experimental readout might have contributed to the notion that 3′ UTRs have evolved mainly as means of gene expression regulation. Furthermore, vector-based reporter systems do not faithfully recapitulate the endogenous control of 3′ UTR isoform expression and function by promoters, enhancers, 5′ UTRs and introns14,36,152,153,154; therefore, whenever possible, functional characterization of 3′ UTRs should be performed in their endogenous genomic contexts. Such approaches have become feasible with several recently developed genome editing technologies.
Shifting APA through use of small hairpin RNAs and ASOs
Small hairpin RNAs or small interfering RNAs that target the unique sequence of long 3′ UTR isoforms can be used to specifically deplete these isoforms49 (Fig. 7a). To downregulate short 3′ UTR isoforms, modified antisense oligonucleotides (ASOs) that bind to sequences surrounding proximal PASs can be used. These ASOs can physically block access of the CPA machinery to the nascent RNA, thus preventing 3′-end formation at the blocked PAS155,156,157 (Fig. 7b). Similarly, ASO targeting of intronic PASs was used to shift mRNA processing towards splicing of the intron, resulting in increased expression of the full-length protein152.
a | Small hairpin RNAs or small interfering RNAs (siRNAs) targeting the extended part of 3′ untranslated region (UTR) isoforms exclusively downregulate the expression of long 3′ UTR (LU) isoforms. b | Antisense oligonucleotides (ASOs) that are complementary to a specific proximal polyadenylation site (PAS) prevent binding of the cleavage and polyadenylation (CPA) machinery and exclusively downregulate expression of short 3′ UTR (SU) isoforms. c | CRISPR–iPAS. An enzymatically dead Cas13 (dCas13) is targeted by a guide RNA (dCas13 RNP) to a region slightly upstream of a PAS of interest, where it prevents CPA factors from recognizing the PAS. d | CRISPRpas. An enzymatically dead Cas9 (dCas9) is targeted by a guide RNA to a region downstream of a PAS of interest, where it acts as a roadblock for RNA polymerase II (Pol II) elongation, thereby increasing CPA at the PAS. e | Permanent removal of a specific PAS or entire 3′ UTR isoforms using a pair of Cas9 RNPs to delete the region of interest. f | PAS mutagenesis through homologous recombination can be achieved by CRISPR–Cas9-mediated DNA cleavage in the presence of a homologous repair template that contains the mutation. Cas RNP, Cas protein–guide RNA complex; KO, knockout.
Shifting APA using CRISPR–Cas systems
PAS choice can be manipulated using CRISPR–Cas systems. For example, with CRISPR–iPAS, site-specific recruitment of catalytically dead RNA-specific Cas13 to proximal PASs seems to prevent access of CPA factors and shifts APA towards distal PAS use158 (Fig. 7c). Alternatively, with CRISPRpas, proximal PAS use is enhanced through recruitment of the DNA-specific catalytically dead Cas9 to sequences downstream of the PAS54 (Fig. 7d). It is thought that the Cas9 roadblock leads to pausing and disengagement of the transcribing Pol II complex, which appears to be particularly suitable for increasing the use of weak PASs.
In addition to the above-mentioned tools for transient modulation of PAS use, the CRISPR–Cas technology can also permanently change APA. CRISPR–Cas9 was used to insert exogenous PASs into 3′ UTRs. For example, a preprint reports overexpression of the neurotrophic factor GDNF in mice by introduction of a strong PAS downstream of its stop codon159. A similar approach in induced pluripotent stem cells serves as example for therapeutic genome editing160. Myotonic dystrophy type I is caused by a toxic CUG repeat expansion in the 3′ UTR of DMPK. CRISPR–Cas-mediated insertion of two strong PASs upstream of the repeat permanently eliminated transcripts with the repeat expansion and led to reversal of the disease phenotype in skeletal muscle fibres derived from induced pluripotent stem cells160.
Deletion of 3′ UTRs or individual PASs using CRISPR–Cas
Several laboratories have used pairs of guide RNAs to generate defined genomic deletions to eliminate entire 3′ UTRs or specific PASs in cell lines or model organisms29,48,53,64,75,154,161,162 (Fig. 7e). In a related approach, when a homologous DNA template is provided, individual point mutations can be introduced into the PAS hexamer to generate precise sequence perturbations75 (Fig. 7f).
If the sequence context of the PAS contains a site for guide-RNA binding, a single guide RNA–Cas9 complex can be used to disrupt an individual PAS. DNA polymerase-η (Polη), which is encoded by POLH, performs DNA translesion synthesis and contributes to drug resistance in cancer; POLH deletion prevents the drug resistance163. POLH generates three 3′ UTR isoforms, and single guide RNAs were used to disrupt either the proximal PAS or the distal PAS. Each disruption had substantial effects on Polη expression, nearly fully recapitulating the effects of the gene deletion163. This result suggests that elimination of an individual PAS of a multi-UTR gene may be a strategy for downregulating gene and protein expression in experimental or therapeutic settings.
