Abstract
The neurovascular unit (NVU) is a complex multicellular structure that helps maintain cerebral homeostasis and blood-brain barrier (BBB) integrity. While extensive evidence links NVU alterations to cerebrovascular diseases and neurodegeneration, the underlying molecular mechanisms remain unclear. Here, we use zebrafish embryos carrying a mutation in Scavenger Receptor B2, a highly conserved endolysosomal protein expressed predominantly in Radial Glia Cells (RGCs), to investigate the interplay among different NVU components. Through live imaging and genetic manipulations, we demonstrate that compromised acidification of the endolysosomal compartment in mutant RGCs leads to impaired Notch3 signaling, thereby inducing excessive neurogenesis and reduced glial differentiation. We further demonstrate that alterations to the neuron/glia balance result in impaired VEGF and Wnt signaling, leading to severe vascular defects, hemorrhages, and a leaky BBB. Altogether, our findings provide insights into NVU formation and function and offer avenues for investigating diseases involving white matter defects and vascular abnormalities.
Similar content being viewed by others
Introduction
In recent years, the concept of the neurovascular unit (NVU) has gained prominence in the fields of neuroscience and vascular biology. The NVU refers to a complex multicellular structure within the brain comprising neurons, astrocytes, pericytes, and specialized endothelial cells (ECs), which work in concert to help maintain cerebral homeostasis, by controlling blood flow and ensuring the proper functioning of the blood-brain barrier (BBB)1,2. ECs forming the BBB vessel wall exhibit a distinctive array of characteristics that set them apart from ECs found in other vascular beds3. Their remarkable organotypicity stems from their ability to tightly control the passage of substances between the bloodstream and the brain parenchyma due to the presence of tight junctions, which form a nearly impermeable barrier that shields the delicate neural environment from potential toxins and fluctuations in systemic composition, as well as a specific set of transporters. Recent studies have also revealed the unique gene expression repertoire of BBB-ECs4,5. While this EC specialization is fundamental for maintaining brain homeostasis and safeguarding neurological integrity, its underlying molecular basis has only recently begun to be elucidated.
Brain vascular abnormalities can lead to various, sometimes life-threatening forms of cerebrovascular disease6, including ischemic and hemorrhagic stroke, vascular malformations, vascular cognitive impairment, and dementia. Within this category, cerebral small vessel diseases (CSVDs) encompass a range of sporadic or hereditary pathological alterations of cerebral arterioles, capillaries, and venules, occurring primarily within the white matter. These changes contribute to demyelination, BBB dysfunction, glial activation, and axonal loss7. Despite the growing recognition of the crucial roles played by neuro-vascular and glia-vascular interfaces in the formation and functioning of the NVU and BBB, most of the research on cerebrovascular diseases and CSVD, in particular, has primarily focused on characterizing pericyte and EC dysfunction. In contrast, our understanding of how cells of the neural parenchyma contribute to CSVD and the molecular signaling pathways underlying their pathological interactions with the vasculature is poorly understood.
During embryonic development, multipotent radial glia cells (RGCs) serve as the primary source of neurons and glia (i.e., astrocytes and oligodendrocytes)8. Oligodendrocytes (OL)—the myelinating cells of the central nervous system (CNS)—, and astrocytes, star-shaped glial cells with specialized endfeet that ensheath blood vessels, arise from RGCs and undergo proliferation, migration, and differentiation to generate the diverse population of glial cells. The timing and spatial distribution of glial cell development are tightly regulated by a combination of intrinsic genetic programs, extrinsic signals from neighboring cells, and environmental factors. Among them, the Notch signaling pathway, particularly the Notch3 receptor, has been shown to play critical roles in promoting RGC differentiation toward glial lineages9,10,11.
