Abstract
Accelerated materials development with machine learning (ML) assisted screening and high throughput experimentation for new photovoltaic materials holds the key to addressing our grand energy challenges. Data-driven ML is envisaged as a decisive enabler for new perovskite materials discovery. However, its full potential can be severely curtailed by poorly represented molecular descriptors (or fingerprints). Optimal descriptors are essential for establishing effective mathematical representations of quantitative structure-property relationships. Here we reveal that our persistent functions (PFs) based learning models offer significant accuracy advantages over traditional descriptor based models in organic-inorganic halide perovskite (OIHP) materials design and have similar performance as deep learning models. Our multiscale simplicial complex approach not only provides a more precise representation for OIHP structures and underlying interactions, but also has better transferability to ML models. Our results demonstrate that advanced geometrical and topological invariants are highly efficient feature engineering approaches that can markedly improve the performance of learning models for molecular data analysis. Further, new structure-property relationships can be established between our invariants and bandgaps. We anticipate that our molecular representations and featurization models will transcend the limitations of conventional approaches and lead to breakthroughs in perovskite materials design and discovery.
Similar content being viewed by others
Introduction
Organic-inorganic halide perovskites (OIHPs) are hailed as a wonder cost-effective photovoltaic material with record efficiencies (exceeding 25%) that are now on par with the ubiquitous Si solar cells. These exceptional optoelectronic properties are attributed to their large absorption cross-sections, long balanced electron-hole diffusion lengths and excellent defect tolerance1,2,3. OIHPs possess the general ABX3 crystal structure, where A is a monovalent (organic or inorganic) cation (e.g., Cs, CH3NH3− etc), B is a divalent cation (e.g., Pb, Sn) and X is a halide anion (e.g, Cl, I, Br). Nevertheless, high performance OIHP solar cells contain Pb and suffer from poor ambient stability. Hence, there is an urgency to find new perovskites with advanced and improved properties that could circumvent these shortcomings. Consequently, machine learning (ML) models have found widespread application in the analysis of single perovskites4,5,6,7,8,9,10, double perovskites8,11,12,13, Pb-free perovskites14,15,16,17, perovskite solar cells device performance and fabrication18,19,20,21,22,23 as well as combined with high throughput experimentation24. A common framework in these models is that they consider a series of geometric and physical properties as perovskite molecular features. Among these features, several important descriptors (e.g., bandgap, absorption maxima, carrier mobility, reorganization energy, etc.) are selected or identified through statistical analysis or learning-based importance analysis. These descriptors are then employed in regression models or ML models to establish a quantitative structure-activity/property relationship (QSAR/QSPR). Subsequently, these models are further used in the screening of a large pool of perovskites, that are generated by the combination of possible monovalent organic molecular cations for A-site, divalent metal cations for B-site and halogen (or non-halogen) atoms for X-sites.
Despite the inroads, one of the central challenges in ML (beyond perovskite materials and relevant to all molecular data) is the construction of appropriate and suitable descriptors or fingerprints, known as featurization or feature engineering25,26,27,28. Efficient featurization should preserve the most important intrinsic structure information and remain invariant to rigid transformations. Traditional material descriptors consider structure information, including atomic radius, ion radius, orbital radius, tolerance factor, octahedral factor, packing factor, crystal structure measurements, and other surface/volume features, and physical properties, such as ionization potential, ionic polarizability, electron affinity, Pauling electronegativity, valence orbital radii, HOMO and LUMO, etc. Even though these perovskite descriptors can detail some general structural and physical properties, they are inadequate in representing the complicated intrinsic connection topologies underpinning materials formation and the rich interplay of fundamental interactions.
Recently, topological data analysis (TDA) based topological invariants and persistent Ricci curvature based geometric invariants applied as molecular descriptors have achieved good success in material data analysis (e.g., amorphous solids, granular and porous materials etc)29,30,31 and drug design (e.g., organic molecules and proteins)32,33,34,35,36. Nonetheless, applications to inorganic crystalline materials are few and far between, which is more challenging to encode compared to molecules. Different from traditional molecular descriptors, multiscale intrinsic invariant-based molecular descriptors characterize the fundamental structure information and have better transferability to ML models. Here, we are greatly inspired by the simple yet elegant algebraic topology that affords unique local and global structure encoding without needing any assumptions to describe the actual physics. For OIHPs, we posit that multiscale intrinsic structural descriptors afford a new paradigm in representing the rich physics and complex interplay of various interactions (i.e., electron-phonon coupling, Rashba effects, Jahn Teller effects, Van der Waals (VDW) interactions, hydrogen bonding (HB) effects, etc) within the organic and inorganic sub-lattices. Could one simply engineer unified and optimal descriptors based on multiscale intrinsic structural features underpinned by the rich physics in this hybrid organic-inorganic material system as efficient inputs for ML models?