Screens for functional elements in 3′ UTRs
CRISPR–Cas technology was also applied to functionally interrogate 3′ UTR sequences. A proof-of-concept study in Drosophila melanogaster showed that tiling libraries of guide RNAs targeting a 3′ UTR can be used to identify cis-regulatory elements164. However, owing to redundancy of regulatory elements, small insertions or deletions in non-coding sequences often have limited effects on gene expression. Tiling screens in organisms with large genomes, such as humans or mice, currently do not provide sufficient coverage because of limited availability of unique targeting sequences. However, a recent innovative approach overcame the limitation of low coverage55. The study authors screened C. elegans for mutations in non-coding regulatory regions, including 3′ UTRs, that are associated with fitness or morphological phenotypes. Through random pairing of guide RNAs in their regions of interest, they generated many diverse sets of deletions. Deletion mapping in worms with specific phenotypes generated a mutagenesis profile and allowed the identification of hotspots with regulatory potential55. This novel approach identifies non-coding cis-regulatory elements and their interdependency.
APA therapeutics
The implication of aberrant 3′ UTR isoform expression patterns in human disease, in particular cancer, has created a strong interest in manipulating APA pharmacologically. In this section, we discuss new chemical compounds and sequence-based therapeutics that have the potential to be used clinically to correct both global and gene-specific APA dysregulation.
Compounds that shift APA
With the discovery of disease-associated changes in 3′ UTRs26,33,34,111,129,151, APA deregulation presents a new target for therapeutic intervention. Reporter assays identified compounds that broadly shift APA towards use of proximal PASs165. In addition to new molecules, the compounds included topoisomerase inhibitors that were previously reported to modulate APA166. This result suggests that some of the currently used drugs may have a substantial impact on APA. However, so far, large-scale studies that evaluate drug-induced changes in APA are lacking.
The recent development of computational methods to analyse APA from scRNA-seq data may help to identify small molecules that predominantly target isoform expression instead of gene expression. For example, the compound JTE-607 was developed two decades ago, and in preclinical studies caused selective lethality in acute leukaemia and Ewing sarcoma cells167. Recently, it was recognized that the small molecule binds CPSF3, the endonuclease of the CPSF complex, which causes extensive readthrough transcription18,167.
Other known drugs, including molecules that alter DNA structure, chromatin or Pol II processivity, such as inhibitors of topoisomerases, histone deacetylases, or CDK12 and CDK13, were shown to cause shifts in APA33,34,80,166,168. However, in most cases, the mechanism of action by which these compounds alter APA is not known. Owing to their DNA damage-inducing capacity, these compound classes are used as anticancer drugs, but it is currently unclear whether the changes in APA enhance or counteract the intended treatment effects170.
RNA therapeutics
A more gene-specific therapeutic approach involves the use of ASOs to modulate APA in specific disease genes. Promising proof-of-concept studies in neuroblastoma, prostate cancer and muscular dystrophy effectively suppressed PAS use in cell culture and in mice152,155,156,157. For example, ASOs targeting the only PAS in DUX4 diminish overall transcript maturation and can be used to correct DUX4 overexpression in muscular dystrophy155,156.
DNA-based and RNA-based strategies for vaccines, cancer immunotherapies and gene replacement therapies are intensely explored. For example, 3′ UTR sequences optimized for stability have been identified and used in the Pfizer–BioNTech mRNA vaccine against severe acute respiratory syndrome coronavirus 2 (ref.169). However, better knowledge of the function of 3′ UTR-dependent gene regulation will inspire improved sequence design to include isoform-specific and cell compartment-specific targeting.
Conclusion and future perspective
In this Review, we have discussed how the integration of different regulatory mechanisms gives rise to highly specific and dynamic APA landscapes. Further research is required to gain more insights into gene-specific isoform regulation in different conditions, which could help to address some remaining open questions, such as how the activity of APA-regulating enzymes is confined to particular gene loci.
Although only a handful of APA events have been thoroughly investigated on a functional level, they provide a glimpse of the potential contribution of APA regulation to phenotypic complexity. Resolving the expression patterns of 3′ UTR isoforms will help to elucidate gene functions and their impact on human health. Importantly, changes in APA can also modify the cellular response to external cues. Although it is known that cancer is frequently characterized by 3′ UTR shortening, the extent of cooperativity between APA and oncogenic pathways needs to be further characterized, which could lead to the development of new therapeutic approaches that target APA regulators.
