Abstract
Automated particle segmentation and feature analysis of experimental image data are indispensable for data-driven material science. Deep learning-based image segmentation algorithms are promising techniques to achieve this goal but are challenging to use due to the acquisition of a large number of training images. In the present work, synthetic images are applied, resembling the experimental images in terms of geometrical and visual features, to train the state-of-art Mask region-based convolutional neural networks to segment vanadium pentoxide nanowires, a cathode material within optical density-based images acquired using spectromicroscopy. The results demonstrate the instance segmentation power in real optical intensity-based spectromicroscopy images of complex nanowires in overlapped networks and provide reliable statistical information. The model can further be used to segment nanowires in scanning electron microscopy images, which are fundamentally different from the training dataset known to the model. The proposed methodology can be extended to any optical intensity-based images of variable particle morphology, material class, and beyond.
Similar content being viewed by others
Introduction
Understanding the design rules that dictate materials chemistry is critical to enabling the rational design of energy storage systems. Moreover, connecting single-entity and ensemble measurements is paramount to understanding how structure–function relationships propagate across length scales and dictate the performance of hierarchical systems in battery materials1,2. The ability to probe a multitude of contrast mechanisms from a single measurement has enabled many insights into the working principles of electrochemically active materials3. Spectromicroscopy techniques such as scanning transmission X-ray microscopy and X-ray ptychography, for example, leverage X-absorption and scattering events to capture morphological and electronic structure information which can be colocalized at the nanometer level to provide chemical maps for a region of interest4,5,6,7. The application of such information-rich measurements to particle networks has been limited, in part, due to the complexity of extracting morphological and chemical features from large and complex datasets. Dataset dimensionality reduction techniques such as principal component analysis considerably improve the ease of deciphering chemical markers often contained within spectra8. Nevertheless, there is a need for more efficient and effective workflows to obtain size and shape descriptors that can be utilized with chemical information to explore physio-chemical phenomena as a function of various descriptors7,9,10,11,12.
In recent years, image segmentation algorithms that leverage the parallel processing capability of neural networks have garnered significant attention because of their potential to enable automated image analysis13,14. For example, the well-received Mask region-based convolutional neural networks (Mask R-CNN) algorithm15 is now utilized routinely for general segmentation tasks. Common Object in Context (COCO)16 and PASCAL Visual Object Classes (VOC)17 have been developed in concert to train and benchmark the performance of algorithms for computer vision studies. However, the requirement of large datasets to train deep-learning algorithms has been challenging to meet with experimental microscopy data in the material-chemistry community due to an inherent complexity in generation and the time-consuming nature of human annotation. Nevertheless, deep-learning algorithms based on empirical data and human annotation have been developed for several material classes, such as graphene flakes18 imaged by optical microscopy, carbon nano-fibers19 imaged by SEM and a further collection of electron microscopy images for various material class20. Similarly, attempts to augment real image datasets of polycrystalline grains21, or usage of image rendering techniques on nano-particles images22,23 to counteract the prohibitive data acquisition step, have been proven to be successful.
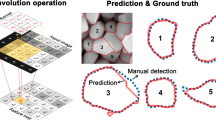
To overcome the challenges associated with limited training data, especially, with emphasis on instance segmentation, we have developed a deep learning model, based on the Mask R-CNN algorithm, which has been trained entirely on synthetically generated microstructures. By applying an optical density compression step, the algorithm can segment and obtain statistical information from both electron and X-ray microscopy images of vanadium pentoxide (V2O5), a canonical cathode material. From an instance segmentation perspective, the V2O5 nanoparticle dispersions shown in Fig. 1 represent a counterpoint that stands in contrast to the case of “ideal” monodispersion of nanoparticles characterized by a nearly spherical geometry for which the automated size determination process is documented20,24,25,26. In many cases, the task of segmenting nanoparticles is exacerbated by the diversity of particle shapes (e.g., nanospheres, nanorods, nanobelts, tetrapods, 2D hexagons, nanocubes, and nanodiscs) and dispersions found in the literature and industrial applications24,27. The synthesis method for the nanowires featured in this work combined with their non-spherical geometry leads to considerable irregularity in thickness, length-to-width aspect ratios, and edge profiles, which collectively result in a significant degree of variability in particle shape despite the all-encompassing nanowire classification. This shape variability combined with particle agglomeration makes for an ambitious instance segmentation task that closely resembles the needs of many applications today28,29. Through the lens of chemistry-mechanics coupling, V2O5 appears as a fascinating case study of image analysis on the role of particle size, shape, and geometry5,6,30. The promise of V2O5 as a cathode material stems from a high theoretical capacity, the potential for ’beyond lithium’ batteries, and the geographical diversity of vanadium deposits which reduces criticality concerns that have throttled cobalt battery chemistries10,31,32. A well-documented bottleneck for the widespread adoption of the thermodynamically stable α-V2O5 phase is the emergence of intercalation-induced structural transformations, which create significant phase inhomogeneities and stress gradients that have been linked to deleterious impacts on performance and longevity33,34. It has been shown that the patterns of lithiation in these systems are strongly modified by dimensional and morphological features such as particle size, particle geometry, curvature, and interconnects—the coupling of high-dimensional X-ray imaging methods, ensemble measurements, and image segmentation holds considerable promise for identifying optimal crystalline sizes and geometries to overcome inherent material bottlenecks that give rise to lithiation heterogeneities resulting ultimately in stress accumulation5,6,9.
In the present work, we consider three different types of microscopy images generated by X-ray ptychography, scanning transmission X-ray microscopy (STXM) and scanning electron microscopy (SEM) techniques. While each imaging technique is fundamentally different, the model developed in this work demonstrates a remarkable robustness when segmenting nanorod-like structures. The present work is organized as follows: in section “Human annotation of the V2O5 dataset,” we briefly introduce the experimental datasets, which are later utilized to validate the performance of the model. For the model training, we generate a series of synthetic images. The workflow of data generation is explained in section “Synthetic dataset generation,” whereas the details on the training of the Mask R-CNN model are presented in section “Application and training of the Mask R-CNN model”. In section “Instance segmentation results of microscopy images”, results on training, evaluation, and segmentation of considered microscopy images from the above mentioned imaging techniques are provided and discussed. Further, towards the reproducibility of the model, an interactive web-based segmentation application has been developed and can be found at: https://share.streamlit.io/linbinbin92/V2O5_app/V2O5_app.py for testing and further image analysis.