Geometry and topology provide the structural foundation for all physical models. In general, geometry studies the shapes of subjects while topology analyzes structural connections (see Fig. 1). Geometrical and topological descriptors are mathematical quantities and invariants, that not only characterize detailed structural information, but also provide the basis for physical functions and properties. Structure-function connections are the underlying principles for physical models, QSAR/QSPR models, and machine learning models. However, traditional geometrical and topological descriptors provide descriptive, phenomenological, and superficial structural properties, such as atomic radius, ion radius, orbital radius, tolerance factor, and others as stated above. Such descriptors are deficient in high-level abstraction and generalization, thus they are usually problem-specific with a lower transferability for ML models. In contrast, the intrinsic geometrical and topological invariant based molecular descriptors are highly-abstract and generalized quantities that characterize the most intrinsic and fundamental structural properties, and more suitable for learning models. Figure 1 illustrates a comparison between traditional geometrical and topological descriptors and the intrinsic descriptors.
Multiscale representations are of key importance for physical models. Molecular interactions exist at various scales, ranging from covalent bonds, ionic bonds and hydrogen bonds at shorter distances, to long-range interactions, such as Van der Waals and electrostatic interactions. To characterize the various multiscale interactions that occur in perovskite structures, we consider atom-specific multiscale representations for perovskites. A perovskite supercell is decomposed into a series of atom-sets, each containing certain atom types. For instance, an OIHP supercell is decomposed into three atom sets with atoms from A-site, B-site and X-site, respectively. Since A-site is made of organic compounds, the A-site atom set is further decomposed into element-specific atom sets with C, N, O, and H respectively. Structural and interactional information is embedded in different combinations of these atom sets. In our model, a total of 30 combinations from the above atom-sets are considered (see Methods). Multiscale simplicial complex representations are generated for each atom set.
Generally speaking, simplicial complex is the generalization of graph, and it comprises of 0-simplexes (vertices), 1-simplexes (edges), 2-simplexes (triangles), 4-simplexes (tetrahedrons), and higher dimensional simplexes. Based on each atom-set point cloud data, a series of simplicial complexes at different scales are generated through a filtration process (see Methods). At each scale, a simplex is formed when distances between every two atoms are smaller than the cutoff distance, i.e., the filtration parameter. Under a smaller filtration value, connections only exist between atoms close to each other, characterizing local scale information. For a larger filtration value, even atoms that are far away from each other can be connected, indicating a global scale representation (see Methods). Figure 2 illustrates a multiscale atom-specific representation for OIHP structure.
A An illustration of atom-specific representation of OIHP structure. The perovskite supercell is decomposed into a series of atom-sets. B The atom radius is chosen as the filtration parameter. The systematically increasing filtration value generates a series of topological spaces at different scales. C The corresponding simplicial complexes from the filtration process.
Here we consider two types of persistent functions (PFs), including persistent homology (PH) and persistent Ricci curvature (PRC), for perovskite structure characterization. PFs study the persistence and variation of mathematical invariants within the multiscale simplicial complexes, that is the behavior of these invariants during the filtration process. PH uses the topological invariance of Betti number, which geometrically is the number of components, loops, holes, and other high dimensional cycle structures. PRC studies the geometric invariance of the Ricci curvature, which has two discrete forms, i.e., Olliver Ricci curvature and Forman Ricci curvature (FRC). We consider persistent Forman curvature (PFC) and use it to characterize the “curvedness“ of simplexes in simplicial complexes. Molecular descriptors are generated from PH through a binning approach on persistent barcodes. PFC-based molecular descriptors are obtained from persistent attributes, which are statistical and combinatorial properties of FRCs on the multiscale simplicial complexes. A total of 10 persistent attributes are used in our PFC model. Note that PF-based molecular descriptors or fingerprints have a much larger size than traditional perovskite descriptors (TPDs). Previously, 32 TPDs derived from structural and physical properties are considered16. In contrast, our PF-based perovskite fingerprints can have a size as large as several thousands, depending on the discretization schemes. In general, PF-based fingerprints are information-rich and provide a more thorough description of perovskite intrinsic structural and interaction information.
