Abstract
Complex natural and synthetic materials, such as subcellular organelles, device architectures in integrated circuits, and alloys with microstructural domains, require characterization methods that can investigate the morphology and physical properties of these materials in three dimensions (3D). Electron tomography has unparalleled (sub-)nm resolution in imaging 3D morphology of a material, critical for charting a relationship among synthesis, morphology, and performance. However, electron tomography has long suffered from an experimentally unavoidable missing wedge effect, which leads to undesirable and sometimes extensive distortion in the final reconstruction. Here we develop and demonstrate Unsupervised Sinogram Inpainting for Nanoparticle Electron Tomography (UsiNet) to correct missing wedges. UsiNet is the first sinogram inpainting method that can be realistically used for experimental electron tomography by circumventing the need for ground truth. We quantify its high performance using simulated electron tomography of nanoparticles (NPs). We then apply UsiNet to experimental tomographs, where >100 decahedral NPs and vastly different byproduct NPs are simultaneously reconstructed without missing wedge distortion. The reconstructed NPs are sorted based on their 3D shapes to understand the growth mechanism. Our work presents UsiNet as a potent tool to advance electron tomography, especially for heterogeneous samples and tomography datasets with large missing wedges, e.g. collected for beam sensitive materials or during temporally-resolved in-situ imaging.
Similar content being viewed by others
Introduction
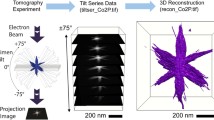
Electron tomography has attracted extensive attention in three-dimensional (3D) characterization of nanomaterials due to its unpaired nanometer and even atomic spatial resolution. For example, electron tomography has been used to obtain 3D maps of atomic coordinates in crystals to understand their defect and grain structures1,2,3,4,5, of polymer film morphologies to build the morphology‒function relationship in applications such as molecular separation and optoelectronic devices6,7, and of nanoparticle (NP) shapes to guide their synthesis and self-assembly8,9. In electron tomography, a sample is tilted over a range of angles, and the projected (scanning) transmission electron microscopy ((S)TEM) images which record the material thickness integrated over the electron beam paths at the pixel level are reconstructed to generate the 3D volumetric images. The 3D images not only capture the surface morphology of samples, but reveal internal voids, which are inaccessible by surface characterization methods of a similar resolution such as scanning probe microscopy and scanning electron microscopy7.
However, due to geometric constraints of electron microscopes and sample holders, electron tomography is fundamentally limited by the missing wedge effect. Using the simplest inverse Radon transform as an example, a typical 3D reconstruction in electron tomography can be decomposed into multiple elementary steps of 2D reconstructions. When the tilt series (projection images in x and y coordinates at different tilt angle α, Fig. 1a, b) are transposed to switch the tilt axis (y-axis in this example) with angular axis (α in Fig. 1b), sinusoidal wave-like patterns (in x and α coordinates at different locations on y-axis, Fig. 1c) are yielded, known as sinograms. In other words, tilt series and sinogram stacks are the same data but viewed from different axes. For each sinogram (in x and α coordinates) in a sinogram stack, a mathematical operation of inverse Radon transform is used to reconstruct an image in x and z coordinates (Fig. 1d). Finally, the reconstruction from each sinogram is restacked along the tilt axis (y-axis in this example) to give the final 3D reconstruction in x, y, z coordinates (Fig. 1e). Meanwhile, due to instrument limitations, the tilt range is usually limited to ‒60° to +60° (hereinafter referred to as ±60°)6,7 or ±70° (Fig. 1b, c)8,9. The lack of tilt images at higher angles leads to band-like missing patterns when transposed into sinograms (Fig. 1c). As a result, during the reconstruction, the lack of information at those missing angles causes feature distortion with wedge-like shapes (Fig. 1d,e)10, which is called as the missing wedge artifacts11.
a Schematic showing the tilt-and-project process during the tilt series acquisition. The beam (black arrows) never reaches within the missing angles annotated by the red lines due to the beam blocking from the holder. b The tilt series, which is a stack of projection images taken at different tilt angle α. The missing angles appear as missing projection images at certain tilt angles shown as red slices. c The sinogram stack, which contains the same data as (b) but is just simply transposed to switch the α- and y-axis. Due to the transpose, the missing projections in the tilt series appear as a band-shaped missing angle mask in the sinogram stack as denoted by the red dashed box. d Separating the sinogram stack gives individual sinograms. Each individual sinogram undergoes inverse Randon transform to yield the reconstruction slice. Due to the missing angle, wedge-shaped distortion appears in the reconstruction slices, as denoted by the red dashed boxes. e The reconstruction slices are restacked to give the final 3D reconstruction, which also shows the missing wedge artifact. f UsiNet inpaints each individual sinogram, leaving no missing wedge artifact in the reconstruction slices. g Restacking the UsiNet inpainted reconstructions yields no missing wedge artifact.
Various data analysis algorithms have been developed to correct the missing wedge effect. Different from the simple inverse Radon transform12, iterative reconstruction algorithms including Simultaneous Iterative Reconstruction Technique (SIRT)13, Model Based Iterative Reconstruction (MBIR)14, Discrete Algebraic Reconstruction Technique (DART)15, and Low-tilt Tomographic Reconstruction (LoTToR)16 have been shown to suppress the missing wedge artifacts in electron tomography. Among them, SIRT is versatile but cannot correct all missing wedge-related distortion as discussed in the comparison with our approach below, while other methods require assumptions such as homogeneous material density throughout the sample and are prone to complicated fine choices of input parameters. More recently, machine learning methods such as neural networks have emerged to contribute to high-performance information recovery of missing wedge artifacts in electron tomography. On one hand, neural networks have been shown to correct missing wedge distortions in existing 3D reconstructions produced by conventional reconstruction algorithms such as weighted back projection (WBP, a discretized version of inverse Radon transform) and MBIR11,17,18. On the other hand, neural networks can also generate the missing contents in the sinograms at the unmeasured tilt angles to fundamentally prevent the formation of the missing wedge artifact19. Such generation of missing information can be achieved by the algorithm known as image inpainting, where an incomplete or partially masked image serves as the input to retrieve the missing regions. This method has been applied to tasks such as corrupted photograph restoration20 and removal of obscure features in satellite images21 or self-driving22. However, in the existing machine learning-based missing wedge correction methods for electron tomography11,19, supervised training is consistently required. As a result, an ideal training dataset fully tilted over ±90°, without missing wedge artifacts, must be provided for model training. Such dataset is impractical to obtain experimentally in electron tomography. Thus, simulated images, X-ray computed tomography (CT) images, digital photos, and random polygon images have been used as the training dataset with ground truth for model training in electron tomography11,19. In these studies, the inevitable discrepancy between training dataset and experimental datasets limits the extent of missing wedge correction.
Here we present Unsupervised Sinogram Inpainting for Nanoparticle Electron Tomography (UsiNet), the first sinogram inpainting method that can be realistically used for experimental electron tomography by circumventing the need for ground truth (i.e., no training data at the full tilt range is needed). This method is inspired by the unsupervised image inpainting models used in photography restoration and medical CT sinogram inpainting tasks, including several generative adversarial network (GAN) models23,24,25 such as AmbientGAN26 and MisGAN27. Instead of using the GAN architecture, where a generator network and a discriminator network have to be trained jointly, only one simple U-Net is employed in our UsiNet training. U-Net, which is an image-to-image convolutional neural network (CNN), can directly predict the restored sinogram (Fig. 1c, f) from the experimental sinogram stack with missing angles, by minimizing the objective function (loss) between the restored images and the experimental images28. Here missing angles are defined as the angles at which features or projections of the sample are missed. Compared with these existing supervised machine learning methods, UsiNet can fundamentally avoid the difficulty in obtaining the missing wedge-free ground truth that shares similar feature with the sample of interest and the paradox of simulating the training dataset without knowing the sample morphology in advance. The unsupervised inpainting in UsiNet is achieved by collectively learning from the sinograms of every NP in the sample. Meanwhile, no spatial averaging is needed, making the method compatible with heterogeneous and polydisperse samples while still resolving the 3D shapes of every NP.