Results
Human annotation of the V2O5 dataset
Manual annotation of the V2O5 datasets was facilitated by the web-based annotation tool Makesense.ai35 in a polygon format, where points along a particle border are set to form the shape of the particles (see Fig. 1). This step was performed for every particle in the present images, and the annotated file was saved in JavaScript Object Notation (JSON) format to serve the validation purpose. It is worth noting that some limitations of the manual annotation process such as a sensitivity to human error and a dependence on spatial resolution of the native images naturally exist. Further sources of error stem from the inherent complexity of the dispersion, which results in many instances where particles are overlapped.
Synthetic dataset generation
To generate synthetic image datasets reminiscent of the V2O5 experimental dataset particles, we have developed a random nanowire generator using the software Geodict®36. In the generation step, for each training sample, the number of particles, length, shape distribution was specified to create 3D voxel-based structures, Fig. 2. The chosen size of the domain was 512 × 512 × 200 (W × B × H). For the present work, the number of particles, diversity of morphology, and resolution, approximate the experimental information contained in the experimental X-ray ptychography data in Fig. 1b. Higher resolution i.e. larger domain size, can be chosen at cost of longer generation time and image file size. The height of the domain for the synthetic 3D microstructure was chosen such that it exceeded the total height of the overlapped nanowires. Further, the nanowires were internally enumerated and deposited one after another. This workflow ensures that the labels are predetermined, thus bypassing the need for human annotation at a later phase. In order to mirror the transmission intensity generated by X-ray ptychography, an optical density compression step was applied to emulate thickness information. Here, voxels were first compressed along the out-of-plane direction then summed and divided by the total thickness of the nanostructure. The pixel values comprising the projected optical density image map in the in-plane directions were therefore normalized, and further transformed to a gray color scale, expressed as a value from 0 to 255. Accordingly, regions where two or more particles are overlapped can be distinguished by a sudden change in optical density (i.e. pixel intensity). In a subsequent step, a standard Gaussian filter (filter size of 2) was applied to the images to account for blurring of the particle edges. As shown in Fig. 2, the ground truth sample was split into individual binary masks for each nanowire contained in the synthetic ground truth image. Therefore, for each training sample, the dataset consists of one input image and N output binary mask images with N as the number of particles in that image. These binary mask images can be further used to obtain statistical information about the morphology descriptors. Before evaluation, initial dataset size of 250 images were generated for training purposes. After an initial evaluation, additional images were generated in recursive steps involving a greater diversity of particle morphology to closely replicate the experimental data. A final dataset of 1250 synthetic images were obtained with a 80/20 split for the training and validation steps. In order to introduce further variability in the dataset, standard augmentation techniques have been applied as the dataset is fed forward to the dataloader. Note here, we do not create an additional dataset in the training process. To account for intrinsic variability in the contrast and brightness of experimental microscopy datasets, we applied a random brightness and a random contrast filter ranging from 0.7 to 1.2. A random flip in both horizontal and vertical direction with a probability of 0.5 has been applied. Details on the training process and dataset can be found in the model training and data section. We note that it is possible to enhance the synthetic dataset with further real images. However, addition of experimental data to the training set did not yield a notable improvement in performance of the model as discussed in further detail in the Supplementary Note 3.
Application and training of the Mask R-CNN model
To date, numerous deep learning models for instance segmentation can be found in the computer vision literature. Ever since 2017, the Mask R-CNN and its variants (Mask scoring R-CNN37, TensorMask38) have been solid options for the task of instance segmentation due to their high accuracy of instance mask predictions. Other famous object detectors, such as YOLO (You only look once)39 has been also found in the segmentation of nanowire-like materials40. However, YOLO algorithms generally aim at higher training and prediction speed without providing prediction masks in the first place. To this end, recent extensions of YOLO, such as YOLACT41 have been developed with mask predictions towards real-time application. Other similar deep-learning based methods with specific modifications applied to fibrous structure segmentation can be found in the literature42,43. While we believe that other instance segmentation algorithms can also be applied for the present instance segmentation study, we decided to apply the well-established Mask R-CNN algorithm within the Detectron2 framework44 due to its great built-in API and well-structured online documentation. Details on the essential components of the Mask R-CNN model can be found in the methods section. It is important to note that while the synthetic dataset was modeled after experimental microscopy images, the training steps for the model developed in this work were performed solely on the synthetic dataset without any real images. The stochastic gradient descent method was used with the default setting provided by the Detectron2 implementation of Mask R-CNN algorithm44. The hyperparameters modified in the parameter study can be found in Table 1. For the present study, the effect of synthetic dataset size, as well as hyperparameters as learning rate, region of interest batch size per image (ROI HEAD) and Non-Maximum-Suppression (NMS) were studied. ROI HEAD is a subset of the proposed ROIs bounding boxes from Region Proposal Networks (RPN), so the loss can be calculated on this subset rather than on all of the box proposals. Intersection over Union (IOU) threshold (IOU THR) for RPN defines the ratio of overlaps between the ground truth boxes and proposed boxes during the training. NMS mainly acts as a filter to remove overlapping proposed boxes. More details and default hyperparameter settings can be found in44. We further note that our model leverages pre-trained weights on the COCO dataset and the chosen learning rates were therefore larger than training a completely new network from scratch. Results on model prediction performances trained with smaller learning rates can be further found in Supplementary Note 4. The training was performed on four A100/V100 Nvidia GPUs at Lichtenberg Cluster, TU Darmstadt for a total time of 5–10 h with a batch size of 8 images per GPU. The training time here should only provide an approximation, additional information can be found in the Supplementary Note 7. Detailed study and optimization of the performance speed was not the objective of this work.