Results and Discussion
PF-based OIHP clustering
To demonstrate the capabilities of our PF-based fingerprints in the characterization of OIHP structures, we consider the clustering of three phases of Methylammonium lead halides (MAPbX3, X = Cl, Br, I), i.e., orthorhombic, tetragonal, and cubic phase of MAPbX3. It is known that the stability of OIHP is one of the major challenges to commercial application. The Van der Waals (VDW) interactions and hydrogen bonding (HB) effects, which are all closely related to the distances between organic molecules and inorganic ions, play an important role in OIHP structure stabilization. To test the sensitivity of our PF-based fingerprints to the variations of VDW interactions and HB effects, we simulate the structural fluctuations for three phases of MAPbX3 using molecular dynamics (MD). The MD simulations are carried out to stabilize the initial configurations (see Methods). For each OIHP structure, 1000 configurations are equally sampled from its MD simulation trajectory, thus a total of 9000 OIHP structures are generated. The clustering of these OIHP structures is studied using unsupervised learning models, in particular t-distributed stochastic neighbor embedding (t-SNE). Figure 3 illustrates the results for t-SNE clustering models with different input features, including atom XYZ-coordinates, TPDs, PFC-based fingerprints, and PH-based fingerprints. For atom XYZ-coordinates, even though they are in fact structural information, their simple direct transformation into features fails to provide any discrimination between the different types of OIHPs. Next, we examine TPDs. The 32 TPDs are listed in Methods. Since TPDs incorporate information from different atom types and unit cell volume, they can clearly separate the 9 structural types. However, they fail to characterize the configuration variations within each trajectory. In other words, no detailed structure variation is captured by these descriptors. In contrast, both PF-based fingerprints possess better capabilities in characterizing the intrinsic structure information and discriminating the different types of OIHPs. It should be noticed that both PF-based fingerprints require structure information, i.e., atom types and their coordinates, while TPDs do not require that.
Four feature generation schemes are considered, including XYZ-coordinates (A), traditional perovskite descriptors (TPD) (B), persistent Forman curvature (PFC)-based fingerprint (C), and persistent homology (PH)-based fingerprint (D). Each trajectory contains 1000 configurations and t-SNE model is used for clustering (of the last 500 configurations at equilibrium). The x-axis and y-axis are the two principal dimensions in the t-SNE model.
PF-based OIHP formation energy and bandgap prediction
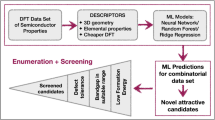
Another major focus of artificial intelligence (AI)-based perovskite design is the prediction of the formation energy and bandgap. In fact, learning models with traditional perovskite descriptors have already achieved good performance with Pearson correlation coefficient (PCC) between prediction results and density functional theory (DFT) results larger than 0.9516,17. However, the accuracy of these learning models in bandgap prediction can be further improved. Here we consider a well-maintained OIHP dataset with 1346 DFT-based structure and bandgap results37 to validate our PF-fingerprints. Previously, Wu et al.16 utilized this 1346 OIHP database to train their gradient boosting regression (GBR), support vector regression (SVR) and kernel ridge regression (KRR) models. In their models, 32 molecular descriptors are considered. Lu et al.17 have used only 212 OIHP structures from this dataset for training their GBR models with 30 molecular descriptors. In contrast, our PH- and PFC-based molecular fingerprints are of size 4319 and 10800, respectively (See Methods). More importantly, our fingerprints provide a systematic characterization of intrinsic molecular structures and can naturally incorporate the physical properties of the molecules. To avoid the overfitting problem, we consider the gradient boost tree (GBT) models and develop two model types, i.e., PH-GBT and PFC-GBT. Figure 4 illustrates the flowchart of our PF-based machine learning models. The material structures are represented as simplicial complexes, from which geometric and topological features are extracted. The PH- and PFC-based features are combined with GBT models for OIHP bandgap prediction. Figure 5A shows a comparison of the results from our PF-based molecular descriptors and the ones from traditional molecular descriptors16. To ensure fair comparison, a 10-fold cross validation is considered. Three indices are used for error estimation, including Coefficient of determination (R2), mean absolute error (MAE), and mean square error (MSE). It can be seen that our PF-based learning models can significantly reduce the errors, with R2 ≈ 0.94 and MSE ≈ 0.073 for PH-GBT. Detailed results can be found in Supplementary Table 2. Note that a 5-fold cross validation for a much