References
Tian, B. & Manley, J. L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 (2017).
Gruber, A. J. & Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 20, 599–614 (2019).
Lianoglou, S., Garg, V., Yang, J. L., Leslie, C. S. & Mayr, C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396 (2013).
Hoque, M. et al. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods 10, 133–139 (2013).
Fansler, M. M., Zhen, G. & Mayr, C. Quantification of alternative 3′UTR isoforms from single cell RNA-seq data with scUTRquant. Preprint at bioRxiv https://doi.org/10.1101/2021.11.22.469635 (2021).
Singh, I. et al. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nat. Commun. 9, 1716 (2018).
Gruber, A. J., Gypas, F., Riba, A., Schmidt, R. & Zavolan, M. Terminal exon characterization with TECtool reveals an abundance of cell-specific isoforms. Nat. Methods 15, 832–836 (2018).
Mayr, C. Regulation by 3′-untranslated regions. Annu. Rev. Genet. 51, 171–194 (2017).
Mayr, C. What are 3′ UTRs doing? Cold Spring Harb. Perspect. Biol. 11, a034728 (2019).
Chen, X. & Mayr, C. A working model for condensate RNA-binding proteins as matchmakers for protein complex assembly. RNA 28, 76–87 (2022).
Higgs, D. R. et al. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature 306, 398–400 (1983).
Gehring, N. H. et al. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat. Genet. 28, 389–392 (2001).
Stacey, S. N. et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat. Genet. 43, 1098–1103 (2011).
Kwon, B. et al. Enhancers regulate 3′ end processing activity to control expression of alternative 3′UTR isoforms. Nat. Commun. 13, 2709 (2022).
Shi, Y. et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33, 365–376 (2009).
Schonemann, L. et al. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 28, 2381–2393 (2014).
Chan, S. L. et al. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev. 28, 2370–2380 (2014).
Liu, H. & Moore, C. L. On the cutting edge: regulation and therapeutic potential of the mRNA 3′ end nuclease. Trends Biochem. Sci. 46, 772–784 (2021).
Boreikaite, V., Elliott, T., Chin, J. & Passmore, L. A. RBBP6 activates the pre-mRNA 3′-end processing machinery in humans. Genes Dev. 36, 210–224 (2022).
Bogard, N., Linder, J., Rosenberg, A. B. & Seelig, G. A deep neural network for predicting and engineering alternative polyadenylation. Cell 178, 91–106 (2019).
Derti, A. et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 22, 1173–1183 (2012).
Jenal, M. et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538–553 (2012).
Gruber, A. R., Martin, G., Keller, W. & Zavolan, M. Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol. 9, 1405–1412 (2012).
Li, W. et al. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 11, e1005166 (2015).
Lackford, B. et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 33, 878–889 (2014).
Masamha, C. P. et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510, 412–416 (2014).
Brumbaugh, J. et al. Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell 172, 629–631 (2018).
Ogorodnikov, A. et al. Transcriptome 3′end organization by PCF11 links alternative polyadenylation to formation and neuronal differentiation of neuroblastoma. Nat. Commun. 9, 5331 (2018).
Kamieniarz-Gdula, K. et al. Selective roles of vertebrate PCF11 in premature and full-length transcript termination. Mol. Cell 74, 158–172 (2019).
Marini, F., Scherzinger, D. & Danckwardt, S. TREND-DB-a transcriptome-wide atlas of the dynamic landscape of alternative polyadenylation. Nucleic Acids Res. 49, D243–D253 (2021).
Tang, H. W. et al. The TORC1-regulated CPA complex rewires an RNA processing network to drive autophagy and metabolic reprogramming. Cell Metab. 27, 1040–1054 (2018).
Zheng, D. et al. Cellular stress alters 3′UTR landscape through alternative polyadenylation and isoform-specific degradation. Nat. Commun. 9, 2268 (2018).
Dubbury, S. J., Boutz, P. L. & Sharp, P. A. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 564, 141–145 (2018).
Krajewska, M. et al. CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat. Commun. 10, 1757 (2019).
LaBella, M. L. et al. Casein kinase 1delta stabilizes mature axons by inhibiting transcription termination of ankyrin. Dev. Cell 53, 130 (2020).
Oktaba, K. et al. ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol. Cell 57, 341–348 (2015).
Legnini, I., Alles, J., Karaiskos, N., Ayoub, S. & Rajewsky, N. FLAM-seq: full-length mRNA sequencing reveals principles of poly(A) tail length control. Nat. Methods 16, 879–886 (2019).
Wang, X. et al. Full-length transcriptome reconstruction reveals a large diversity of RNA and protein isoforms in rat hippocampus. Nat. Commun. 10, 5009 (2019).