Specifically, in this study, we first benchmark UsiNet using the sinograms of simulated 2D polygons and 3D polyhedra, representing the shape diversity of NPs to facilitate direct comparison with ground truth. UsiNet is evaluated and compared with WBP and SIRT by a set of quantitative metrics including mean squared error (MSE), peak signal-to-noise ratio (PSNR), and structural similarity index measure (SSIM). UsiNet demonstrates consistently higher performance in the comparisons, even given small training datasets of sinograms containing only 20 NPs in the view and of narrow tilt ranges ( ± 45°), highlighting its robustness in practical settings. UsiNet is then applied to experimentally obtained electron tomography tilt series, containing 126 gold NPs with sizes ranging from 40 nm to 80 nm and various shapes including regular polyhedrons such as decahedra and octahedra, and other irregular shapes. Comparing with SIRT reconstruction without inpainting, we show that UsiNet generates more reasonable NP shapes without missing wedge-caused shape distortion and with much sharper boundaries. Morphology sorting is further used to elucidate a synthesis‒morphology relationship. Our work presents UsiNet as a potent tool to advance electron tomography with special advantages in accommodating sample heterogeneity and electron tomography datasets with large missing wedges such as those of beam sensitive materials and collected during temporally-resolved in-situ imaging for applications in battery, catalysis, self-assembly, and composite manufacturing.
Results
Overview of the UsiNet workflow
In Fig. 1, we show that the tilt image stack, which has a dimensionality of M × N × D (M and N are the height and width of each tilt image, and \(D\) is the number of tilt angles), can be regarded as N stacked sinograms with a size of M × D, which are generated by slicing the image stack along the tilt axis direction. Taking one sinogram as an example (Fig. 2a), the missing angles are present in the sinograms as a band-like mask (Fig. 2b). The most straightforward sinogram inpainting can be achieved by training a CNN model to learn from the ground truth (Fig. 2a) of the measured sinogram with missing angles (Fig. 2b). But in practice, the ground truth of experimental tomographic dataset is not known. Thus in UsiNet training, a randomly initialized U-Net is first asked to make a noisy prediction of inpainting from the original sinogram with missing angles (Fig. 2b, c). We refer to this prediction as the first inpainting result. Then, another same-sized band-like mask is applied to the first inpainting result at a random position, resulting in a masked first inpainting result (Fig. 2d). The same U-Net model is asked to fill the masked first inpainting result (Fig. 2d) by using the first inpainting result (Fig. 2c) as the ground truth. The MSE loss is calculated between the consequent second inpainting result (Fig. 2e) and the first inpainting result (Fig. 2c) and backpropagated to update the U-Net model. In Fig. 2, steps c–e are performed iteratively until the loss converges (Fig. 2f). This training workflow design bypasses the need for a ground truth sinogram and thus achieves the unsupervised sinogram inpainting solely relying on the experimental tilt series with missing angles. The architecture of the U-Net model used in this work is shown in Fig. 2g.
a A ground truth sinogram, which exists but cannot be obtained experimentally. b The measured sinogram with missing angles. c The first inpainting result produced by the U-Net model. d The inpainting result in (c) masked by a mask with the same size as the missing region in (b) but at a random location. e The second inpainting result produced by the same U-Net model using (d) as the input. Next, (c) serves the ground truth to optimize the inpainting (e) in each training iteration. f The final inpainting result of (b) after 100 training epochs. g Schematic of the U-Net model used in this work.
In this training procedure, UsiNet learns and predicts the masked regions from the experimental sinograms containing samples (NPs here or other nanoscale features to be reconstructed) that are similar but exhibiting random orientations. Namely, the recovery of the missing features in sinogram A relies on the similarity between sinogram A and sinogram B and the fact that the same feature masked in A is not masked in sinogram B. Such assumption is similar to the parameterized, stochastic measurement process used in other unsupervised image inpainting studies23,26, where the positions of the masked regions need to be random in each incomplete image to allow inpainting. In practice, we satisfy this assumption by shifting the missing angles, i.e., the masks, to random positions (Methods). To do so, the NPs collectively need to sample the full range of out-of-plane orientations. Otherwise, certain features will be always masked by missing angles and will not be learned by the model. The fully infilled sinograms (Fig. 2f) will be used as inputs for reconstruction with missing wedge artifacts corrected.
Note that the NPs of different out-of-plane orientations do not need to be identical. UsiNet works well for heterogeneous samples as shown below. NPs with extended facets tend to sit flat on the TEM grid to maximize van der Waals force between the NPs and the TEM grid (Supplementary Fig. 1). We develop a polymer coating technique which facilitates random out-of-plane orientation of NPs, proved effective even for triangular prisms of extended flat planes (Methods and Supplementary Figs. 1, 2).
Validation of UsiNet on simulated 2D polygonal shapes
As discussed in Fig. 1, the reconstruction of single sinograms serves as the elementary step of reconstructing 3D shapes. Thus we first validate the applicability of UsiNet on the inpainting of single sinograms, which can be generated from 2D polygons. Simulated sinograms of 2D triangles with known ground truth are used for validation. First, the sinograms of a full tilt range of ±90° at 3° interval are simulated for a total of 3,000 triangles with different sizes, corner truncations, orientations, and centroid positions (Fig. 3a, left). Next, masks covering a 60° range positioning at random tilt angles are applied to the sinograms (the angular axis is labeled in Fig. 3a) to represent the missing angles in tilt series (Fig. 3a, middle). The training process described above is then implemented. During one epoch, defined as one iteration starting from Fig. 2c to Fig. 2e, a randomly sampled subset (1,000 images here) of the training dataset is fed into the U-Net. We define epoch size as the number of sinogram images used in the subset, which is similar to the concept of batch size but with two differences. First, every image updates the neural network weights. When epoch size = 1000, the neural network is updated 1,000 times in an epoch. Second, the images are randomly sampled instead of evenly split. This random sampling of a fixed number of images (as the epoch size) is needed to fix the number of forward and backpropagation for regulating the convergence in each iteration so that the progress of convergence in each epoch is affected by the abundance instead of the total size of training dataset. The performance is then evaluated by the MSE between the first inpainting result and second inpainting result (Fig. 3b, blue line, the actual loss we optimize during training). Convergence of this training MSE is effectively minimizing the MSE between the first inpainting result and the ground truth sinogram (Fig. 3b, orange line, hereto after referred to as validation MSE), although the model does not use ground truth sinogram throughout the training. With the intensity values of the sinograms normalized in the range of 0–255, both the training and validation MSE losses converge to values less than one, in comparison with the baseline MSE (~570) representing the difference between the ground truth and the freshly masked sinograms (Fig. 3b, black line). The mean PSNR and SSIM between the ground truth (Fig. 3a, left) and final prediction (Fig. 3a, right) of all 3,000 simulated sinograms are measured to be 50.19 and 0.9993 respectively, indicating high fidelity of the inpainting results. As to the impact of epoch size, it turns out larger epoch size combined with lower learning rate leads to more stable convergence and lower final MSE, which is favorable (Supplementary Note 1 and Supplementary Fig. 3) at the cost of longer training time. For our experimental data, an epoch size of 2,000 is shown as sufficient to balance the training efficiency and desired training loss.
a From left to right: the ground truth sinogram at ±90° tilt range, the masked sinogram at ±60° tilt range, and the prediction sinogram at ±60° tilt range after inpainting. b The MSE loss evolution during the training. Training loss is calculated between the first and second inpainting results as indicated in Fig. 2. Validation loss is calculated between the first inpainting results and the ground truth. The baseline is the loss between the ±60° and ±90° sinograms. c From left to right, the 2D triangle image which gives the sinogram in (a); the ±60° sinogram reconstructed by WBP; the ±60° sinogram reconstructed by WBP after inpainting; the ±60° sinogram reconstructed by SIRT; the ±60° sinogram reconstructed by SIRT after inpainting. d The IoU and BF scores of the images reconstructed by WBP with and without inpainting. e The IoU and BF scores of the images reconstructed by SIRT with and without inpainting. Error bars represent the standard deviation of images in the training dataset.