smaller-sized data with only 212 OIHP structures is considered in Lu et al’s paper, where their results are R2 = 0.97 and MSE = 0.086. Hence, our PF-based approach compares favorably and performs much better than traditional materials descriptors-based machine learning models. Further, we have compared with the state-of-the-art deep learning models, in particular, SchNet38 and MEGnet39. Supplementary Table 2 lists 10-fold cross-validation results of SchNet and MEGnet on the OIHP dataset. In general, our PHa-GBT model can outperform SchNet but is slightly inferior to MEGnet. However, our models have better interpretability than deep learning models. New structure-property relationships can be obtained through the feature importance analysis. Supplementary Figs. 3 and 4 show the feature importance results for PH-based descriptors and PRC-based descriptors, respectively. The top three most important features and their correlation with the bandgap values are illustrated in Supplementary Figs. 3b and 4b. It can be seen that high (inverse) correlations can be found between our descriptors and bandgap values. In particular, PRC-based persistent mean (0-simplex) for atom-set ACOHBX at filtration value 3.0Å has the best correlation of −0.849. Geometrically, a higher Ricci-curvature indicates regions with more connections (such as clusters and communities), while a lower Ricci-curvature indicates the regions with less connections (such as links). A lower persistent mean value indicates less connections in the simplicial complexes generated (at 3.0 Å) from the material data, which corresponds to a higher bandgap value. Note that all our PH-based and PRC-based descriptors are intrinsic invariants that characterize general global structural information, and they can be used in the establishment of new structure-property relationships.
A Results for the 10-fold cross-validation for OIHP bandgaps from 1346 OIHP database16. Three TPD-based machine learning models (blue bars), including TPD-based kernel ridge regression (TPD-KRR), TPD-based support vector regression (TPD-SVR) and TPD-based gradient boosting tree (TPD-GBT) are used in comparison with persistent function-based models (green bars), including PFC-GBT and PH-GBT. B Results for Leave One Group Out (LOGO) cross-validation test for A-site, B-site and X-site.
To further assess the performance of PF-based descriptors and traditional molecular descriptors, we consider the Leave One Group Out (LOGO) cross-validation test for A-site, B-site and X-site groups. More specifically, in the dataset with 1346 OIHP structures, there are 16 types of A-site organic cations, 3 types of B-site group-IV cations and 4 types of X-site halide anions. In the LOGO test, one type of OIHP structure based on the site atoms is used as the test set, and the rest of the data is used as the training set. For instance, all structures in the entire dataset can be classified into 16 groups based on A-site organic cations. A total of 16 LOGO validation tests will be conducted by repeating the process of using one group as the test set and the rest of the 15 groups as the training set. The average result over 16 tests is used as an indication of the model for the prediction of A-site atoms. Similar LOGO tests are considered for B-site and X-site atoms. The traditional molecular descriptor-based GBR models are used for comparison. In each study, the LOGO processes are repeated for 10 times and the median values are chosen as the final outcome. Figure 5B illustrates the results for R2, MAE and MSE from the LOGO tests for A-, B-, and X-sites. It can be seen that in all LOGO tests, PF-GBT models (i.e., PH/PFC-GBT) have better performance than TPD-GBT models. For all models, their prediction accuracy for B-site and X-site is significantly lower than A-site. In particular, the model accuracy for X-site is lowest with negative R2 for all three models. Note that LOGO cross validation has never been used in all the previous ML models for perovskites. This indicates a potential problem for model validation in all these ML models. Nonetheless, the MAE and MSE from our PF-GBT models are lower than that of the TPD-GBT model for X-site. In general 5-fold or 10-fold validation with all training data may significantly overestimate the prediction power of the model for X-site atom prediction. Note that lesser training data and more test data also contribute to the lower accuracies for the B- and X-site LOGO models. More detailed results can be found in Supplementary Tables 3 to 5. Apart from the bandgap, we also tested the models with the refractive index (n) and frequency-dependent dielectric constants (i.e., electronic (ϵelec), ionic (ϵion) and total (ϵelec + ϵion)). The 10-fold cross-validation results of the three models are presented in Supplementary Tables 3 to 5. It is obvious that all three models have higher accuracy in the prediction for n and ϵelec; while having lower accuracy for ϵion as well as that for the total. For all the predictions, our PFC-GBT and PH-GBT models consistently yield better results than TPD-GBT.