Here, we report the identification of a zebrafish mutant featuring impaired RGC differentiation and aberrant gliogenesis, which lead to defective cerebral angiogenesis, early onset of micro-hemorrhages, BBB dysfunction, and compromised myelination, all highly reminiscent of CSVD phenotypes. The mutation affects Lysosome Membrane Protein II (Limp2)/Scavenger Receptor Class B Member 2 (SCARB2), a protein found in the membrane of late endosomes and lysosomes12, highly conserved in vertebrates, including humans (Supplementary Fig. 1a). Using live imaging and genetic manipulations, we identify compromised acidification of the endolysosomal compartment in mutant RGCs as one of the major outcomes of Scarb2a depletion and illustrate how this phenotype leads to disrupted Notch3 processing and impaired Notch3 signaling, thereby resulting in excessive neurogenesis and aberrant glia differentiation. Finally, we demonstrate that alterations to the neuron/glia balance result in impaired vascular endothelial growth factor (VEGF) and Wnt signaling activation, leading to severe vascular defects, hemorrhages, and a leaky BBB. Overall, our findings highlight the link between aberrant glial cell differentiation and cerebrovascular defects and demonstrate that hindering Notch intracellular processing in non-vascular cells elicits pronounced defects throughout the CNS vasculature, further impacting the formation and functionality of the BBB.
Results
scarb2a mutants feature defective CNS angiogenesis, a dysfunctional BBB, and sporadic intracranial hemorrhage
We generated scarb2a mutants as part of a CRISPR/CAS-based mutagenesis screen aimed at identifying genes controlling CNS angiogenesis. The mutation induces a premature stop codon at amino acid 73, resulting in the absence of the conserved C-terminal transmembrane domain and cholesterol binding site (Supplementary Fig. 1a, b).
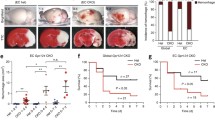
At 48 h post-fertilization (hpf), scarb2a mutants are readily distinguishable from their wt siblings based on their short body length and curved trunk (Fig. 1a, b). In addition, around 25–30% of the mutants display intraventricular hemorrhage, primarily in the hindbrain, starting at ~40 hpf (Fig. 1b, arrow, Supplementary Fig. 1c). Homozygous larvae die between ~8–14 days post-fertilization (dpf), independently of the presence of hemorrhage (Supplementary Fig. 1d) indicating that the hemorrhage per se is not the main cause of lethality. Heterozygous animals, in contrast, reach adulthood and are viable and fertile. At 4 dpf, behavioral abnormalities previously associated with epileptic seizures, such as burst activity and whole-body trembling13, were also observed.
a, b Brightfield images of wild-type (wt) and scarb2a−/− embryos at 48 hpf depicting morphological defects and intraventricular hemorrhage (b, arrow) in the mutants (Nexperiments=3, for each nwt = 10, nscarb2a−/−= 10). c Schematic diagram illustrating dorsal views of the developing zebrafish hindbrain vasculature and its different components, as shown in d–i. d–i Confocal images of Tg(kdrl:mCherry) wt and scarb2a−/− at 34 (d, e), 38 (f, g) and 52 (h, i) hpf, showing increasing defects in the morphology and patterning of mutant CtAs, starting at 38 hpf (for each developmental stage Nexperiments = 3, nwt = 12, nscarb2a−/−=12). j Schematic reconstruction of wt and scarb2a−/− hindbrain vasculature at 60 hpf depicting parameters quantified in k. k Quantification at 60 hpf shows no differences in CtA sprouting from the PHBCs or CtA connections to the BA, but increased numbers of CtAs interconnections (n = 13 embryos, two-tailed Student’s t-test, PPHBC = 0.46, PBA = 0.1214, PCTAs = 0.0038) in mutant hindbrains. l, m Selected confocal images from a time-lapse series of Tg(fli1:EGFP;gata1a:DsRed) wt and scarb2a−/− show extravasation of red blood cells (RBCs) from the CtAs in the mutants, starting at ~44 hpf (m, arrow, Nexperiments = 3, for each nwt = 1, nscarb2a−/−=1). n, o Intravascular injection of Rhodamine Dextran 2000 KDa at 60 hpf demonstrates compromised integrity and enhanced permeability of BBB vessels in mutant embryos (* mark unspecific dye accumulation in the skin, Nexperiments = 3, for each nwt = 5, nscarb2a−/−=5). p, q' Confocal images of Tg(kdrl:EGFP;glut1b:mCherry) show strong downregulation of glut1b expression in scarb2a mutants (co-localization channel shown in white, Nexperiments = 3, for each nwt = 6, nscarb2a−/−=6). CtAs central arteries, PHBC Primordial hindbrain channel, BA basal artery. Error bars are mean ± s.e.m. *P < 0.05, ** P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not statistically significant. Scale bars, a, b = 100 μm; d–i; k, l; m, n; p, q‘= 50 μm. Source data are provided as a Source Data file.