Next, the inpainted sinograms are forwarded to reconstruction algorithms (WBP, SIRT) and compared with the reconstructions from the sinograms without inpainting. As shown in Fig. 3c, for the sinograms without inpainting, the edge of the ground truth triangle is reconstructed as a wedge shape by the WBP algorithm due to the missing angles. Although the SIRT algorithm shows a more effective suppression of the missing wedge artifact than WBP, it predicts a blurry edge. In contrast, when the UsiNet inpainted sinograms are used as inputs, the reconstructions generated by both WBP and SIRT restore the sharp edges in the ground truth triangle and shows no distortion, demonstrating superior correction of the missing wedge artifacts. Quantitative evaluations of the reconstruction results are listed in Fig. 3d, e, where the reconstructions of all 3,000 sinograms in the training dataset are binarized and compared with the ground truth using the intersection-over-union (IoU) and boundary F1 (BF) scores. For WBP reconstruction, our UsiNet improves the IoU from 0.8848 to 0.9738 and BF from 0.7233 to 0.9995, with >10% improvement in both metrics. For SIRT reconstructions, although missing wedge correction mechanism is already included, our UsiNet still improves the IoU from 0.9739 to 0.9927 and BF from 0.9897 to 0.9995.
Validation of UsiNet on simulated 3D polyhedral shapes
For electron tomography, inpainting and reconstruction are implemented on 3D tilt series (sinogram stacks) instead of 2D projections (single sinograms). Here we adopt the 2D inpainting method discussed above to every 2D sinogram slice in the 3D tilt series along the tilt axis. Although this approach could sacrifice some spatial correlation information in 3D, it is chosen over the direct inpainting of 3D tilt series using a 3D U-Net29 due to much smaller computation cost. For validation, we build 3D models of triangular nanoprisms with different edge lengths, thicknesses, corner truncations, orientations, and centroid positions, and project these models in a tilt angle range of ±90° with a 3° interval as the ground truth (Fig. 4a, b). Each ground truth tilt series is sliced along the tilt axis (y-axis in Fig. 4a) to generate the sinogram stack, and a randomly selected mask of a 60° range (red dashed box in Fig. 4c) is applied to the sinograms, which will be used as the training dataset. In a typical training dataset, tilt series of 300 particles are simulated and around 32,000 sinogram images are sliced. After the training process, the missing angles in the sinogram are filled by the U-Net model as shown in Fig. 4c. Slicing the sinogram stack along the angle axis (α-axis in Fig. 4a) reproduces each tilt image in the tilt series. The comparison between the ground truth image and the U-Net prediction is shown as Fig. 4b, d, demonstrating the capability of this method to ‘fabricate’ previously non-existing tilt image. Quantitatively, the training MSE and validation MSE converge to around 4 and 2 after 200 epochs, respectively (Supplementary Fig. 4a). These MSE values are slightly higher than those of 2D images (Fig. 3b), which can be attributed to the complication that in 3D inpainting, different intersectional shapes are generated from different slicing of the 3D object, increasing the diversity of sinogram patterns. The mean PSNR and SSIM are 44.00 and 0.9967 respectively, showing high similarity between the ground truth and predicted sinograms.
a The simulated 3D sinogram stack at a ± 90° tilt range, which is simply 2D sinograms stacked along the y-direction. Viewing the sinogram stack through slicing at certain tilt angle α gives individual tilt images. b An example of individual tilt image from the ground truth. c Sinogram inpainting results stacked along the y-direction. The region annotated by the red dashed box is removed from (a) and then predicted by the model. d The inpainting result from the tilt image view the same as (b). e The SSIM and PSNR evaluating the sinogram inpainting quality with different training dataset sizes and tilt ranges. Baselines are scores of the sinograms without inpainting. f The MSE and IoU evaluating the WBP reconstruction quality from sinograms under different training conditions indicated in the legends in (e). Baselines are scores of the reconstructions from sinograms without inpainting. g The SSIM and PSNR evaluating the sinogram inpainting quality under different amounts of Poisson noise. h The MSE and IoU evaluating the WBP reconstruction quality from sinograms under different training conditions indicated in the legends in (g). i The SSIM and PSNR evaluating the sinogram inpainting quality with different numbers of polyhedron shapes included in the training dataset. Single shape: triangular prism; two shapes: triangular prism and cube; three shapes: triangular prism, cube, and octahedron; six shapes: triangular prism, cube, octahedron, tetrahedron, tetrahexahedron, and random polyhedron. j The MSE and IoU evaluating the WBP reconstruction quality from sinograms under different training conditions indicated in the legends in (i).
The training of UsiNet relies on learning from many NPs sampling random out-of-plane orientations. To systematically evaluate the number of NPs needed for training, we decrease the total number of NPs included in the training dataset while keeping the epoch size (number of sinograms sampled in each epoch) the same. Thus the total number of backpropagations remains the same and the only difference is the size of the training dataset. The SSIM and PSNR of the models trained on 300, 100, 50, and 20 NPs are shown in Fig. 4e (see Supplementary Fig. 4 for their MSE evolution during epochs). We find that decreasing the training dataset size down to 20 can still produce inpainting sinograms with high similarity to the ground truth (the mean SSIM and PSNR above 0.9800 and 40.00, respectively). 3D reconstructions from the inpainted dataset based on the models trained by different numbers of NPs all consistently show improvements, with their MSE < 1.000 × 10−3 and IoU > 0.9500 (Fig. 4f). In comparison, the reconstructed 3D images from the missing angle sinograms have higher MSE and lower IoU of 3.263 × 10−3 and 0.8251 respectively. Meanwhile, we observe that there is a chance of non-convergence during training with 10 tilting series or unstable training process leading to divergence of loss since every round of training is random due to weight initialization, stochastic gradient descends, sample shuffling, and other factors. In experiments, combination of non-ideal conditions such as tilt series misalignment and complex shape composition can make more tilt series in the training dataset desirable for reliable training. As for the tilt angle range of the training dataset, other than the ±60° tilt angle range conventionally accessible in most electron tomography studies, the tilt angle range can be narrower in practice given the choice of sample holder, the relative position of the sample on the TEM grid, and the total dose that the sample can tolerate with. To test the extreme of the method, we simulate tilt series with a ± 45° tilt range at the same 3° tilt interval. As shown in Fig. 4e, the inpainting results are still improved a lot and are much closer to the ground truth compared with the tilt series with missing angles (see Supplementary Fig. 4 for the MSE evolution during epochs). Moreover, the ±45° model produces reconstructions with MSE and IoU close to those of the ±60° models (Fig. 4f). Model training with training dataset of even smaller number of NPs and narrower tilt range is difficult (Supplementary Notes2 and 3). During the practical application, the existence of noise could affect the performance of UsiNet inpainting. To evaluate the effect of imaging noises in experimental images on the performance of UsiNet, Poisson noises are introduced into the sinograms during model training. As shown in Fig. 4g and Supplementary Fig. 5, UsiNet is still able to generate good inpainting results with PSNR of 36.52 and SSIM of 0.9785 when the signal-to-noise ratio (SNR) decreases to 5 in the training dataset, which is similar to the experimental condition. Supplementary Fig. 5 clearly shows that the inpainting results are noise-free and do not exhibit visible artifact at the SNR values tested. All resulting reconstructions achieve an IoU around 0.95 while the high MSE should be explained by the introduction of noise itself (Fig. 4h). Besides that, training datasets containing shape mixtures of polyhedrons (e.g., octahedron, tetrahedron, cube, tetrahexahedron, irregular polyhedron) are simulated to represent the diverse nanoparticles one might analyze in experiments (Supplementary Fig. 6). Consistently, the models show good inpainting and reconstruction quality on these training datasets (Fig. 4i, j and Supplementary Fig. 6).