Instance segmentation results of microscopy images
In this section, the segmentation results are evaluated considering the metrics defined in the section “model evaluation metrics”. It is important to re-emphasize that the deep learning model has been trained solely on synthetic image datasets modeled after the X-ray ptychography and scanning transmission X-ray microscopy data. Consequently, the SEM image, which is distinctive in terms of contrast generation is foreign to the trained model. All images are, obtained from mentioned microscopy techniques as they are, and were not pre-processed or filtered for the evaluation purpose. The only pre-processing step applied to the ptychography and STXM images involves the conversion from transmission data to absorbance (optical density)8. The results obtained from the deep learning model are compared to manually annotated results, which are subject to uncertainty to certain level due to visual limitation. We again note that the current model leverages pre-trained model weights and can be re-trained and extended to multiple object classes of further morphologies, such as, e.g., nano-spheres, see Supplementary Note 1 for further information.
Before applying the model to real microscopy images, 20 models were trained with various hyperparameters to examine their influence on the synthetic images, see Table 1. Subsequently, all 20 trained models were applied to segment all three types of microscopy image. The best performer in mask segmentation AP for each image type was selected to visualize the segmentation masks in Figs. 3, 4, 6, and 8b. For a full list of all AP results, readers are referred to Supplementary Table 1. We start to evaluate the model segmentation accuracy at a semantic level to estimate to what degree the model can segment the actual nanowires from the background. The inherent strength of instance segmentation is that it includes the subordinate functionality of semantic segmentation, where the semantic results can be immediately extracted from the mask predictions. As shown in Figs. 3, 4, 6, and 8b, fairly good segmentation results were obtained. These object masks were then used to evaluate semantic segmentation accuracy presented in Figs. 3, 4, 6, and 8c. The color blue denotes the true positive (TP) pixels, to which the model predicts correctly as given in the ground truth provided by human annotation. The green color indicates false positive (FP) pixels, which means the model has inaccurately predicted that these pixels belong to a particular nanowire. The red color denotes the false negatives (FN), which depict the pixels that belong to a nanowire (based on the ground truth) but were not identified as such by the model. The performance of the trained model is discussed in more detail in the following sections.
a Test image. b Predicted instance masks with lower opacity plotted on the test image. c Semantic binary mask. Blue: TP, Red: FN, Green: FP; every nanowire instance in the present synthetic image has been successfully segmented with highest accuracy and negligible deviation in the overlapping regions and particle boundaries.
Note that to make the model generally accessible as a segmentation tool of nanorod-like structures, we have further developed a web-based interactive application for readers to access and data-mining their own image datasets. Upon uploading the data and initializing the prediction model, statistics on predicted masks can be obtained and visualized accordingly. Details and access on the web-based interactive application can be found in data and code availability section.
Synthetic nanowire image
As for the synthetic nanowires shown in Fig. 3, not surprisingly, the deep learning model correctly segments particles contained within the synthetic nanowire datasets. The obtained AP of the tested 20 models reach a AP score of around 90 for bounding box regression and mask segmentation, respectively, see Table 1. As the synthetic image type is basically known to the model, the good accuracy is expected. Tuning the hyperparameters, test dataset would generally result in a better AP in bounding box prediction and segmentation masks, however, not influence the results on synthetic images significantly. To demonstrate the power of our model in instance segmentation of overlapped, optical density-based nanowire images, we additionally compared the prediction capabilities of our model to a traditional routine algorithm—the distance map-based Watershed algorithm—for a simple exemplary synthetic image. Supplementary Figs. 3–12 in Supplementary Note 2 demonstrated the superiority of our model, while the Watershed algorithm generally fails to isolate individual nanowires in the overlapped regions.
X-ray ptychography image
Although the model has been trained solely by synthetic datasets, good segmentation results are observed on experimental datasets. For the presented X-ray ptychography image, the model predicts the overall binary mask with good accuracy and scores a segmentation score of 86.6, see Table 2. The AP, AP75 for both the bounding box, and the segmentation mask are comparatively high and score around 40, respectively. AP50 reaches a score of around 62. From the metrics, it is mentionable that APl are greater than APs and APm, indicating that the image contains larger particles and they were segmented to a greater degree than smaller ones. At the instance level, two false-positive nanowires have been identified and are (shown in green) indicated by white arrows in Fig. 4c; the origin of this false-positive result is low pixel intensity near the threshold that separates particles from the background. For the same reason, these particles were not manually annotated but nevertheless were identified by the model thus demonstrating its performance, which is competitive with careful human annotation (while being much more accurate). The two particles shown in red (indicated by yellow arrows) are missed by the model, presumably by the noise in the corresponding region. But overall, considering the optical density input, the individual particles are extracted with good accuracy. In general, the mask predictions consistently perform extremely well when particles are well separated in space while overlapping regions (notoriously more difficult to segment) are still identified with good accuracy. Some issues arise when the optical density gradient is low with less clear transitions in the overlapped region. Some issues arise when the optical density gradient is low with less clear transitions in the overlapped region. Further, the model tends to find smaller particles within larger instances as shown by the cyan arrow. The origin of this limitation stems from the broad range of particle aspect ratios and thicknesses in the experimental data (ca. 50–500 nm), thus resulting in a less continuous optical density distribution with individual particles. In the training data, particle morphologies were generated assuming a prismatic structure with little-to-no variation in the cross-sectional shape. In contrast, the experimentally synthesized V2O5 are subject to defect formation, particle sintering, and intrinsic variations in the crystal growth during synthesis. This leads to a particle dispersion that is highly complex, non-prismatic and an ambiguous optical density mapping and further complicates the detection in the overlapped area and create artifacts which would possibly mislead trained models. This also introduces additional challenges during synthetic data generation and the segmentation tasks. Nevertheless, this complexity in particle size, shape, and extent of curvature has pronounced effects on the emergent properties of these cathode particles so their correct identification remains important5,6,30. Here, over-predicted particle masks inside the larger ones can be easily removed in a post-processing step. This step was not performed here to preserve the originality of the model prediction. However, to enhance the general prediction capability of the model and avoid post-processing procedures, the generation of non-prismatic structures for training datasets can be instrumental and will remain as future work. Lastly, statistical information on particle area size, aspect ratio and orientation are compared in Fig. 5 in form of histogram with kernel density estimation (KDE). The area size is determined by summation of the pixels belonging to different particle masks. The aspect ratio is calculated as the ratio between image coordinates of the longer edge to the shorter edge of the corresponding predicted mask. The orientation is considered as the angle between the particle alignment to the horizontal axis and ranging from 0 to 180 degree. As can be found in Fig. 5, the statistical information from the X-ray ptychography data is in a qualitatively good agreement. The main discrepancy is contributed by the number of additionally detected smaller particles inside the larger particles and as explained previously, leading to a higher density of the histogram for area size of 2000–4000 pixels and aspect ratio 2–4. This perturbation can be also observed for orientation for particles with 75 to 100 degrees and 150 to 175 degrees. As the number of particles in the image is comparably small, the feature distribution becomes sensitive to the number of detected particles.