Spies, N., Burge, C. B. & Bartel, D. P. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 23, 2078–2090 (2013).
Gruber, A. R. et al. Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat. Commun. 5, 5465 (2014).
Wang, R., Zheng, D., Yehia, G. & Tian, B. A compendium of conserved cleavage and polyadenylation events in mammalian genes. Genome Res. 28, 1427–1441 (2018).
Mariella, E., Marotta, F., Grassi, E., Gilotto, S. & Provero, P. The length of the expressed 3′ UTR is an intermediate molecular phenotype linking genetic variants to complex diseases. Front. Genet. 10, 714 (2019).
Mittleman, B. E. et al. Alternative polyadenylation mediates genetic regulation of gene expression. eLife 9, e57492 (2020).
Li, L. et al. An atlas of alternative polyadenylation quantitative trait loci contributing to complex trait and disease heritability. Nat. Genet. 53, 994–1005 (2021).
Pinto, P. A. et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 30, 2431–2444 (2011).
Berkovits, B. D. & Mayr, C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367 (2015).
Ma, W. & Mayr, C. A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein-protein interactions. Cell 175, 1492–1506 (2018).
Chen, H. F., Hsu, C. M. & Huang, Y. S. CPEB2-dependent translation of long 3′-UTR Ucp1 mRNA promotes thermogenesis in brown adipose tissue. EMBO J. 37, e99071 (2018).
Lee, S. H. & Mayr, C. Gain of additional BIRC3 protein functions through 3′-UTR-mediated protein complex formation. Mol. Cell 74, 701–712 (2019).
Patrick, R. et al. Sierra: discovery of differential transcript usage from polyA-captured single-cell RNA-seq data. Genome Biol. 21, 167 (2020).
Li, W. V., Zheng, D., Wang, R. & Tian, B. MAAPER: model-based analysis of alternative polyadenylation using 3′ end-linked reads. Genome Biol. 22, 222 (2021).
Agarwal, V., Lopez-Darwin, S., Kelley, D. R. & Shendure, J. The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nat. Commun. 12, 5101 (2021).
Mitschka, S., Fansler, M. M. & Mayr, C. Generation of 3′UTR knockout cell lines by CRISPR/Cas9-mediated genome editing. Methods Enzymol. 655, 427–457 (2021).
Shin, J. et al. CRISPRpas: programmable regulation of alternative polyadenylation by dCas9. Nucleic Acids Res. 50, e25 (2022).
Froehlich, J. J. et al. Parallel genetics of regulatory sequences using scalable genome editing in vivo. Cell Rep. 35, 108988 (2021).
Yaffe, D., Nudel, U., Mayer, Y. & Neuman, S. Highly conserved sequences in the 3′ untranslated region of mRNAs coding for homologous proteins in distantly related species. Nucleic Acids Res. 13, 3723–3737 (1985).
Takagaki, Y., Seipelt, R. L., Peterson, M. L. & Manley, J. L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87, 941–952 (1996).
Taliaferro, J. M. et al. Distal alternative last exons localize mRNAs to neural projections. Mol. Cell 61, 821–833 (2016).
Lee, S. H. et al. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature 561, 127–131 (2018).
Ciolli Mattioli, C. et al. Alternative 3′ UTRs direct localization of functionally diverse protein isoforms in neuronal compartments. Nucleic Acids Res. 47, 2560–2573 (2019).
Kaida, D. et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468, 664–668 (2010).
Cugusi, S. et al. Heat shock induces premature transcript termination and reconfigures the human transcriptome. Mol. Cell 82, 1573–1588 (2022).
Luo, W. et al. The conserved intronic cleavage and polyadenylation site of CstF-77 gene imparts control of 3′ end processing activity through feedback autoregulation and by U1 snRNP. PLoS Genet. 9, e1003613 (2013).
Wang, R., Zheng, D., Wei, L., Ding, Q. & Tian, B. Regulation of intronic polyadenylation by PCF11 impacts mRNA expression of long genes. Cell Rep. 26, 2766–2778 (2019).
Yu, M. et al. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 350, 1383–1386 (2015).
Gregersen, L. H. et al. SCAF4 and SCAF8, mRNA anti-terminator proteins. Cell 177, 1797–1813 (2019).
Gromak, N., West, S. & Proudfoot, N. J. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell Biol. 26, 3986–3996 (2006).
Sheridan, R. M., Fong, N., D’Alessandro, A. & Bentley, D. L. Widespread backtracking by RNA Pol II is a major effector of gene activation, 5′ pause release, termination, and transcription elongation rate. Mol. Cell 73, 107–118 (2019).