Of late, CsGeX3 perovskites have emerged as “greener“ alternatives for lead-based perovskites. By replacing the B-site lead atom with germanium, these perovskites offer environmental friendly substitutes that can also provide low-cost and high efficiency in photovoltaic devices40,41,42. As a special test of the predictive capabilities of our models, we consider the three completely inorganic lead-free perovskite solar cells (CsGeX3, X = Br, Cl, I) in these emergent Ge systems. Our PH-GBT model predicts the three bandgaps with an RMSE of 0.461eV and MAE of 0.393eV as compared to the traditional molecular descriptors, which have both RMSE and MAE as 0.873eV. This further validates the potential of our approach. Note that for this prediction test, 48 perovskites with formula ABX3 were specifically added to the existing 1346 OIHPs to boost the predictive power of our models. The details of the 48 perovskites can be found in Supplementary Table 11.
The ability to efficiently encode material structures into well-defined descriptors readable by ML models is critical for maximizing their predictive power. Ideally, descriptors should be uncorrelated as excessive correlation between features will affect the accuracy and efficiency of the model. In which case, further feature selection is necessary43,44. Traditional perovskite descriptors are usually obtained from descriptive, phenomenological, and superficial structural properties. They lack the in-depth and intrinsic information and have a lower transferability and generalizability. Thus, they understandably perform poorly at perovskite bandgap prediction in ML models. Here, we present PF-based fingerprints that are generated from intrinsic geometrical and topological invariants, and enable a high-level abstraction and generalization of perovskite molecular structural properties. This approach not only circumvents the shortcomings of the traditional descriptors, but also provides a better representation of the underlying Physics. Hence, PF-GBTs can achieve significantly lower errors and better predictive accuracy in bandgap estimation. Importantly, our PF-based learning models can be universally applied in material data analysis and can potentially lead to new materials design and discovery breakthroughs.
Methods
Topological representations
Mathematical representations provide an elegant means for visualizing material topological structures at the molecular level. They are also the foundation for the physical and machine learning models. Graph (or network) models play a dominant role in the representation of material molecular structures. They describe the pair-wise interactions within and/or between molecular structures. As a generalization of graphs, simplicial complexes are proposed for the modeling of higher-dimensional interactions. Triangle meshes and tetrahedron meshes are special types of simplicial complexes. Based on graph and simplicial complex representations, geometric and topological invariants can be derived and used in the structure characterization.
Graph and simplicial complex
Graph models are the most widely used topological models for material, chemical and biological systems. In these models, atoms and (covalent) bonds are usually represented as vertices and edges, respectively. Graphs are special cases of simplicial complexes, which are constructed from simplexes under certain rules. A k-simplex σk = {v0, v1, v2, ⋯ , vk} is the convex hull formed by k + 1 affinely independent points v0, v1, v2, ⋯ , vk as follows,
Geometrically, a 0-simplex is a vertex, a 1-simplex is an edge, a 2-simplex is a triangle, and a 3-simplex represents a tetrahedron. Simplices are the building block for the simplicial complex. In general, a simplicial complex K is a finite set of simplices that satisfy two essential conditions. Firstly, any face of a simplex from K is also in K. Secondly, the intersection of any two simplices in K is either empty or shares faces.
Filtration-based multiscale representation
Molecules usually have different characteristic scales, ranging from covalent bond scale, hydrogen bond scale, to VDW bond scale and electro-static interaction scale. Multiscale representations are of essential importance for molecular characterization. Mathematically, a multiscale representation can be naturally generated from a filtration process45. Filtration parameter, denoted as f and key to the filtration process, is usually chosen as sphere radius (or diameter) for point cloud data, edge weight for graphs, and isovalue (or level set value) for density data. A consistent increase (or decrease) of the filtration value will induce a sequence of hierarchical topological representations.
Persistent homology
Persistent homology is the key model in topological data analysis. It studies the persistence and variation of homology generator during a filtration process. Different from geometry and topology models, PH manages to incorporate geometrical measurements into topological invariants, thus it provides a balance between geometric complexity and topological simplification.