The 3D spatial correlation along the tilt axis direction can be important. Although the inpainting of each split sinogram is expected to be independent, in reality the existence of noise and misalignment of tilt images can make the 3D spatial correlation along the tilt axis direction valuable for correctly predicting the missing sinogram patterns. For example, in Supplementary Fig. 7, we show that in the presence of Poisson noise, the synthetic tilt image shows wavy boundary and line-like features because each slice is independently inpainted. While such imperfection could only cause minor wavy features on the reconstructed surface, as shown in Supplementary Fig. 7e, by simply inputting three consecutive neighboring slices instead of the middle one only, the 3D spatial correlation is restored and the inpainted tilt image is much smoother. Note that this modification of input channels otherwise only leads to neglectable improvement on the evaluation metrics of inpainting and reconstruction. Such modification only involves adding two more channels in the input layer of the U-Net model, which does not significantly increase the computational cost. The restoration of 3D spatial correlation is expected to be more complete with more input layer channels, i.e., the number of consecutive neighboring slices. Meanwhile, note that inpainting is independent from the following-up reconstruction algorithms. The inpainted sinograms could be stacked back together and used as tilt images instead of split sinograms, just like those used for conventional tomography reconstruction process. As a result, the 3D correlation along the tilt axis can also be considered by advanced reconstruction algorithms such as SIRT and MBIR.
Importantly, missing wedge effects tend to be more pronounced for the NPs with large flat facets oriented orthogonal to the beam direction, which can still be addressed by UsiNet. Using triangular prisms as an example, we simulate the sample tilt series in the range of ±60° with a 60° missing angle range at different out-of-plane particle orientation θ relative to the substrate (Fig. 5a). When the sinograms with missing angles are used as the inputs, both WBP and SIRT reconstructions show the θ-dependent MSE and IoU (Fig. 5b, c). The reconstruction quality (of low MSE and high IoU) drops dramatically with decreasing θ, i.e., the NP changing from standing to lying on the TEM grid. This θ-dependence can be visualized by the 3D grayscale rendering (Fig. 5d–h) as well as slicing of the reconstructions along the tilt axis (Fig. 5i–r). In the sliced view, when the orientation of a feature deviates from the beam direction more than the tilt range (90°–60° = 30°, which means for any line segments along the contour within ±30° angle to the horizontal axis), the missing wedge artifact shows up in the WBP reconstruction, leading to feature spreading along horizontal contours while leaving the vertical contours undisturbed. As shown in Fig. 5i, j and Fig. 5n, o, for a lying nanoprism (θ = 0°), it contains more horizontal contours than a standing one (θ = 90°), leading to more distorted reconstruction with spreading features. The SIRT reconstruction in general suppresses the spreading features appeared in the WBP reconstruction and produces smoother NP contours at small θ angles. However, it fails to extract the true shape of the particle. Expansion of the shape along z-axis can still be observed (highlighted in Fig. 5d, f, Fig. 5i, k and Fig. 5n, p). This distortion can lead to significant errors when measuring the prism thickness (Fig. 5i, k). On the other hand, reconstructions from the inpainting tilt series show higher quality using both WBP and SIRT reconstruction algorithms and at all possible out-of-plane particle orientations (θ = 0°‒90°, Fig. 5b, c) under both MSE and IoU metrics. Figure 5g, h, l, m, q, r show that the missing wedge artifacts are eliminated.
a Schematic showing the definition of particle orientation θ (white), which is different from the tilt angle α (black). θ measures the dihedral angle between the basal plane of the prism and the substrate. b MSE of triangular prism particles at different θ reconstructed by different algorithms. The inpainting reconstructions consistently have lower MSE at all θ from 0° to 90°. c IoU of triangular prism particles at different θ reconstructed by different algorithms (after binarization). The inpainting reconstructions consistently have higher IoU at all θ from 0° to 90°. d The 3D grayscale rendering of a triangular prism particle model with θ = 0° (lying on the substrate). e–h The 3D grayscale rendering of the particle in (d) after projection and reconstruction by different methods. The tilt range is ±60°. i A sliced view of the same triangular prism particle model in (d). j–m The same sliced views of the particle in (i) after projection and reconstruction by different methods. The tilt range is ±60°. n A sliced view of a triangular prism particle model with θ = 90° (standing on the substrate). o–r The same sliced views of the particle in (n) after projection and reconstruction by different methods. The tilt range is ±60°. White arrows in (k, m) and (p, r) indicate the discrepancy between particle shape measurements with and without inpainting.
The validation using simulated 3D NP dataset demonstrates the capability of UsiNet to robustly correct missing wedge effect. The method learns directly from tilt series of multiple particles with missing angles, which do not require any manual labeling30 or image simulation process31 to generate training labels. The inpainted sinograms not only visually resemble the ground truth tilt series, but also show high SSIM, PSNR, and give satisfying reconstruction results. Moreover, UsiNet is shown to work with a training dataset size as small as 20 NPs, which can be easily cropped from a few low-magnification tilt series (<3 tilt series in most cases) in experiments (see Methods for cropping details), and a tilt range as narrow as ±45°. It is also reasonable to anticipate UsiNet’s applicability to multi-component nanoparticle samples. For example, UsiNet can be applied to high-angle annular dark-field (HAADF) STEM32 or bright field TEM33 tomography data containing material components with different contrast. The input of UsiNet can also be STEM-energy-dispersive X-ray spectroscopy (EDX) tomography data34, which applies to multi-metallic nanoparticles and each elemental map can be independently reconstructed. While the former might challenge the inpainting accuracy of the current UsiNet to provide distinguishable material contrast in the final reconstruction, STEM-EDX tomography is known to have very low SNR and thus difficult to inpaint. The implementation of both directions could be achieved by larger training datasets, more kernels in the neural network, and the fine-tuning of the hyperparameters during model training.
Application of UsiNet to experimental heterogenous NPs
After the validation of UsiNet on simulated dataset, we apply UsiNet to experimentally collected electron tomography dataset of gold decahedral NPs. Decahedral NPs consist of large, flat facets which are susceptible to missing wedges as discussed in Fig. 5d–h. The as-synthesized sample is heterogeneous, containing decahedral NPs and byproducts of other polyhedral shapes and irregular impurities (Supplementary Fig. 1). Low magnification HAADF STEM tilt series of the samples are taken over a tilt range of ±60° at an angle increment of 3°. The resulting tilt series are then cropped to generate the tilt series of each individual NP (Methods and Supplementary Fig. 2). Tilt series of 126 individual NPs are obtained and serve as both the training dataset and the testing dataset for inpainting and reconstruction. In Fig. 6a, we show two representative NPs of a decahedron (NP 1, desirable product) and a triangular prism (NP 2, byproduct), both of which happen to lie flat (θ ≈ 0°) on the TEM grid. Reconstructions using WBP and SIRT generates either spreading or blurry contours (Fig. 6b–g). Those missing wedge artifacts are attributed to their missing side views in the experimental tilt series (highlighted by the red dashed boxes in Fig. 6a). Implementing UsiNet on the gold decahedron sample fills the sinograms at high tilt angles (Fig. 6b, e) and thus retrieves the full tilt range of ±90°, leading to smooth side views of the particles (Fig. 6a). Clearly, the trained model well inpaints both two shapes and is applicable to heterogeneous samples. In the sliced reconstructions (Fig. 6c, f) of NPs 1 and 2 in Fig. 6a, non-physical, wedge-like protrusion is clearly shown in the WBP and SIRT reconstructions from the tilt series without inpainting. In contrast, the reconstructions from inpainted tilt series shows solid edge even on the particle surfaces orthogonal to the beam direction, where most severe distortions tend to occur. The line profiles of the image pixel intensity (Fig. 6d, g) across the particle boundaries in the SIRT slices (as denoted by the white dashed boxes) show solid boundary in the inpainted reconstructions, which is revealed by sharp intensity transition as compared to the smooth, gradual intensity increase in the profiles of reconstructed particle without inpainting. Note the dip in the line profile of the inpainted reconstruction is caused by the non-linear damping of the signal intensity of the equipment instead of the sinogram inpainting, which is known as the cupping artifact35. Although this is beyond the scope of the current research, we would like to point it out that the cupping artifact could be solved by recalibrating the intensity values to be linear with material thickness or more complicated reconstruction algorithms, such as MBIR14,35.
a The experimental tilt series of a decahedral NP (top row, NP 1) and a triangular nanoparism (bottom row, NP 2). The experimental tilt range is ±60° and the tilt images in the red dashed box region are synthesized by the model. b The sinogram of NP 1 after inpainting. The red dashed box regions annotate the sinogram pattern generated by UsiNet at high angles. c The sliced views of NP 1 reconstructed by WBP and SIRT, without and with sinogram inpainting. d The line profile of intensity values along the vertical direction suggested by white dashed boxes in (c). e The sinogram of NP 2 after inpainting. The red dashed box regions annotate the sinogram pattern generated by UsiNet at high angles. f The sliced views of NP 2 reconstructed by WBP and SIRT, without and with sinogram inpainting (g). The line profile of intensity values along the vertical direction suggested by white dashed boxes in (e).