a Test image. b Predicted instance masks with lower opacity plotted on the test image. The best performer in AP segmentation mask in Table 2 was used to visualize the prediction masks in b. c Semantic binary mask. Blue: TP, Red: FN, Green: FP; yellow arrows indicate the main FN pixels, i.e., nanowire pixels missed by the model. White arrows indicate the FP pixels, which were overlooked in the manual annotation, but detected by the model. Cyan arrows indicate over-predicted pixels, of which the model predicts them as additional nanowires, but were only regions of higher intensity.
Histograms and their kernel-density estimates (KDE) are illustrated as a function of area (summation of pixels corresponding to each particle mask), aspect ratio (particle length/width), and orientation (angle relative to the horizontal axis) in a–c, respectively. The statistical results show qualitative agreement; however, the main discrepancy represented by over-predicted pixels cause the shift of KDE curve in the respective number ranges.
STXM image
Of the two X-ray microscopy techniques considered in this work, X-ray ptychography offers the greatest spatial resolution (ca. 6 nm), thus, from a purely image segmentation perspective we expect the performance of the model to be greatest for this class of images. Nevertheless, techniques such as scanning transmission X-ray microscopy which offer slightly lower spatial resolution (ca. 25 nm) but enabled more detailed mapping of spectral features (i.e. have richer chemical information) are equally important for cheminformatics. In this work, the original resolution of the STXM image was 100x100 pixels and fewer than in the X-ray pytchography image. To enable sharper manual annotation, we rescaled the STXM image to the size of pytchography image for easier visual access (it is important to note that this does not fundamentally change the resolution enabled by the experimentation). The number of particles, their variations in morphology, and the complexity of their dispersion is noticeably greater than the X-ray ptychography image making the segmentation task considerably more challenging. Nevertheless, the segmentation accuracy scores around 75 and the AP score is around 30/21 for bounding box and masks, respectively. Since there exist relatively no larger particles in the image, APl was not provided. The comparably lower but surprisingly good scores highlight the complexity of segmenting complex particle dispersions with several instances overlap and agglomeration. This is especially noticeable for the false positive green particles in Fig. 6c (see also Table 3), which were generally overlooked in the manual annotation process. The access of human annotation is strictly limited for image of such complexity. However, at the instance level, from a visual perspective, the model performs considerably well. Overlapped particles are consistently identified and agglomerations, while difficult to identify visually, are captured by the deep learning model. The statistical distribution of the features in Fig. 7 agree well both to a qualitative and quantitative extent. The shape of KDE agrees well with the manual ground truth distribution. As the model prediction captured smaller particles, which were not manually labeled, the statistics of the prediction are shown to have a higher density distribution in each feature characteristic, observable as small peak shift of the KDE curve. The results suggest that for complicated particle networks contained in a relatively low-resolutioned and low-contrast image, deep learning models indeed deliver more comprehensive information on the statistical information than human annotations. We note that further comparison of manually annotated STXM images and their corresponding statistics can be found in Supplementary Note 5.
a Test image.b Predicted instance masks with lower opacity plotted on the test image. The best performer in AP segmentation mask in Table 3 was used to visualize the prediction masks in b. c Semantic binary mask, Blue: TP, Red: FN, Green: FP; in c, many FP nanowires are present, which indicate the missing manual labels, but detected by the model. The results demonstrate the robustness of the model in segmenting image of low-resolution and densely packed particles.
Histograms and KDE curves are illustrated as a function of area, aspect ratio, and orientation in a–c, respectively. The statistical results show good qualitative and quantitative agreement. The main reason for the shift of KDE curves are due to the FP pixels, leading to higher density estimates in the respective number range.
SEM image
The deep learning model shows success in segmenting the particles in the ptychography and STXM optical density images, in part, due to the mechanisms of contrast generation, which involves the transmission of an incident X-ray source through the bulk of the material. Here, the degree of transmission is related to the energy-specific elemental absorption cross-section, corresponding to excitation of electrons from core levels to unoccupied or partially occupied states, giving rise to similar absorption contrast for compositionally homogeneous particles and allowing the discernment of overlapped intersections (in form of increased optical density due to thickness effects). As a point of comparison, in scanning electron microscopy, the detection of secondary electrons or backscattered electrons from the surface is sensitive to surface morphology, edge effects, and charge build-up and is fundamentally different from the X-ray ptychography and STXM images shown in previous sections. To demonstrate the versatility of the model, we introduce a scanning electron micrograph for the purposed of segmentation. Despite the fundamental differences in contrast generation between the data used to train the model and the SEM data utilized as an input here, the model performs sufficiently well in the overlapping regions despite the absence of optical density information. We postulate that the deep learning model still captures the contrast gradients at the particle boundaries and utilizes it as a criterion to identify individual fibers in the overlapped regions, independent of the background. Nevertheless, residual errors in the segmentation persist; for instance, as in the previous dataset, agglomerations are not well separated, as shown by the cyan arrow in Fig. 8b (see also Table 4). In addition to unidentified nanowires, (indicated by yellow arrows) minor discrepancies exist in the rightmost region of the image, where oversized masks (green, indicated by white arrows) were predicted for individual wires. Here, the absence of optical density information combined with the distinct mode of contrast relative to STXM and X-ray ptychography contribute to the lower AP shown for the SEM image. Nevertheless, we see strong potential to improve the observed inconsistency. For example, an additional class for agglomerated phase can be introduced and generated in the microstructure generation step, to further differentiate between different foreground phases and, subsequently, nanowires instances. Despite the lower AP score around 13, the statistical results seem to be less sensitive in the presence of more significant particle numbers. The main deviation lies in the number of undetected isolated particles of smaller size and therefore leading to an underestimation of statistical density w.r.t. area size up to 1000 pixels, aspect ratio from 2.5 to 7.5 and orientation from 100–150 degrees, as shown in Fig. 9a–c. We further refer to Supplementary Figs. 19 and 20 for another comparative case of SEM image with highly attached nanowires.