Persistent Ricci curvature
Ricci curvature is one of the fundamental concepts in differential geometry and theoretical physics46,47. It describes manifold local quantities, such as growth of volumes of distance balls, transportation distances between balls, divergence of geodesics, and meeting probabilities of coupled random walks48. In the general theory of relativity, the Einstein field equations relate the geometry of space-time with the distribution of matter by the use of Ricci curvature tensor. Ricci flow, introduced by Hamilton, is key to the Perelman’s proof of Poincaré conjecture49. Two discrete Ricci curvature forms, i.e., Ollivier Ricci curvature (ORC)50,51,52,53,54,55,56 and Forman Ricci curvature (FRC)57,58,59,60, have been developed to characterize different aspects of the classical Ricci curvature. Here we focus on the Forman Ricci curvature. FRC is a defined as a combinatorial property between upper-adjacent, lower-adjacent and parallel simplexes on CW complexes57.
Data Availability
All the data and code used in this paper can be found in the Supplementary Information and are openly available in our GitHub page https://github.com/ExpectozJJ/PF-OIHP and in DR-NTU (Data) at: https://doi.org/10.21979/N9/CVJWZ9.
References
Sum, T. C. & Mathews, N. Advancements in perovskite solar cells: photophysics behind the photovoltaics. Energy Environ. Sci. 7, 2518–2534 (2014).
Xing, G. et al. Long-Range Balanced Electron- and Hole-Transport Lengths in Organic-Inorganic CH3NH3PbI3. Science 342, 344–347 (2013).
Brandt, R. E., Stevanović, V., Ginley, D. S. & Buonassisi, T. Identifying defect-tolerant semiconductors with high minority-carrier lifetimes: beyond hybrid lead halide perovskites. MRS Commun. 5, 265–275 (2015).
Pilania, G., Balachandran, P. V., Kim, C. & Lookman, T. Finding new perovskite halides via machine learning. Front. Mater. 3, 19 (2016).
Xie, T. & Grossman, J. C. Crystal graph convolutional neural networks for an accurate and interpretable prediction of material properties. Phys. Rev. Lett. 120, 145301 (2018).
Balachandran, P. V. et al. Predictions of new ABO3 perovskite compounds by combining machine learning and density functional theory. Phys. Rev. Mater. 2, 043802 (2018).
Xu, Q. C., Li, Z. Z., Liu, M. & Yin, W. J. Rationalizing perovskite data for machine learning and materials design. J. Phys. Chem. Lett. 9, 6948–6954 (2018).
Li, Z., Xu, Q., Sun, Q., Hou, Z. & Yin, W. J. Thermodynamic stability landscape of halide double perovskites via high-throughput computing and machine learning. Adv. Funct. Mater. 29, 1807280 (2019).
Park, H. et al. Exploring new approaches towards the formability of mixed-ion perovskites by DFT and machine learning. Phys. Chem. Chem. Phys. 21, 1078–1088 (2019).
Schmidt, J. et al. Predicting the thermodynamic stability of solids combining density functional theory and machine learning. Chem. Mater. 29, 5090–5103 (2017).
Pilania, G. et al. Machine learning bandgaps of double perovskites. Sci. Rep. 6, 19375 (2016).
Askerka, M. et al. Learning-in-templates enables accelerated discovery and synthesis of new stable double perovskites. J. Am. Chem. Soc. 141, 3682–3690 (2019).
Agiorgousis, M. L., Sun, Y. Y., Choe, D. H., West, D. & Zhang, S. B. Machine learning augmented discovery of chalcogenide double perovskites for photovoltaics. Adv. Theory Simul. 2, 1800173 (2019).
Jacobs, R., Luo, G. & Morgan, D. Materials discovery of stable and nontoxic halide perovskite materials for high-efficiency solar cells. Adv. Funct. Mater. 29, 1804354 (2019).
Im, J. et al. Identifying Pb-free perovskites for solar cells by machine learning. npj Comput. Mater. 5, 37 (2019).
Wu, T. & Wang, J. Global discovery of stable and non-toxic hybrid organic-inorganic perovskites for photovoltaic systems by combining machine learning method with first principle calculations. Nano Energy 66, 104070 (2019).
Lu, S. et al. Accelerated discovery of stable lead-free hybrid organic-inorganic perovskites via machine learning. Nat. commun. 9, 3405 (2018).
Li, J., Pradhan, B., Gaur, S. & Thomas, J. Predictions and strategies learned from machine learning to develop high-performing perovskite solar cells. Adv. Energy Mater. 9, 1901891 (2019).