This sharp intensity transition in the inpainted reconstructions also facilitates segmentation of the NPs by pixel intensity thresholding, which is a foundational step for quantitative analysis of particle morphology. In 3D view (Supplementary Fig. 8), in the reconstructions without inpainting, the missing wedge artifact appears as diffuse intensity values on the particle surfaces and blurs the real particle boundary position, in contrast to either WBP or SIRT reconstructions with inpainting where the particle boundary is clear. We apply the reconstruction to all 126 inpainted sinograms and perform subsequent segmentation of individual NPs for quantitative analysis (Fig. 7 and Supplementary Figs. 9–11). For polyhedral products with well-defined shapes, decahedron accounts for the majority population (26%, Fig. 7a, b), which are reported to be enclosed by {111} facets and can be regarded as an assembly of five tetrahedral units sharing common faces36. It is followed by prism (16%) and icosahedron (13%), with the former characterized by two {111} basal planes and the latter as a packing of 20 tetrahedra. All three types of particles share a twinned structure, either grown from multi-twinned seeds or seeds with stacking faults, resulting from a slow initial reduction rate37. Literatures have reported that icosahedral NPs are stable at smaller particle sizes compared to decahedral NPs, which matches with our observation that the mean volume of icosahedra is smaller than that of decahedra (Fig. 7c)38,39. Aside from twinned NPs, we also observe octahedra and truncated tetrahedra accounting for 11% and 2%, respectively, of the polyhedral NP product. These two shapes are both single-crystalline, enclosed by {111} facets. The dominance of {111} facets in the product suggests their selective stabilization during Au growth in the presence of poly(vinyl pyrrolidone) (PVP), consistent with previous density functional theory calculations that PVP-covered Au(111) surface is thermodynamically more stable relative to Au(100)40.
a 3D rendering of all 126 NPs reconstructed and segmented from the decahedral NP sample and colored according to their shapes. Blue: decahedra; Green: triangular prisms; Purple: icosahedra; Pink: octahedra; Orange: truncated tetrahedra; Red: trigonal bipyramids; Gray: impurities with irregular polyhedral shapes. Two particles denoted by the arrows correspond to the denoted datapoints in (f). b The fractions of the species in (a) among all particles. c The volume distribution of the species in (a). The white circles represent average volumes. d The scree plot of the descriptor PCA. e A plot showing how the descriptors are projected into the PC 1–PC 2 space. L1, L2, and L3 stand for the major axis lengths of a 3D shape. f Descriptors from each NP in the reconstruction projected into the PC 1–PC 2 space. Two datapoints denoted by the arrows correspond to the denoted particles in (a).
To quantify the shape heterogeneity of the NP sample, five dimensionless scalar descriptors—including three major axis length ratios, solidity, and sphericity—are extracted from each segmented NP model41,42 and dimension-reduced via principal component analysis (PCA) to show the shape distribution of the NPs (Fig. 7d–f). Suggested by the scree plot (Fig. 7d), the first two principal components (PCs) containing 99.5% of total variance are plotted for data visualization. As shown in the datapoint distribution in the PC 1–PC 2 space (Fig. 7e, f), the long projection vector lengths of the major axis ratios (L1/L2, L1/L3, and L2/L3) indicate those descriptors contribute more to the sample heterogeneity. Meanwhile, the shortest projection vector length of solidity suggests a small variance of the particle solidity across the sample, which is reasonable considering the highly convex shape of all product NPs, giving solidity values close to 1. The datapoints representing decahedral NPs locate in the 4th quadrant, consistent with the direction of the L2/L3 projection. The flattened and rounded shape of the decahedral NPs leads to similar L1 and L2 (longest and second longest major axis length) and very short L3. In contrast, the elongated shape of trigonal bipyramid causes long L1 but short L2 and L3, which makes their datapoint distribution highly correlate with the direction of L1/L2. The separation of datapoints from these two types of NPs illustrates their distinct shapes.
The shape distinctions in the PC space learnt from known particles can help the identification of unknown irregular impurities. For example, two gray datapoints with arrow annotations located at the top right corner of Fig. 7f turn out to be two plate-like particles with high L1/L2, L1/L3, and L2/L3. Instead, the gray datapoint population spanning in the second quadrant should be particles of shapes between octahedron (pink) and trigonal bipyramid (red) with elongation on one major axis direction, suggested by their high L1/L2 but low L1/L3 and L2/L3. These impurities could be intermediate structures during the growth of decahedron, prism, or octahedron43. Such sorting of the desired shapes and identification of byproducts will contribute to the understanding of the synthesis yield, which also potentially benefits the development of effective purification methods.
Discussion
We purpose UsiNet, an unsupervised sinogram inpainting method to correct the missing wedge effect in electron tomography. The unsupervised training in UsiNet does not require ground truth, manual annotation, or tilt image simulation, and thus is practically applicable to real electron tomography datasets where full angle tilt series are not obtainable. We demonstrate that UsiNet works with a small number of training dataset (down to 20 NPs) and narrow tilt range (±45°), which can be immediately useful for beam sensitive polymeric and biological materials where the tilt range can be limited by accumulated beam damage. The tolerance with a narrow tilt range could be critical for studies involving in-situ electron tomography—for example, on the evolution of the 3D shapes of NPs during chemical reactions such as electrochemical cycling, catalysis, and corrosion—where only scarce tilt series can be collected to ensure temporal resolution. Moreover, UsiNet does not require sample averaging and can thus apply to a broad range of heterogeneous NP systems such as electrode NPs used in rechargeable ion batteries, catalytical NPs, and nanoplastics. The missing wedge effect is otherwise particularly problematic for heterogeneous systems by generating anisotropic distortion. Although our demonstration focuses on colloidal NPs, the principle of unsupervised inpainting is expected to work for other samples containing 3D nanoscale morphology details, such as microstructural domains in alloys and crumples in polyamide separation membranes. UsiNet brings the full potential of electron tomography in charting the relationships of morphology with synthesis and performance of materials. A wide scope of applications can be enabled by UsiNet, such as uncovering degradation mechanisms of battery or catalytical nanomaterials, understanding morphologies and aggregation behaviors of naturally formed nanoplastics, and optimizing synthetic protocols of NPs with varying compositions.
Methods
Chemicals
Gold(III) chloride trihydrate (≥49.0%, HAuCl4·3H2O, Sigma-Aldrich), poly(vinyl pyrrolidone) (PVP, average Mw~55,000, Sigma-Aldrich), diethylene glycol (DEG, 99%, Sigma-Aldrich), polystyrene-block-poly(acrylic acid) (PS-b-PAA) (PS154-b-PAA49, Mn = 16,000 for the PS block and Mn = 3,500 for the PAA block, Mw/Mn = 1.15, Polymer Source Inc.), polystyrene-block-poly(4-aminomethyl styrene) (PS-b-P4AMS) (PS96-b-P4AMS34, Mn = 10,000 for the PS block and Mn = 4500 for the P4AMS block, Mw/Mn = 1.2, Polymer Source Inc.), 2-naphthalenethiol (2-NAT, 99%, Sigma-Aldrich), N,N-dimethylformamide (DMF, anhydrous, 99.8%, Sigma-Aldrich), tetrahydrofuran (THF, AR, Macron), ethanol (200 proof, Decon Laboratories, Inc.) were purchased and used without further purification. Water used in this work was nanopure water (18.2 MΩ·cm at 25 °C) purified by a Milli-Q Advantage A10 system.