Skourti-Stathaki, K., Proudfoot, N. J. & Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011).
Beaudoin, J.-D. & Perreault, J.-P. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 41, 5898–5911 (2013).
Nanavaty, V. et al. DNA methylation regulates alternative polyadenylation via CTCF and the Cohesin complex. Mol. Cell 78, 752–764 (2020).
Soles, L. V. & Shi, Y. Crosstalk between mRNA 3′-end processing and epigenetics. Front. Genet. 12, 637705 (2021).
Bataille, A. R. et al. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol. Cell 45, 158–170 (2012).
Zaborowska, J., Egloff, S. & Murphy, S. The pol II CTD: new twists in the tail. Nat. Struct. Mol. Biol. 23, 771–777 (2016).
Cortazar, M. A. et al. Control of RNA Pol II speed by PNUTS-PP1 and Spt5 dephosphorylation facilitates termination by a “sitting duck torpedo” mechanism. Mol. Cell 76, 896–908 (2019).
Laitem, C. et al. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat. Struct. Mol. Biol. 22, 396–403 (2015).
Krishnamurthy, S., He, X., Reyes-Reyes, M., Moore, C. & Hampsey, M. Ssu72 is an RNA polymerase II CTD phosphatase. Mol. Cell 14, 387–394 (2004).
Huang, K. L. et al. Integrator recruits protein phosphatase 2A to prevent pause release and facilitate transcription termination. Mol. Cell 80, 345–358 (2020).
Cossa, G., Parua, P. K., Eilers, M. & Fisher, R. P. Protein phosphatases in the RNAPII transcription cycle: erasers, sculptors, gatekeepers, and potential drug targets. Genes Dev. 35, 658–676 (2021).
Fan, Z. et al. CDK13 cooperates with CDK12 to control global RNA polymerase II processivity. Sci. Adv. 6, eaaz5041 (2020).
Eaton, J. D. & West, S. Termination of transcription by RNA polymerase II: BOOM! Trends Genet. 36, 664–675 (2020).
Rozenblatt-Rosen, O. et al. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc. Natl Acad. Sci. USA 106, 755–760 (2009).
Yang, Y. et al. PAF complex plays novel subunit-specific roles in alternative cleavage and polyadenylation. PLoS Genet. 12, e1005794 (2016).
Tellier, M. et al. CDK12 globally stimulates RNA polymerase II transcription elongation and carboxyl-terminal domain phosphorylation. Nucleic Acids Res. 48, 7712–7727 (2020).
Chen, F. et al. Context-dependent modulation of Pol II CTD phosphatase SSUP-72 regulates alternative polyadenylation in neuronal development. Genes Dev. 29, 2377–2390 (2015).
Kuehner, J. N., Pearson, E. L. & Moore, C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell Biol. 12, 283–294 (2011).
Xiang, K. et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467, 729–733 (2010).
Dantonel, J. C., Murthy, K. G., Manley, J. L. & Tora, L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389, 399–402 (1997).
McCracken, S. et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385, 357–361 (1997).
Rosonina, E., Bakowski, M. A., McCracken, S. & Blencowe, B. J. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 278, 43034–43040 (2003).
Calvo, O. & Manley, J. L. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 24, 1009–1020 (2005).
Nagaike, T. et al. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol. Cell 41, 409–418 (2011).
Harel-Sharvit, L. et al. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143, 552–563 (2010).
Trcek, T., Larson, D. R., Moldon, A., Query, C. C. & Singer, R. H. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell 147, 1484–1497 (2011).
Bregman, A. et al. Promoter elements regulate cytoplasmic mRNA decay. Cell 147, 1473–1483 (2011).
Zid, B. M. & O’Shea, E. K. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514, 117–121 (2014).
Haimovich, G., Choder, M., Singer, R. H. & Trcek, T. The fate of the messenger is pre-determined: a new model for regulation of gene expression. Biochim. Biophys. Acta 1829, 643–653 (2013).
Calvo, O. & Manley, J. L. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 17, 1321–1327 (2003).
Xiao, R. et al. Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121 (2019).
Baltz, A. G. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012).
Brannan, K. W. et al. SONAR discovers RNA-binding proteins from analysis of large-scale protein-protein interactomes. Mol. Cell 64, 282–293 (2016).
Caudron-Herger, M. et al. R-DeeP: proteome-wide and quantitative identification of RNA-dependent proteins by density gradient ultracentrifugation. Mol. Cell 75, 184–199 (2019).
Caudron-Herger, M., Jansen, R. E., Wassmer, E. & Diederichs, S. RBP2GO: a comprehensive pan-species database on RNA-binding proteins, their interactions and functions. Nucleic Acids Res. 49, D425–D436 (2021).