a Test image. b Predicted instance masks with lower opacity plotted on the test image. The best performer in AP segmentation mask in Table 4 was used to visualize the prediction masks in b. c Semantic binary mask. While sufficiently good segmentation results are found for overlapping nanowires, several FN pixeled nanowires, indicated by yellow arrows in b were missed by the model. Further discrepancies can be found in the region of agglomeration denoted by FP pixels and white arrows. The absence of optical density information combined with the distinct mode of contrast relative to STXM and X-ray ptychography contribute to the comparably lower prediction performance.
Histograms and KDE curves are illustrated as a function of area, aspect ratio, and orientation in a–c, respectively. The statistical results show good qualitative agreement. The main discrepancy exists for smaller FN nanowires missed by the model, thus leading to higher-density estimates in the ground truth (manual annotation) in the respective number ranges, e.g. the peak in area size distribution.
Discussion
Advancements in chemical imaging have enabled improvements in image collection speed, signal-to-noise, and computational power enabling the stronger connections between structure-function relationships and emergent properties. In this work, we have developed a deep learning model for instance segmentation which has been trained solely by synthetic datasets emulated after experimental microscopy data. A benchmarked assessment of the Mask R-CNN algorithm assesses the prediction accuracy of the model on experimental datasets collected by X-ray ptychography, scanning transmission X-ray microscopy, and scanning electron microscopy. Despite variations in spatial resolution, particle dispersion densities, and contrast generation, the model scores well across all the considered microscopy data thus demonstrating the versatility of the neural network in the segmentation tasks. The introduction of optical density values in the synthetic datasets utilized to train the model enables accurate predictions and individual segmentation instances for overlapping particles—a challenge that is strongly relevant to particle dispersions but has seen limited advancement due to inherent complexities. The Mask R-CNN algorithm is designed for instance segmentation, which includes regional proposal networks that can locate instances in a large scale image, and further network components can differentiate the class of objects. Given the one class of nanowires in our current work, further instance classes can be included easily within our synthetic image generation workflow and would provide higher generalization capability and accuracy. The segmentation capability of the model can be further extended towards coaxial or hiearchical nanowires structures, see Supplementary Note 8. A web-based, and interactive segmentation tool based on the model developed in this work has been made publicly available at https://share.streamlit.io/linbinbin92/V2O5_app/V2O5_app.py. Future work will also focus on the introduction of noise to replicate light perturbations, angle dependencies, and variable background characteristics in order to improve the model’s robustness to real-life datasets. In light of considerable ongoing developments in situ X-ray and electron microscopy techniques45, we will further seek to implement the developed methods in real-time process control settings during nanowire growth and battery operation. Here, a continuous stream of image-derived particle statistics will aid identification of change points and targeted processing interventions to control particle geometry during manufacturing; elaboration of the methods to chemical imaging will enable coupling of real-time imaging to control dynamics (through appropriate transfer functions) for early detection of electrode degradation during battery operation46,47,48. Our current model achieves a frame rate per second (fps) of approximately 1 using a conventional graphics processing units (GPU) of type RTX 2070. Further improvements in processing speed are expected using a more performant GPU card with further model optimizations.
Methods
Synthesis of V2O5 nanowires
It is important to note here that two polymorphs of V2O5 are featured in this work. The α-V2O5 polymorph is the thermodynamically favorable phase in the V2O5 system and crystallizes in a layered structure with orthorhombic symmetry. ζ-V2O5 is a metastable 1D tunnel-structured polymorph that preserves the composition of its thermodynamically stable counterpart while exhibiting drastically different structural motifs34. While distinct in terms of their crystal structure, both α-V2O5and ζ-V2O5 produce nearly identical nanoparticle morphologies during a hydrothermal synthesis. Nanowires of α-V2O5, the thermodynamic sink for binary vanadium oxides, were synthesized by a hydrothermal growth process. Briefly, V3O7 ⋅ H2O nanowires were initially prepared and calcined in air to obtain α-V2O5 nanowires crystallized in the orthorhombic phase, as reported previously5. Typical dimensions span from 50 to 400 nm in width and up to several microns in length. Chemical lithiation was achieved via submersion into a 0.01M n-butyllithium solution in heptane. Metastable ζ-V2O5 nanowires were prepared by a series of hydrothermal reactions as described in the previous work49. Briefly, bulk V2O5 and silver acetate were hydrothermally reacted to form an intermediate β-Ag0.33V2O5 product. To create the tunnel-structured ζ-V2O5, β-Ag0.33V2O5 was hydrothermally reacted with HCl in aqueous conditions to leach the Ag from the structure. For electrochemical sodiation, CR2032 coin cells were prepared under an inert argon environment. The working electrode was prepared by casting a mixture of the active material (ζ-V2O5, 70 wt.%), conductive carbon (Super C45, 20 wt.%), and binder [poly(vinylidene fluoride) 10 wt.%] dispersed in N-methyl-2-pyrrolidone onto an Al foil substrate. Sodium metal and glass fiber were used for the counter electrode and separator, respectively. For the electrolyte, 1M NaPF6 solution was prepared using a solvent mixture of ethylene carbonate and diethyl carbonate (1:1 volumetric ratio). The extent of sodiation was controlled by galvanostatic discharging using a LANHE (CT2001A) battery testing system. Cells were disassembled, washed with dimethyl carbonate (DME), and dried for 24 h in an inert argon environment.
Scanning electron microscopy
Before imaging, dispersions of α-V2O5 and ζ-V2O5 nanowires were created by drop-casting onto a silicon nitride substrate5. The SEM image shown in Fig. 1a was collected on a Tescan LYRA-3 instrument equipped with a Schottky field-emission source and a low aberration conical objective lens.