Synthesis of gold decahedral NPs
The gold decahedra were synthesized following a method previously reported, with slight modification44. Typically, 7.0 g of PVP was added to 25 mL of DEG hosted in a 100 mL flask, and the mixture was heated at 180 °C in an oil bath till PVP completely dissolved. Afterwards, 2.0 mL of DEG solution containing 20 mg of HAuCl4 was injected in one shot with a pipette. The reaction continued for 10 min and was then quenched by immersing the flask in an ice−water bath. The solution was split into two portions, with each portion (around 13.5 mL) mixed with 16.5 mL of ethanol. The solid product was collected by centrifugation at 11,000 rpm for 45 min, after which most of supernatant was removed and the remaining 400 μL of sediments was redispersed in 2 mL of water and 18 mL of ethanol. After the second round of centrifugation at 14,000 rpm for 20 min, 19.6 mL of supernatant was removed, and the final product was redispersed in 19.6 mL of water. The resulting solution was measured to give 11.058 OD at its maximum UV-Vis absorption peak at 561 nm.
Before the polymer coating, the solution prepared above was centrifuged to reduce the OD to 5 at its maximum UV-Vis absorption peak at 561 nm. 113.0 µL of solution was transferred into a 1.5 mL centrifuge tube and diluted with water to reach a final volume of 1.5 mL. The diluted solution was then centrifuged at 5800 rpm for 20 min. After centrifugation, 1.48 mL of the supernatant was removed from the tube. The sediments were then diluted with water to reach a final volume of 250 µL (stock solution I).
Polymer coating of gold decahedral NP
To learn the missing wedge information from different particles, different regions on the particles have to be covered in the missing angles. This is hard to realize when the NPs frequently show one preferred orientation on the TEM grid, such as those with a shape of triangular prism or decahedron. To randomize their out-of-plane orientations, we coated polymer layers on the NP surface. Two block copolymers including PS-b-PAA and PS-b-P4AMS were tested. After the polymer coating, we found that the NPs coated with PS-b-P4AMS showed more random orientation on the TEM grid while those coated with PS-b-PAA did not. Thus, the PS-b-P4AMS-coated NPs were finally used for the tomography.
The PS-b-PAA coating on decahedral NPs followed a literature method45. Specifically, a 2-NAT solution (40 µL, 2 mg·mL−1 in DMF) was mixed with 780 µL of DMF in an 8 mL glass vial. 100 µL of stock solution I and 100 µL of water was then sequentially added into the vial dropwise using pipette with vortexing, followed by addition of PS-b-PAA solution (80 µL, 8 mg·mL−1 in DMF) in one shot without vortexing. The vial was capped tightly with a Teflon-lined cap and sonicated for 10 s, parafilm-sealed, heated at 110 °C in an oil bath, and left undisturbed for 2 h. The reaction mixture was then cooled down to the room temperature in the oil bath, which typically took 90 min. The solution was transferred to a 1.5 mL microcentrifuge tube and centrifuged three times (5000 rpm, 25 min; 4,500 rpm, 15 min; and 4500 rpm, 15 min) to remove the unreacted 2-NAT and PS-b-PAA from the NPs. After each centrifugation, 1.45 mL of the supernatant was removed and the remaining 50 µL of sediment was re-dispersed with 1.45 mL of water. After the third round of centrifugation, the 50 µL of sediment containing PS-b-PAA-coated NP was diluted with 50 µL of water. 4 µL of the diluted NPs was drop-casted onto an air plasma-treated TEM grid and left dry in the air.
The PS-b-P4AMS coating process on decahedral NPs was similar to that of PS-b-PAA. A 2-NAT solution (40 µL, 2 mg·mL−1 in DMF) was mixed with 635 µL of DMF in an 8 mL glass vial. 100 µL of stock solution I and 100 µL water was then sequentially added into the vial dropwise using pipette with vortexing, followed by adding PS-b-P4AMS solution (240 µL, 2 mg·mL−1 in a mixture solvent with 18% water, 36% DMF, and 46% THF in volume fraction) in one shot without vortexing. The vial was capped tightly with a Teflon-lined cap and sonicated for 10 s, parafilm-sealed, heated at 110 °C in an oil bath, and left undisturbed for 2 h. The reaction mixture was cooled down to room temperature in the oil bath, which typically took 90 min. The solution was transferred to a 1.5 mL microcentrifuge tube and centrifuged three times (5000 rpm, 25 min; 4500 rpm, 15 min; and 4500 rpm, 15 min) to remove the unreacted 2-NAT and PS-b-P4AMS from the NPs. After each centrifugation, 1.45 mL of the supernatant was removed and the 50 µL of sediment was re-dispersed with 1.45 mL of water. After the third round of centrifugation, the 50 µL of sediment containing PS-b-P4AMS-coated NP was diluted with 50 µL of water. 4 µL of the diluted NPs was drop-casted onto an air plasma-treated TEM grid and left dry in the air.
Tilt series acquisition
A ThermoFisher Scientific Talos F200X G2 (S)TEM at 200 kV was used for taking the tilt series images of polymer-coated gold decahedral NPs at STEM-HAADF mode. The probe spot size was set to 8 and the dwell time was 1 µs, which led to a frame time of 23.56 s and kept the polymer coating on NPs undamaged. A total of 41 tilt images were acquired over a tilt range of ‒60° to +60° with an angle increment of 3°. During the acquisition, the stage was aligned and set to eucentric height automatically by the software Tomography STEM provided by ThermoFisher. In total, 5 tilt series were taken at low magnification (1720 nm field of view) and large image size (4096-by-4096 pixel).
2D image simulation
A customized MATLAB code was used to generate 2D images of triangles at 256-by-256-pixel sizes as well as their corresponding sinograms. The triangles were defined by the convex hull of three circles positioned in an equilateral arrangement. The orientations, edge lengths, vertex circle radii, and triangle pixel intensities were given by random variables following uniform distributions in reasonable ranges. Meanwhile small perturbations were imposed to the triangle centroid and vertice positions following Gaussian distributions in reasonable ranges. The built-in function radon.m was used to generate the rotational projection of each triangle image at the angle range from –90° to +90° at a 3° interval, which resulted in the sinogram containing 61 line-projection slices at a size of 61-by-256 pixel. For the purpose of unsupervised model training, the continuous 20 out of 61 line projections starting from random locations were replaced by zero in each sinogram. This corresponded to a tilt range of ±60° as used in the experimental acquisition.
3D image simulation
Similar to 2D image simulation, a customized MATLAB code was used to generate volumetric images of triangular nanoprisms as well as other polyhedron shapes at 256-by-256-by-256-pixel sizes defined by the convex hull of several spheres positioned at the vertices of the polyhedrons. The orientations, sizes, and tip sphere radii were given by random variables following uniform distributions in reasonable ranges. The MATLAB version of ASTRA Toolbox46,47 was used to generate the tilt series of each triangular prism at the angle ranging from –90° to +90° with a 3° interval, which resulted in the tilt series containing images at 61 different angles at a size of 61-by-256-by-256. For the unsupervised model training, a 3D mask with size of 20-by-256-by-256 pixel was first applied to random locations to replace the corresponding pixels by zero, which resembled the ±60° tilt range as used in the experimental acquisition. Each tilt series were then split along the tilt axis direction to give 256 individual sinograms with sizes of 61-by-256 pixel ready for serving as the training data.
Tilt series alignment
Although the images were aligned during the tilt series acquisition by mechanically shifting the stage, the resulting alignment still needed to be refined. The low magnification images were aligned using the patch tracking module in the open-source software IMOD 4.9.3 (University of Colorado, http://bio3d.colorado.edu/)48. During the patch tracking, 16 patches with a 512-by-512-pixel size were tracked and pixels within a 205-pixel margin near the boundary were trimmed. The aligned low magnification tilt series were then cropped to give the tilt series of individual NPs.