Lu, D. et al. CREPT accelerates tumorigenesis by regulating the transcription of cell-cycle-related genes. Cancer Cell 21, 92–104 (2012).
Tan-Wong, S. M. et al. Gene loops enhance transcriptional directionality. Science 338, 671–675 (2012).
Lamas-Maceiras, M., Singh, B. N., Hampsey, M. & Freire-Picos, M. A. Promoter-terminator gene loops affect alternative 3′-end processing in yeast. J. Biol. Chem. 291, 8960–8968 (2016).
Miki, T. S., Carl, S. H. & Großhans, H. Two distinct transcription termination modes dictated by promoters. Genes Dev. 31, 1870–1879 (2017).
Trott, A. J. & Menet, J. S. Regulation of circadian clock transcriptional output by CLOCK:BMAL1. PLoS Genet. 14, e1007156 (2018).
Greenwell, B. J. et al. Isoform-specific regulation of rhythmic gene expression by alternative polyadenylation. Preprint at bioRxiv https://doi.org/10.1101/2020.12.12.422514 (2020).
Zhu, Y. et al. Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol. Cell 69, 62–74 (2018).
Yang, S. W. et al. A cancer-specific ubiquitin ligase drives mRNA alternative polyadenylation by ubiquitinating the mRNA 3′ end processing complex. Mol. Cell 77, 1206–1221 (2020).
Yang, S. W. et al. Structural basis for substrate recognition and chemical inhibition of oncogenic MAGE ubiquitin ligases. Nat. Commun. 11, 4931 (2020).
Mayr, C. & Bartel, D. P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 (2009).
Xia, Z. et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat. Commun. 5, 5274 (2014).
Sandberg, R., Neilson, J. R., Sarma, A., Sharp, P. A. & Burge, C. B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647 (2008).
Neve, J. et al. Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res. 26, 24–35 (2016).
Katahira, J. et al. Human TREX component Thoc5 affects alternative polyadenylation site choice by recruiting mammalian cleavage factor I. Nucleic Acids Res. 41, 7060–7072 (2013).
Muller-McNicoll, M. et al. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 30, 553–566 (2016).
Chen, S. et al. The mRNA export receptor NXF1 coordinates transcriptional dynamics, alternative polyadenylation, and mRNA export. Mol. Cell 74, 118–131 (2019).
Silla, T. et al. The human ZC3H3 and RBM26/27 proteins are critical for PAXT-mediated nuclear RNA decay. Nucleic Acids Res. 48, 2518–2530 (2020).
Tang, P. et al. Alternative polyadenylation by sequential activation of distal and proximal PolyA sites. Nat. Struct. Mol. Biol. 29, 21–31 (2022).
Cheng, L. C. et al. Alternative 3′ UTRs play a widespread role in translation-independent mRNA association with the endoplasmic reticulum. Cell Rep. 36, 109407 (2021).
Battle, A., Brown, C. D., Engelhardt, B. E. & Montgomery, S. B. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Ghosh, S. et al. CFIm-mediated alternative polyadenylation remodels cellular signaling and miRNA biogenesis. Nucleic Acids Res. 50, 3096–3114 (2022).
Sun, M. et al. NUDT21 regulates 3′-UTR length and microRNA-mediated gene silencing in hepatocellular carcinoma. Cancer Lett. 410, 158–168 (2017).
Tan, S. et al. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene 37, 4887–4900 (2018).
Chu, Y. et al. Nudt21 regulates the alternative polyadenylation of Pak1 and is predictive in the prognosis of glioblastoma patients. Oncogene 38, 4154–4168 (2019).
Xing, Y. et al. Downregulation of NUDT21 contributes to cervical cancer progression through alternative polyadenylation. Oncogene 40, 2051–2064 (2021).
Subramanian, A. et al. Alternative polyadenylation is a determinant of oncogenic Ras function. Sci. Adv. 7, eabh0562 (2021).
Haigis, K. M., Cichowski, K. & Elledge, S. J. Tissue-specificity in cancer: the rule, not the exception. Science 363, 1150–1151 (2019).
Bassell, G. J. et al. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 18, 251–265 (1998).
Miller, S. et al. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36, 507–519 (2002).
Wu, K. Y. et al. Soluble adenylyl cyclase is required for netrin-1 signaling in nerve growth cones. Nat. Neurosci. 9, 1257–1264 (2006).
An, J. J. et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134, 175–187 (2008).
Cox, L. J., Hengst, U., Gurskaya, N. G., Lukyanov, K. A. & Jaffrey, S. R. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat. Cell Biol. 10, 149–159 (2008).