X-ray ptychography
X-ray ptychography measurements were performed at the coherent scattering and microscopy beam line of the Advanced Light Source in Berkeley, CA. An optic with a 60 nm outer zone width, and a 40 nm step-size of the field of view was utilized. The image shown in Fig. 1b depicts the ratio between the X-ray absorption intensities at 527 and 529.8 eV, which correspond to known excitations to t2g and eg*, respectively, which are indicative of the extent of intercalation. The symmetry labels herein indicate transitions to final vanadium 3d-O 2p hybrid states; the line shapes, peak positions, and relative intensities of these absorption features reflect the specifics of vanadium reduction, electronic structure, and chemical bonding in these compounds and have been interpreted with the help of first-principles density functional theory calculations in past work5,6,50.
Scanning transmission X-ray microscopy
The STXM measurements were performed at the spectromicroscopy beam line 10D-1 of the Canadian Light Source in Saskatoon, SK utilizing a 7 mm generalized Apple II elliptically polarizing undulator source (EPU). Here, a focused beam spot was raster-scanned across the field of view with a 35 nm step size (thus determining the spatial resolution). A series of images were collected from 508 eV to 560 eV in 0.2 eV increments. The STXM image shown in Fig. 1(c) depicts the average absorption (optical density) contrast from 508 eV to 560 eV5.
Model architecture of the Mask R-CNN algorithm
In the following section, the basic structure of Mask R-CNN model and its workflow are briefly explained. The model architecture can be divided into 3 main parts15, as illustrated in Fig. 10:
-
Feature extraction. This step is usually referred to as the backbone of the model and is constructed with multiple CNN layers. The input image is introduced and passed through the CNNs to extract representative features of the entire image. The CNN layers are usually deep and contain most of the model weights updated during the training steps. The backbone used here can easily be tailored to the desired segmentation task in order to improve speed and performance. The backbone used for the training in this work is a Feature Pyramid Network (FPN) with the ResNet-50 network14 pre-trained on COCO-dataset15. FPN addresses one of the main challenges in object detection which is detecting objects at different scales51. FPN is constructed of two pathways, a bottom-up and a top-down pathway so it can capture features at different scales. See also Supplementary Note 6 for more details on FPN structures52.
-
Region of interest proposal and alignment. This step in the model is designed to identify and extract instances from feature maps produced by the backbone. The Region proposal network (RPN) achieves this by generating a series of region of interest (ROIs), each encapsulating a single instance. The model generates hundreds of ROIs and an associated a confidence score to quantify the probability of encompassing an object for the given ROI. After filtering and modifying the coordinates of each ROI, RPN advances portions of the feature map (corresponding to each ROI) with a fixed size to the model prediction head in order to determine the properties (e,g, bounding box, and instance mask, etc.) of each instance.
-
Overhead for mask and bounding box prediction and classification. The so-called prediction heads are functions that predicts the characteristics of that proposed instance. For object detection purpose, most common R-CNN structures typically provide two instance heads, namely bounding box regression head, which draws a bounding box around an instance, and the instance classification head to classify the object class. The typical prediction head of Mask R-CNN is therefore a extension of R-CNN models with the mask segmentation, in which a binary mask is generated to label the predicted instance.
Model loss functions
During training, the difference between model prediction and ground truth should be minimized. This optimization procedure requires the definition of a function to perform this calculation, usually referred to a loss function or a cost function. Typically, in neural networks, the optimization goal is to minimize this loss function. Different loss functions can be used for different tasks based on the input data and the desired output of the model. In the Mask R-CNN model used in this work, the defined loss function is based on the summation of 3 individual loss functions15:
where \({{{{\mathscr{L}}}}}_{{{{\rm{bbox}}}}}\) is the smooth L1 loss function used for predicting bounding box coordinates, with the advantage of having steady gradients for large loss numbers and fewer oscillations during updates for smaller loss values. \({{{{\mathscr{L}}}}}_{{{{\rm{cls}}}}}\) is a cross entropy loss function to measure the classification of multiple classes and returns a probability between 0 and 1. In the case of a binary segmentation (in this case, the nanowire and the background), the cross-entropy loss function can be written as:
where the \(\log\) is the natural log, y is a binary value (0 or 1) indicating the class of observation, and p is the predicted probability for the given observation. As for \({{{{\mathscr{L}}}}}_{{{{\rm{mask}}}}}\), it is a binary cross-entropy for the generated binary mask of size m × m for each ROI.
Model evaluation metrics
To evaluate the segmentation results, we made use of three metrics. The segmentation accuracy defined in Eq. (3) is used here as simple pixel-wise correctness, which only examines the true predicted pixels with predictions made from all the objects in the foreground. TP denotes the true positive, FP the false positive, and FN the false negative predictions. Note that this metric is only applied to the foreground, which differs from the common use considering the background pixels likewise.
The second evaluation scheme is according to COCO dataset16 based on mean average precision (mAP). This is introduced briefly in the following section. Firstly, to confirm a correct prediction of bounding box or mask, intersection over union is used (IoU). It is defined by the area of intersection between bounding boxes divided by their union as shown in Fig. 11. Predictions are true positive, if IoU is higher than a given threshold, and false negative if that is lower than that threshold. The most common thresholds used are IoU > 50 (AP50) and IoU > 75 (AP75). To further understand mAP, precision and recall are defined as follows:
Recall is considered as true positive prediction rate, i.e., the ratio between true positive predictions and all ground truths. Precision is defined as the ratio between true positive predictions and all predictions that are made. Further, the obtained precision and recall are plotted to obtain the so-called precision-recall (PR) curve with the area under it referred to as average precision (AP). In VOC201017, a modified PR curve was introduced, where precision for a given recall r is set to the maximum precision for any \(\tilde{r}\ge r\). Afterward, the AP can be computed by numerical integration for the area under the curve (AUC) as shown in Fig. 12. In COCO, mAP is defined as the average of AP for all classes in each image. In this work, a single class of nanowires is segmented, but it is worth noting that additional classes can be accounted for with relative ease.
Exemplary precision values corresponding to given recall values, shown by red dots. According to VOC2010, to avoid the wiggly representation of PR curve, shown by dashed green lines, precision values take the previous maximum values to the left to generate the normalized curve, shown by solid blue lines. The final AP is calculated as the area under the normalized curve.