Cropping individual tilt series
A customized MATLAB code was used to crop the individual NP images in the tilt series at all angles in three steps. (1) Centroid translation. NPs at zero-tilt angle were first detected by a simple thresholding (Supplementary Fig. 2a). The particles within a 1024-pixel margin near the lateral image boundaries were eliminated because they would be out of focus at high tilt angle and resulted in poor model training and reconstruction quality (Supplementary Fig. 2a). For every NP, the tilt images at zero-tilt angle were translated to shift the particle centroid at zero-tilt angle to the center of the images, where Lx and Ly were used to annotate the translation on x-axis and y-axis (tilt axis), respectively (Supplementary Fig. 2b). Here the Ly was also directly applied to tilt images at all tilt angle α and Lx was further corrected in step 2. (2) Tilt axis translation on x-axis. The lateral translation of each NP at nonzero-tilt angles Lx(α) was determined through geometric modeling. As shown in Supplementary Fig. 2c, the translation Lx(α) was given by Lx(0)cos(α), where Lx(0) is the zero-tilt Lx as in step 1. This translation moved the tilt axis so it can pass individual NP centroids in the projection. After this step, all individual particle images should be correctly aligned and ready for reconstruction. (3) Tilt axis translation on z-axis. Different height (z-coordinate) of NPs causes lateral translation in the tilt series, which still gives correct reconstructions but increases the diversity of the sinograms and makes the training more difficult. In this step, the tilt series were further aligned according to their z-coordinates. Here all individual tilt series were first reconstructed by a SIRT algorithm (see Reconstruction algorithms) and filtered by a Gaussian filter with excessive sigma, which discarded all shape details and only kept a rough 3D centroid coordinate of the particle. Then the z-coordinate (h in Supplementary Fig. 2d) of each NP was extracted through binarization of the reconstruction. The final lateral translation also followed the geometric modeling as shown in Supplementary Fig. 2d and was given by Lx(0)cos(α) − hsin(α), where h stands for the distance from the particle centroid to the substrate. For each NP, the surrounding area with a size of 320-by-320 pixel was finally cropped and resized to 256-by-256 pixel to match the size of the neural network. After cropping, thresholding and masking were used to remove other particles in the field of view at all tilt angles. However, if particle overlapping was found through manual inspection at any α, this tilt series would be discarded.
Angle shifting of experimental tilt series
One requirement for the unsupervised inpainting method to work is that the mask region (or the missing angles) has to be random instead of fixed. Benefited from the random orientations of NPs on the substrate, the tilt series can be shifted to move the missing region to random angles. As shown in Supplementary Fig. 2e, the real full tilt range is actually ±180° instead of ±90°. But because of the nature of the orthogonal projection, any tilt images with a 180° angular difference are just a mirrored image of each other. As a result, ±90° tilt range is enough to reconstruct all the 3D information. Thus, we first combine the missing angle regions separated at +60°− +90° and –90°− –60° into one region with regular size. Specifically, the sampled tilt series at ±60° (green region in Supplementary Fig. 2e) were first mirrored to give the tilt series at +120°− +180° and –180°− –120° (blue region in Supplementary Fig. 2e). Then a continuous subset spanning 180° range was sampled from the angle range annotated by the red arrows in Supplementary Fig. 2e to create the sinograms with missing angles located in the middle instead of two ends. Similar to the simulated tilt series, the experimental tilt series after missing angle shifting were also split along the tilt axis direction to give 256 individual sinograms with sizes of 61-by-256 pixel, which later served as the model training or predicting inputs. For making the experimental tilt series training dataset, a mini augmentation was done by repeating the random angle shifting mentioned above for three times on each tilt series to increase the diversity of the training data.
Unsupervised training workflow
The training procedure followed the flow chart in Fig. 2. A 2D U-Net with an architecture given by Fig. 2g, an Adam optimizer, and MSE loss were used for the unsupervised image inpainting, and the sinograms either produced from image simulation or cropped from experimental tilt series were used as the training datasets. Using the experimental dataset as an example, splitting all 126 tilt series along the tilt axis (y-axis) after the augmentation gives 126 × 256 × 3 = 96,768 sinograms (three for the mini augmentation mentioned in the last paragraph). However, because at the high- and low-ends of the y-axis, a lot of sinograms turned to be purely black, we removed 99% of the black sinograms to speed up the training, which finally resulted in 44495 sinograms in the training dataset. In each training epoch, a certain number of sinograms (epoch size) were first sampled from the training dataset and predicted by the U-Net model. Each prediction was then randomly masked at an angle range corresponding to 60° (or a belt-like mask with 20-pixel height). Next, the 1st round predictions and their masking results were used as the ground truth and input for U-Net training, respectively, for one epoch of training. The updated U-Net was used in the next epoch and such process was iterated until the loss converged. The epoch size used in experimental data training is 2000. For the effect of epoch size (sinograms sampled in each epoch), learning rate of the Adam optimizer, and number of epochs, please refer to Supplementary Note 1. The training was done on Google Colab platform using the TensorFlow library.
Reconstruction algorithms
The U-Net predictions on sinogram stacks (tilt series) were directly reconstructed by the WBP and SIRT algorithms. For the WBP reconstructions, the MATLAB built-in function iradon.m was used to perform the inverse radon transform on every slice in the sinogram stacks and the results were stacked together to give the reconstructions. For the SIRT reconstructions, the SIRT3D_CUDA algorithm in the MATLAB version of ASTRA Toolbox was directly applied to reconstruct the 3D sinogram stacks. This algorithm used GPU acceleration. The SIRT iteration number was set to be 500.
Binarization and analysis
For the binarization of the reconstructions of simulated data, to make fair comparisons between different algorithms, the binarization threshold of each reconstruction was searched by comparing with its corresponding ground truth to give the highest IoU. For the binarization of experimental data, all reconstructions were first processed by a 3D median filter with a 5-by-5-by-5-pixel size and a 3D Gaussian filter with 2-pixel sigma. The filtered 3D images were then binarized with the Otsu thresholding algorithm49 to give the binary NP models. The volume, major axis length, solidity, and sphericity were all measured by the MATLAB built-in function regionprops3.m. The PCA was done by the MATLAB built-in function pca.m with an input matrix containing 5 descriptors as columns and each NP as rows.
Data availability
The simulated and experimental datasets can be found at https://doi.org/10.13012/B2IDB-7963044_V1.
Code availability
The codes used for UsiNet model training can be found at https://github.com/chenlabUIUC/UsiNet. Additional codes for data preprocessing and analysis are available from the corresponding author upon request.
References
Scott, M. C. et al. Electron tomography at 2.4-ångström resolution. Nature 483, 444–447 (2012).
Chen, C.-C. et al. Three-dimensional imaging of dislocations in a nanoparticle at atomic resolution. Nature 496, 74–77 (2013).
Winckelmans, N. et al. Multimode electron tomography as a tool to characterize the internal structure and morphology of gold nanoparticles. J. Phys. Chem. C. 122, 13522–13528 (2018).
Wang, C. et al. Three-dimensional atomic structure of grain boundaries resolved by atomic-resolution electron tomography. Matter 3, 1999–2011 (2020).
Tian, X. et al. Correlating the three-dimensional atomic defects and electronic properties of two-dimensional transition metal dichalcogenides. Nat. Mater. 19, 867–873 (2020).
Song, X. et al. Unraveling the morphology–function relationships of polyamide membranes using quantitative electron tomography. ACS Appl. Mater. Interfaces 11, 8517–8526 (2019).
An, H. et al. Mechanism and performance relevance of nanomorphogenesis in polyamide films revealed by quantitative 3D imaging and machine learning. Sci. Adv. 8, eabk1888 (2022).
Choueiri, R. M. et al. Surface patterning of nanoparticles with polymer patches. Nature 538, 79–83 (2016).
Chen, G. et al. Regioselective surface encoding of nanoparticles for programmable self-assembly. Nat. Mater. 18, 169–174 (2019).
Wolf, D., Lubk, A. & Lichte, H. Weighted simultaneous iterative reconstruction technique for single-axis tomography. Ultramicroscopy 136, 15–25 (2014).