Moretti, F. et al. Growth cone localization of the mRNA encoding the chromatin regulator HMGN5 modulates neurite outgrowth. Mol. Cell Biol. 35, 2035–2050 (2015).
Terenzio, M. et al. Locally translated mTOR controls axonal local translation in nerve injury. Science 359, 1416–1421 (2018).
Bauer, K. E. et al. Live cell imaging reveals 3’-UTR dependent mRNA sorting to synapses. Nat. Commun. 10, 3178 (2019).
Tushev, G. et al. Alternative 3′ UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron 98, 495–511 (2018).
Hafner, A. S., Donlin-Asp, P. G., Leitch, B., Herzog, E. & Schuman, E. M. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 364, eaau3644 (2019).
Haase, G. et al. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron 35, 893–905 (2002).
Bethune, J., Jansen, R. P., Feldbrugge, M. & Zarnack, K. Membrane-associated RNA-binding proteins orchestrate organelle-coupled translation. Trends Cell Biol. 29, 178–188 (2019).
Cheng, L. C. et al. Widespread transcript shortening through alternative polyadenylation in secretory cell differentiation. Nat. Commun. 11, 3182 (2020).
Fernandes, N. & Buchan, J. R. RPS28B mRNA acts as a scaffold promoting cis-translational interaction of proteins driving P-body assembly. Nucleic Acids Res. 48, 6265–6279 (2020).
Beug, S. T., Cheung, H. H., LaCasse, E. C. & Korneluk, R. G. Modulation of immune signalling by inhibitors of apoptosis. Trends Immunol. 33, 535–545 (2012).
Shepard, P. J. et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17, 761–772 (2011).
Diag, A., Schilling, M., Klironomos, F., Ayoub, S. & Rajewsky, N. Spatiotemporal m(i)RNA Architecture and 3’ UTR regulation in the C. elegans germline. Dev. Cell 47, 785–800 (2018).
Shulman, E. D. & Elkon, R. Cell-type-specific analysis of alternative polyadenylation using single-cell transcriptomics data. Nucleic Acids Res. 47, 10027–10039 (2019).
Burri, D. & Zavolan, M. Shortening of 3′ UTRs in most cell types composing tumor tissues implicates alternative polyadenylation in protein metabolism. RNA 27, 1459–1470 (2021).
Velmeshev, D. et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689 (2019).
Göpferich, M. et al. Single cell 3′UTR analysis identifies changes in alternative polyadenylation throughout neuronal differentiation and in autism. Preprint at bioRxiv https://doi.org/10.1101/2020.08.12.247627 (2020).
Van Etten, J. L. et al. Targeting a single alternative polyadenylation site coordinately blocks expression of androgen receptor mRNA splice variants in prostate cancer. Cancer Res. 77, 5228–5235 (2017).
Theil, K., Herzog, M. & Rajewsky, N. Post-transcriptional regulation by 3′ UTRs can be masked by regulatory elements in 5′ UTRs. Cell Rep. 22, 3217–3226 (2018).
Mitschka, S. & Mayr, C. Endogenous p53 expression in human and mouse is not regulated by its 3′UTR. eLife 10, e65700 (2021).
Chen, J. C. et al. Morpholino-mediated knockdown of DUX4 toward facioscapulohumeral muscular dystrophy therapeutics. Mol. Ther. 24, 1405–1411 (2016).
Marsollier, A. C. et al. Antisense targeting of 3′ end elements involved in DUX4 mRNA processing is an efficient therapeutic strategy for facioscapulohumeral dystrophy: a new gene-silencing approach. Hum. Mol. Genet. 25, 1468–1478 (2016).
Naveed, A. et al. NEAT1 polyA-modulating antisense oligonucleotides reveal opposing functions for both long non-coding RNA isoforms in neuroblastoma. Cell. Mol. Life Sci. 78, 2213–2230 (2021).
Tian, S. et al. CRISPR-iPAS: a novel dCAS13-based method for alternative polyadenylation interference. Nucleic Acids Res. 50, e26 (2022).
Mätlik, K. et al. Gene knock up via 3′UTR editing to study gene function in vivo. Preprint at bioRxiv https://doi.org/10.1101/775031 (2019).
Wang, Y. et al. Therapeutic genome editing for myotonic dystrophy type 1 using CRISPR/Cas9. Mol. Ther. 26, 2617–2630 (2018).
Zhao, W. et al. CRISPR-Cas9-mediated functional dissection of 3′-UTRs. Nucleic Acids Res. 45, 10800–10810 (2017).
Bae, B. et al. Elimination of Calm1 long 3′-UTR mRNA isoform by CRISPR-Cas9 gene editing impairs dorsal root ganglion development and hippocampal neuron activation in mice. RNA 26, 1414–1430 (2020).