For VOC, usually IoU > 50 is considered considered true positive prediction, which results in true predictions with any IoU higher than 0.5 contributing equally to the AP. To rectify this problem, COCO uses different thresholds for IoU ranging from 0.5 to 0.95 with a step size of 0.05 and then reports the average of all computed APs to varying thresholds as mAP, see Eq. (6). In this work, we use AP for mAPcoco and assume the difference is clear from the context.
Evaluation based on COCO also reports more detailed results based on the scale of the detected objects. With reported APsmall for objects with area smaller than 322 pixels, APmedium for objects with area between 322 and 692 pixels and APlarge for objects with area greater than 692 pixels, one can evaluate the model performance on segmenting objects in different scales. Finally, given the fundamental motivation to extract particle statistics from image datasets, the performance of the model is further evaluated based on the accuracy of the predicted statistics. Computation is made based on the segmented masks of each particle. Statistical information obtained from the predictions are then compared to the manually annotated results.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data can be further found in the Supplementary Information. The synthetic image dataset for training has been made publicly accessible in https://github.com/linbinbin92/V2O5_app/tree/master.
Code availability
The code required to reproduce the findings is available under https://github.com/linbinbin92/V2O5_app/tree/master. The interactive web-based segmentation application can be found at: https://share.streamlit.io/linbinbin92/V2O5_app/V2O5_app.py.
References
Baker, L. A. Perspective and prospectus on single-entity electrochemistry. J. Am. Chem. Soc. 140, 15549–15559 (2018).
Li, W. et al. Peering into batteries: electrochemical insight through in situ and operando methods over multiple length scales. Joule 5, 77–88 (2021).
Wolf, M., May, B. M. & Cabana, J. Visualization of electrochemical reactions in battery materials with x-ray microscopy and mapping. Chem. Mater. 29, 3347–3362 (2017).
Yu, Y.-S. et al. Three-dimensional localization of nanoscale battery reactions using soft x-ray tomography. Nat. Commun. 9, 1–7 (2018).
Santos, D. A. et al. Bending good beats breaking bad: phase separation patterns in individual cathode particles upon lithiation and delithiation. Mater. Horiz. 7, 3275–3290 (2020).
Andrews, J. L. et al. Curvature-induced modification of mechano-electrochemical coupling and nucleation kinetics in a cathode material. Matter 3, 1754–1773 (2020).
Mandic, M., Todic, B., Zivanic, L., Nikacevic, N. & Bukur, D. B. Effects of catalyst activity, particle size and shape, and process conditions on catalyst effectiveness and methane selectivity for Fischer–Tropsch reaction: a modeling study. Ind. Eng. Chem. Res. 56, 2733–2745 (2017).
Lerotic, M., Mak, R., Wirick, S., Meirer, F. & Jacobsen, C. Mantis: a program for the analysis of x-ray spectromicroscopy data. J. Synchrotron Radiat. 21, 1206–1212 (2014).
Luo, Y. et al. Effect of crystallite geometries on electrochemical performance of porous intercalation electrodes by multiscale operando investigation. Nat. Mater. 21, 217–227 (2022).
De Jesus, L. R., Andrews, J. L., Parija, A. & Banerjee, S. Defining diffusion pathways in intercalation cathode materials: some lessons from V2O5 on directing cation traffic. ACS Energy Lett. 3, 915–931 (2018).
Zhao, Y. et al. Modeling of phase separation across interconnected electrode particles in lithium-ion batteries. RSC Adv. 7, 41254–41264 (2017).
Reske, R., Mistry, H., Behafarid, F., Roldan Cuenya, B. & Strasser, P. Particle size effects in the catalytic electroreduction of CO2 on cu nanoparticles. J. Am. Chem. Soc. 136, 6978–6986 (2014).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
He, K., Gkioxari, G., Dollár, P. & Girshick, R. Mask r-cnn. In Proc. IEEE International Conference on Computer Vision 2961–2969 (IEEE, 2017).
Lin, T.-Y. et al. Microsoft coco: common objects in context. In European Conference on Computer Vision 740–755 (2014).
Everingham, M., Van Gool, L., Williams, C. K., Winn, J. & Zisserman, A. The Pascal visual object classes (VOC) challenge. Int. J. Comput. Vis. 88, 303–338 (2010).
Masubuchi, S. et al. Deep-learning-based image segmentation integrated with optical microscopy for automatically searching for two-dimensional materials. npj 2D Mater. Appl. 4, 1–9 (2020).
Frei, M. & Kruis, F. E. Fiber-cnn: expanding mask r-cnn to improve image-based fiber analysis. Powder Technol. 377, 974–991 (2021).
Yildirim, B. & Cole, J. M. Bayesian particle instance segmentation for electron microscopy image quantification. J. Chem. Inf. Model. 61, 1136–1149 (2021).
Ma, B. et al. Data augmentation in microscopic images for material data mining. npj Comput. Mater. 6, 1–9 (2020).
DeCost, B. L. & Holm, E. A. Characterizing powder materials using keypoint-based computer vision methods. Comput. Mater. Sci. 126, 438–445 (2017).
Mill, L. et al. Synthetic image rendering solves annotation problem in deep learning nanoparticle segmentation. Small Methods 5, 2100223 (2021).
Rühle, B., Krumrey, J. F. & Hodoroaba, V.-D. Workflow towards automated segmentation of agglomerated, non-spherical particles from electron microscopy images using artificial neural networks. Sci. Rep. 11, 1–10 (2021).
De Temmerman, P.-J. et al. Measurement uncertainties of size, shape, and surface measurements using transmission electron microscopy of near-monodisperse, near-spherical nanoparticles. J. Nanoparticle Res. 16, 1–22 (2014).
Laramy, C. R., Brown, K. A., O’Brien, M. N. & Mirkin, C. A. High-throughput, algorithmic determination of nanoparticle structure from electron microscopy images. ACS Nano 9, 12488–12495 (2015).