Odabaşí, Ç. & Yíldírím, R. Assessment of reproducibility, hysteresis, and stability relations in perovskite solar cells using machine learning. Energy Technol. 8, 1901449 (2020).
Odabaşí, Ç. & Yíldírím, R. Machine learning analysis on stability of perovskite solar cells. Sol. Energy Mater. Sol. Cells 205, 110284 (2020).
Howard, J. M., Tennyson, E. M., Neves, B. R. A. & Leite, M. S. Machine learning for perovskites’ reap-rest-recovery cycle. Joule 3, 325–337 (2018).
Li, F. et al. Machine learning (ML)-assisted design and fabrication for solar cells. Energy Environ. Mater. 2, 280–291 (2019).
Yu, Y., Tan, X., Ning, S. & Wu, Y. Machine learning for understanding compatibility of organic–inorganic hybrid perovskites with post-treatment Amines. ACS Energy Lett. 4, 397–404 (2019).
Sun, S. J. et al. Accelerated Development of Perovskite-Inspired Materials via High-Throughput Synthesis and Machine-Learning Diagnosis. Joule 3, 1437–1451 (2019).
Schütt, K. T. et al. How to represent crystal structures for machine learning: Towards fast prediction of electronic properties. Phys. Rev. B 89, 205118 (2014).
Ramprasad, R., Batra, R., Pilania, G., Mannodi-Kanakkithodi, A. & Kim, C. Machine learning in materials informatics: recent applications and prospects. npj Comput. Mater. 3, 54 (2017).
Isayev, O. et al. Materials cartography: representing and mining materials space using structural and electronic fingerprints. Chem. Mater. 27, 735–743 (2015).
Huan, T. D., Mannodi-Kanakkithodi, A. & Ramprasad, R. Accelerated materials property predictions and design using motif-based fingerprints. Phys. Rev. B 92, 014106 (2015).
Hiraoka, Y. et al. Hierarchical structures of amorphous solids characterized by persistent homology. Proc. Natl Acad. Sci. USA 113, 7035–7040 (2016).
Saadatfar, M., Takeuchi, H., Robins, V., Francois, N. & Hiraoka, Y. Pore configuration landscape of granular crystallization. Nat. Commun. 8, 15082 (2017).
Lee, Y. et al. Quantifying similarity of pore-geometry in nanoporous materials. Nat. Commun. 8, 15396 (2017).
Nguyen, D. D., Gao, K. F., Wang, M. L. & Wei, G. W. MathDL: Mathematical deep learning for D3R Grand Challenge 4. J. Comput. Aided Mol. Des. 34, 131–147 (2020).
Cang, Z. X., Mu, L. & Wei, G. W. Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLoS Comput. Biol. 14, e1005929 (2018).
Cang, Z. X. & Wei, G. W. TopologyNet: Topology based deep convolutional and multi-task neural networks for biomolecular property predictions. PLoS Comput. Biol. 13, e1005690 (2017).
Wee, J. J. & Xia, K. L. Ollivier Persistent Ricci Curvature-Based Machine Learning for the Protein-Ligand Binding Affinity Prediction. J. Chem. Inf. Model. 61, 1617–1626 (2021).
Wee, J. J. & Xia, K. L. Forman persistent Ricci curvature (FPRC)-based machine learning models for protein-ligand binding affinity prediction. Brief. Bioinform. 22, bbab136 (2021).
Kim, C., Huan, T. D., Krishnan, S. & Ramprasad, R. A hybrid organic-inorganic perovskite dataset. Sci. data 4, 170057 (2017).
Schütt, K. T., Sauceda, H. E., Kindermans, P. J., Tkatchenko, A. & Müller, K. R. SchNet-a deep learning architecture for molecules and materials. J. Chem. Phys. 148, 241722 (2018).
Chi, C., Weike, Y., Yunxing, Z., Zheng, C. & Shyue Ping, O. Graph networks as a universal machine learning framework for molecules and crystals. Chem. Mater. 31, 3564–3572 (2019).
Zhang, F. et al. Recent Advances and Opportunities of Lead-Free Perovskite Nanocrystal for Optoelectronic Application. Energy Mater. Adv. 2021, 5198145 (2021).
Yang, D. et al. Germanium-lead perovskite light-emitting diodes. Nat. Commun. 12, 4295 (2021).