Zhang, J. et al. A PolH transcript with a short 3′UTR enhances PolH expression and mediates cisplatin resistance. Cancer Res. 79, 3714–3724 (2019).
Wu, Q. et al. In situ functional dissection of RNA cis-regulatory elements by multiplex CRISPR-Cas9 genome engineering. Nat. Commun. 8, 2109 (2017).
Araki, S. et al. Decoding transcriptome dynamics of genome-encoded polyadenylation and autoregulation with small-molecule modulators of alternative polyadenylation. Cell Chem. Biol. 25, 1470–1484 (2018).
Dutertre, M. et al. A recently evolved class of alternative 3′-terminal exons involved in cell cycle regulation by topoisomerase inhibitors. Nat. Commun. 5, 3395 (2014).
Ross, N. T. et al. CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing’s sarcoma. Nat. Chem. Biol. 16, 50–59 (2020).
Xiang, Y. et al. Comprehensive characterization of alternative polyadenylation in human cancer. J. Natl Cancer Inst. 110, 379–389 (2018).
Orlandini von Niessen, A. G. et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3′ UTRs identified by cellular library screening. Mol. Ther. 27, 824–836 (2019).
Ji, X., Wan, J., Vishnu, M., Xing, Y. & Liebhaber, S. A. αCP poly(C) binding proteins act as global regulators of alternative polyadenylation. Mol. Cell Biol. 33, 2560–2573 (2013).
Gruber, A. J. et al. A comprehensive analysis of 3′ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 26, 1145–1159 (2016).
Hilgers, V., Lemke, S. B. & Levine, M. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 26, 2259–2264 (2012).
Carrasco, J. et al. ELAV and FNE determine neuronal transcript signatures through EXon-activated rescue. Mol. Cell 80, 156–163 (2020).
Chen, X., Lloyd, S. M., Kweon, J., Gamalong, G. M. & Bao, X. Epidermal progenitors suppress GRHL3-mediated differentiation through intronic polyadenylation promoted by CPSF-HNRNPA3 collaboration. Nat. Commun. 12, 448 (2021).
Shen, T. et al. Alternative polyadenylation dependent function of splicing factor SRSF3 contributes to cellular senescence. Aging 11, 1356–1388 (2019).
Schwich, O. D. et al. SRSF3 and SRSF7 modulate 3′UTR length through suppression or activation of proximal polyadenylation sites and regulation of CFIm levels. Genome Biol. 22, 82 (2021).
Chatrikhi, R. et al. RNA binding protein CELF2 regulates signal-induced alternative polyadenylation by competing with enhancers of the polyadenylation machinery. Cell Rep. 28, 2795–2806 (2019).
Masuda, A. et al. Position-specific binding of FUS to nascent RNA regulates mRNA length. Genes Dev. 29, 1045–1057 (2015).
Masuda, A. et al. tRIP-seq reveals repression of premature polyadenylation by co-transcriptional FUS-U1 snRNP assembly. EMBO Rep. 21, e49890 (2020).
Moreira, A. et al. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 12, 2522–2534 (1998).
Danckwardt, S. et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 26, 2658–2669 (2007).
Berg, M. G. et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150, 53–64 (2012).
Bava, F. A. et al. CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature 495, 121–125 (2013).
Batra, R. et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell 56, 311–322 (2014).
Liu, Y. et al. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci. Rep. 3, 2054 (2013).
Licatalosi, D. D. et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 (2008).
Acknowledgements
The authors thank all members of the Mayr laboratory for helpful discussions and critical reading of the manuscript. This work was funded by the NIH Director′s Pioneer Award (DP1-GM123454), a grant from the G. Herold and Leila Y. Mathers Foundation, the Pershing Square Sohn Cancer Research Alliance and an NCI Cancer Center Support Grant (P30 CA008748).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Yongsheng Shi, Bin Tian and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mitschka, S., Mayr, C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat Rev Mol Cell Biol 23, 779–796 (2022). https://doi.org/10.1038/s41580-022-00507-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-022-00507-5
- Springer Nature Limited
This article is cited by
-
Structured 3′ UTRs destabilize mRNAs in plants
Genome Biology (2024)
-
Cleavage and polyadenylation machinery as a novel targetable vulnerability for human cancer
Cancer Gene Therapy (2024)
-
Co-transcriptional gene regulation in eukaryotes and prokaryotes
Nature Reviews Molecular Cell Biology (2024)
-
InPACT: a computational method for accurate characterization of intronic polyadenylation from RNA sequencing data
Nature Communications (2024)
-
Rational design of an artificial tethered enzyme for non-templated post-transcriptional mRNA polyadenylation by the second generation of the C3P3 system
Scientific Reports (2024)