Wang, C., Ding, G., Liu, Y. & Xin, H. L. 0.7 Å resolution electron tomography enabled by deep-learning-aided information recovery. Adv. Intell. Syst. 2, 2000152 (2020).
Wang, G., Ye, J. C. & De Man, B. Deep learning for tomographic image reconstruction. Nat. Mach. Intell. 2, 737–748 (2020).
Gilbert, P. Iterative methods for the three-dimensional reconstruction of an object from projections. J. Theor. Biol. 36, 105–117 (1972).
Venkatakrishnan, S. V. et al. Model-based iterative reconstruction for bright-field electron tomography. IEEE Trans. Comput. Imaging 1, 1–15 (2015).
Batenburg, K. J. & Sijbers, J. DART: A practical reconstruction algorithm for discrete tomography. IEEE Trans. Image Process. 20, 2542–2553 (2011).
Zhai, X. et al. LoTToR: An algorithm for missing-medge correction of the low-tilt tomographic 3D reconstruction of a single-molecule structure. Sci. Rep. 10, 10489 (2020).
Han, Y. et al. Deep learning STEM-EDX tomography of nanocrystals. Nat. Mach. Intell. 3, 267–274 (2021).
Liu, Y.-T. et al. Isotropic reconstruction for electron tomography with deep learning. Nat. Commun. 13, 6482 (2022).
Ding, G., Liu, Y., Zhang, R. & Xin, H. L. A joint deep learning model to recover information and reduce artifacts in missing-wedge sinograms for electron tomography and beyond. Sci. Rep. 9, 12803 (2019).
Bertalmio, M., Sapiro, G., Caselles, V. & Ballester, C. Image inpainting. In Proc. 27th Annual Conference Computer Graphics and Interactive Techniques. 417–424 (ACM, 2000).
Sarpate, G. K. & Guru, S. K. Image inpainting on satellite image using texture synthesis & region filling algorithm. In 2014 International Conference on Advances in Communication and Computing Technologies. 1–5 (IEEE, 2014).
Liao, M., et al. DVI: Depth Guided Video Inpainting for Autonomous Driving. https://www.ecva.net/papers/eccv_2020/papers_ECCV/papers/123660001.pdf (2020).
Pajot, A., de Bezenac, E. & Gallinari, P. Unsupervised adversarial image inpainting. https://www.semanticscholar.org/paper/Unsupervised-Adversarial-Image-Inpainting-Pajot-B%C3%A9zenac/7125656ab99381ca98cb1c8d93a1330506a8ccce (2019).
Pajot, A., de Bezenac, E. & Gallinari, P. Unsupervised Adversarial Image Reconstruction. https://openreview.net/forum?id=BJg4Z3RqF7 (2019).
Zhao, J., Chen, Z., Zhang, L. & Jin, X. Unsupervised learnable sinogram inpainting network (SIN) for limited angle CT reconstruction. https://www.semanticscholar.org/paper/Unsupervised-Learnable-Sinogram-Inpainting-Network-Zhao-Chen/68570ee3fd882b5b33514051d83d8f79e2c7296f (2018).
Bora, A., Price, E. & Dimakis, A. AmbientGAN: Generative Models From Lossy Measurements. https://www.cs.utexas.edu/~ecprice/papers/ambientgan.pdf (2018).
Li, S. C.-X., Jiang, B. & Marlin, B. MisGAN: learning from incomplete data with generative adversarial networks. https://openreview.net/forum?id=S1lDV3RcKm (2019).
Yan, Z., Li, X., Li, M., Zuo, W. & Shan, S. Shift-Net: Image inpainting via deep feature rearrangement. In Computer Vision – ECCV 2018. Lecture Notes in Computer Science. 11218, 3–19 (Springer Cham, 2018).
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T. & Ronneberger, O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016: 19th International Conference, Athens, Greece, October 17–21, 2016, Proceedings, Part II. 424–432 (ACM, 2016).
Moebel, E. et al. Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms. Nat. Methods 18, 1386–1394 (2021).
Skorikov, A., Heyvaert, W., Albecht, W., Pelt, D. M. & Bals, S. Deep learning-based denoising for improved dose efficiency in EDX tomography of nanoparticles. Nanoscale 13, 12242–12249 (2021).
Rajabalinia, N. et al. Coupling HAADF-STEM tomography and image reconstruction for the precise characterization of particle morphology of composite polymer latexes. Macromolecules 52, 5298–5306 (2019).
Galati, E. et al. Shape-specific patterning of polymer-functionalized nanoparticles. ACS Nano 11, 4995–5002 (2017).
Slater, T. J. A. et al. STEM-EDX tomography of bimetallic nanoparticles: a methodological investigation. Ultramicroscopy 162, 61–73 (2016).
Van den Broek, W. et al. Correction of non-linear thickness effects in HAADF STEM electron tomography. Ultramicroscopy 116, 8–12 (2012).
Zhou, S., Zhao, M., Yang, T.-H. & Xia, Y. Decahedral nanocrystals of noble metals: synthesis, characterization, and applications. Mater. Today 22, 108–131 (2019).
Wang, Y., Peng, H.-C., Liu, J., Huang, C. Z. & Xia, Y. Use of reduction rate as a quantitative knob for controlling the twin structure and shape of palladium nanocrystals. Nano Lett. 15, 1445–1450 (2015).
Barnard, A. S., Young, N. P., Kirkland, A. I., van Huis, M. A. & Xu, H. Nanogold: A quantitative phase map. ACS Nano 3, 1431–1436 (2009).
Xia, Y., Xiong, Y., Lim, B. & Skrabalak, S. E. Shape-controlled synthesis of metal nanocrystals: simple chemistry meets complex physics? Angew. Chem. Int. Ed. 48, 60–103 (2009).
Liu, S.-H., Saidi, W. A., Zhou, Y. & Fichthorn, K. A. Synthesis of {111}-faceted Au nanocrystals mediated by polyvinylpyrrolidone: insights from density-functional theory and molecular dynamics. J. Phys. Chem. C. 119, 11982–11990 (2015).
Lee, B. et al. Statistical characterization of the morphologies of nanoparticles through machine learning based electron microscopy image analysis. ACS Nano 14, 17125–17133 (2020).
Wang, X. et al. AutoDetect-mNP: an unsupervised machine learning Algorithm for automated analysis of transmission electron microscope images of metal nanoparticles. JACS Au 1, 316–327 (2021).
Langille, M. R., Zhang, J., Personick, M. L., Li, S. & Mirkin, C. A. Stepwise evolution of spherical seeds into 20-fold twinned icosahedra. Science 337, 954–957 (2012).
Seo, D. et al. Shape adjustment between multiply twinned and single-crystalline polyhedral gold nanocrystals: decahedra, icosahedra, and truncated tetrahedra. J. Phys. Chem. C. 112, 2469–2475 (2008).
Kim, A. et al. Tip-patched nanoprisms from formation of ligand islands. J. Am. Chem. Soc. 141, 11796–11800 (2019).
van Aarle, W. et al. The ASTRA Toolbox: A platform for advanced algorithm development in electron tomography. Ultramicroscopy 157, 35–47 (2015).
Palenstijn, W. J., Batenburg, K. J. & Sijbers, J. Performance improvements for iterative electron tomography reconstruction using graphics processing units (GPUs). J. Struct. Biol. 176, 250–253 (2011).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979).
Acknowledgements
This study was funded by the Office of Navy Research (MURI - N00014-20-1-2419).
Author information
Authors and Affiliations
Contributions
L.Y. and Q.C. conceptualized the work. L.Y., Z.L., J.L., and Q.C. performed the experiments. L.Y. and Q.C. performed the machine learning and data analysis. All authors contributed to the writing of the paper. Q.C. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yao, L., Lyu, Z., Li, J. et al. No ground truth needed: unsupervised sinogram inpainting for nanoparticle electron tomography (UsiNet) to correct missing wedges. npj Comput Mater 10, 28 (2024). https://doi.org/10.1038/s41524-024-01204-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-024-01204-x
- Springer Nature Limited