Duan, C., Zhao, Z. & Yuan, L. Lead-Free Cesium-Containing Halide Perovskite and Its Application in Solar Cells. IEEE J. Photovolt. 11, 1126–1135 (2021).
Bellman, R. E. Adaptive Control Processes: A Guided Tour. (Princeton University Press, Princeton, NJ, 2015).
Schmidt, J. et al. Predicting the thermodynamic stability of solids combining density functional theory and machine learning. Chem. Mater. 29, 5090–5103 (2017).
Edelsbrunner, H., Letscher, D. & Zomorodian, A. Topological persistence and simplification. Discret. Comput. Geom. 28, 511–533 (2002).
Jost, J. & Jost, J. Riemannian geometry and geometric analysis. vol. 42005, (Springer-Verlag Berlin Heidelberg, 2008).
Najman, L. & Romon, P. Modern approaches to discrete curvature. vol. 2184. (Springer International Publishing AG, Switzerland, 2017).
Samal, A. et al. Comparative analysis of two discretizations of ricci curvature for complex networks. Sci. Rep. 8, 8650 (2018).
Perelman, G. Ricci flow with surgery on three-manifolds. Preprint at https://arxiv.org/abs/math/0303109 (2003).
Bakry, D. & Émery, M. Diffusions hypercontractives. In Séminaire de Probabilités XIX 1983/84, 177-206 (Springer-Verlag, Berlin Heidelberg New York, 1985).
Chung, F. R. K. & Yau, S. T. Logarithmic harnack inequalities. Math. Res. Lett. 3, 793–812 (1996).
Sturm, K. T. On the geometry of metric measure spaces. Acta Math. 196, 65–131 (2006).
Ollivier, Y. Ricci curvature of metric spaces. C. R. Math. 345, 643–646 (2007).
Lott, J. & Villani, C. Ricci curvature for metric-measure spaces via optimal transport. Ann. Math. 169, 903–991 (2009).
Ollivier, Y. Ricci curvature of markov chains on metric spaces. J. Funct. Anal. 256, 810–864 (2009).
Bonciocat, A. I. & Sturm, K. T. Mass transportation and rough curvature bounds for discrete spaces. J. Funct. Anal. 256, 2944–2966 (2009).
Forman, R. Bochner’s method for cell complexes and combinatorial Ricci curvature. Discret. Comput. Geom. 29, 323–374 (2003).
Sreejith, R. P., Mohanraj, K., Jost, J., Saucan, E. & Samal, A. Forman curvature for complex networks. J. Stat. Mech.: Theory Exp. 2016, 063206 (2016).
Saucan, E., Sreejith, R. P., Ananth, R. P., Jost, J. & Samal, A. Discrete ricci curvatures for directed networks. Chaos Solit. Fractals 118, 347–360 (2019).
Saucan, E. & Weber, M. Forman’s ricci curvature-from networks to hypernetworks. In International Conference on Complex Networks and their Applications, 706–717 (Springer Nature Switzerland AG, 2019).
Acknowledgements
This work was supported in part by Nanyang Technological University Startup Grant M4081842.110, Singapore Ministry of Education Academic Research fund Tier 1 grant RG109/19 and Tier 2 grants MOE-T2EP50120-0004 and MOE-T2EP20120-0013, as well as the National Research Foundation (NRF), Singapore under its NRF Investigatorship (NRF-NRFI2018-04).
Author information
Authors and Affiliations
Contributions
T.C.S. and K.X. conceived the idea and designed the research. Q.X. performed the DFT and MD simulations. D.V.A. and J.J.W. performed the PF-modelling. All authors analyzed the data, discussed the results and contributed to the manuscript. T.C.S. and K.X. led the project.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Anand, D.V., Xu, Q., Wee, J. et al. Topological feature engineering for machine learning based halide perovskite materials design. npj Comput Mater 8, 203 (2022). https://doi.org/10.1038/s41524-022-00883-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-022-00883-8
- Springer Nature Limited
This article is cited by
-
Prediction of heavy-section ductile iron fracture toughness based on machine learning
Scientific Reports (2024)
-
Geometric data analysis-based machine learning for two-dimensional perovskite design
Communications Materials (2024)
-
Measurement of information content of Perovskite solar cell’s synthesis descriptors related to performance parameters
Emergent Materials (2024)
-
Advances in materials informatics: a review
Journal of Materials Science (2024)
-
Persistent Dirac for molecular representation
Scientific Reports (2023)