Kinnear, C., Moore, T. L., Rodriguez-Lorenzo, L., Rothen-Rutishauser, B. & Petri-Fink, A. Form follows function: nanoparticle shape and its implications for nanomedicine. Chem. Rev. 117, 11476–11521 (2017).
Monchot, P. et al. Deep learning based instance segmentation of titanium dioxide particles in the form of agglomerates in scanning electron microscopy. Nanomaterials 11, 968 (2021).
Mansfeld, U. et al. Towards accurate analysis of particle size distribution for non-spherically shaped nanoparticles as quality control materials. Microsc. Microanal. 25, 2328–2329 (2019).
Horrocks, G. A., Likely, M. F., Velazquez, J. M. & Banerjee, S. Finite size effects on the structural progression induced by lithiation of V2O5: a combined diffraction and Raman spectroscopy study. J. Mater. Chem. A 1, 15265–15277 (2013).
Santos, D. A., Dixit, M. K., Kumar, P. P. & Banerjee, S. Assessing the role of vanadium technologies in decarbonizing hard-to-abate sectors and enabling the energy transition. Iscience 24, 103277 (2021).
Whittingham, M. S. The role of ternary phases in cathode reactions. J. Electrochem. Soc. 123, 315 (1976).
De Jesus, L. R. et al. Mapping polaronic states and lithiation gradients in individual V2O5 nanowires. Nat. Commun. 7, 1–9 (2016).
Luo, Y. et al. Cation reordering instead of phase transitions: origins and implications of contrasting lithiation mechanisms in 1d ζ-and 2d α-V2O5. Proc. Natl Acad. Sci. USA 119, e2115072119 (2022).
Skalski, P., makesense.ai, https://www.makesense.ai/ (2021).
Math2Market GmbH. GrainGeo online manual. https://doi.org/10.30423/userguide.geodict2021-graingeo (2021).
Huang, Z., Huang, L., Gong, Y., Huang, C. & Wang, X. Mask scoring r-cnn. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 6409–6418 (IEEE, 2019).
Chen, X., Girshick, R., He, K. & Dollár, P. Tensormask: a foundation for dense object segmentation. In Proc. IEEE/CVF International Conference on Computer Vision 2061–2069 (IEEE, 2019).
Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You only look once: unified, real-time object detection. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 779–788 (IEEE, 2016).
Bai, H. & Wu, S. Nanowire detection in afm images using deep learning. Microsc. Microanal. 27, 54–64 (2021).
Bolya, D., Zhou, C., Xiao, F. & Lee, Y. J. Yolact: real-time instance segmentation. In Proc. IEEE/CVF International Conference on Computer Vision 9157–9166 (IEEE, 2019).
Konopczyński, T. K., Kröger, T., Zheng, L. & Hesser, J. Instance segmentation of fibers from low resolution CT scans via 3D deep embedding learning. In BMVC (2019).
Aguilar, C., Comer, M., Hanhan, I., Agyei, R. & Sangid, M. 3d fiber segmentation with deep center regression and geometric clustering. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 3746–3754 (IEEE, 2021).
Wu, Y., Kirillov, A., Massa, F., Lo, W.-Y. & Girshick, R. Detectron2. https://github.com/facebookresearch/detectron2 (2019).
Taheri, M. L. et al. Current status and future directions for in situ transmission electron microscopy. Ultramicroscopy 170, 86–95 (2016).
Park, C. & Ding, Y. Data Science for Nano Image Analysis, Vol. 308 (Springer, 2021).
Wang, J., Chen-Wiegart, Y.-cK., Eng, C., Shen, Q. & Wang, J. Visualization of anisotropic-isotropic phase transformation dynamics in battery electrode particles. Nat. Commun. 7, 1–7 (2016).
Lo, Y. H. et al. In situ coherent diffractive imaging. Nat. Commun. 9, 1–10 (2018).
Andrews, J. L. et al. Reversible mg-ion insertion in a metastable one-dimensional polymorph of V2O5. Chem 4, 564–585 (2018).
Maganas, D. et al. First principles calculations of the structure and V L-edge x-ray absorption spectra of V2O5 using local pair natural orbital coupled cluster theory and spin–orbit coupled configuration interaction approaches. Phys. Chem. Chem. Phys. 15, 7260–7276 (2013).
Adelson, E. H., Anderson, C. H., Bergen, J. R., Burt, P. J. & Ogden, J. M. Pyramid methods in image processing. RCA Engineer 29, 33–41 (1984).
Lin, T.-Y. et al. Feature pyramid networks for object detection. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 2117–2125 (IEEE, 2017).
Acknowledgements
This work is supported by German Research Foundation (DFG) B.L. and B.-X.X. acknowledge the financial support under the grant agreement No. 405422877 of the Paper Research project (FiPRe) and the Federal Ministry of Education and Research (BMBF) and the state of Hesse as part of the NHR Program. The authors gratefully acknowledge the computing time granted by the NHR4CES Resource Allocation Board and provided on the supercomputer Lichtenberg II at TU Darmstadt as part of the NHR4CES infrastructure. The calculations for this research were conducted with computing resources under the project project1020, special0007. We acknowledge Abhishek Parija and Dr. David Schapiro for their assistance with X-ray ptychography experiments. X-ray ptychography measurements were performed at the COherent Scattering and MICroscopy (COSMIC) branch of the Advanced Light Source (ALS). A portion of the STXM measurements utilized in this work was collected at the Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. The research at Texas A&M University was supported by the NSF under DMR 1627197. D.A.S. acknowledges support under a NSF Graduate Research Fellowship under grant No. 1746932.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
B.L. conceived the research. B.L. and N.E. performed the dataset generation and model training and the formal analysis. D.A.S. was involved in the conceptualization of the project. Y.L. prepared the particles shown in the images. B.L., N.E., and D.A.S. drafted the manuscript. B.X.X. and S.B. reviewed and discussed the results and organized the funding.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, B., Emami, N., Santos, D.A. et al. A deep learned nanowire segmentation model using synthetic data augmentation. npj Comput Mater 8, 88 (2022). https://doi.org/10.1038/s41524-022-00767-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-022-00767-x
- Springer Nature Limited
This article is cited by
-
Uncertainty-aware particle segmentation for electron microscopy at varied length scales
npj Computational Materials (2024)