In order to ascertain the nature of the bleeding phenotype, we first obtained confocal images of the EC-specific reporter Tg(kdrl:mCherry), which revealed severe defects, particularly in the loop-shaped Central Arteries (CtAs). The CtAs arise as angiogenic sprouts from the dorsal surface of the Primordial Hindbrain Channels (PHBCs) between 32–36 hpf and gradually invade the hindbrain where they anastomose with the Basal Artery (BA)13,14 (Fig. 1c). At 32–34 hpf, no significant defects were observed in the PHBCs, BA, or CtAs of scarb2a mutants (Fig. 1d, e; Supplementary Fig. 1e-g). By ~38 hpf, however, wt CtAs establish connections with the BA, lumenize, and lose filopodia (Fig. 1f), whereas mutant CtAs are greatly disorganized, present numerous filopodia extensions, and establish abnormal interconnections (Fig. 1g). This phenotype worsens by 52 hpf, when wt hindbrains display a well-structured system of stereotypically patterned CtAs (Fig. 1h, j), while scarb2a mutants present a disorganized network of collapsed and hyper-branched capillaries surrounding the BA (Fig. 1i–k). By 60 hpf, we also detected increased numbers of EC nuclei, specifically in the CtAs, suggesting that the phenotypes involve excess EC proliferation as well (Supplementary Fig 1h–j). Notably, these phenotypes were specific to the brain vasculature, as no defects were observed in the trunk intersegmental vessels at similar developmental stages (Supplementary Fig. 1k, l). We then proceeded to evaluate the emergence of the hemorrhagic bleeding by time-lapse imaging Tg(fli1:EGFP;gata1a:DsRed);scarb2a−/− embryos between 40–60 hpf (Fig. 1l, m and Supplementary movie 1). This double transgenic reporter combines the pan-endothelial marker fli1 with the erythrocyte marker gata1 enabling simultaneous visualization of the vascular network and the circulating red blood cells15. We found that in wt embryos, gata1+ erythrocytes remain normally confined within the hindbrain capillaries throughout the course of the experiment. In contrast, increasing hemorrhagic areas are detected in the hindbrains of scarb2a mutants starting at ~44 hpf (Fig. 1m, arrow), resulting in massive bleeding from the CtAs into the ventricles. To investigate if scarb2a mutants also display a dysfunctional BBB, we conducted intravascular injection of 2000 kDa Rhodamine Dextran, a tracer known to be retained within the BBB of zebrafish embryos at 60 hpf14. As seen in Fig. 1n, o, scarb2a mutants presented clear extravasation of the dye from the CtAs, suggesting compromised integrity of the BBB. Moreover, the expression of Glucose transporter 1 (Glut1), a well-established marker of BBB ECs in vertebrates16, was markedly reduced at 60 hpf as evidenced by both Tg(glut1b:mCherry;kdrl:EGFP) confocal images (Fig. 1p, q’) and whole mount in situ hybridization (Supplementary Fig. 1m, n). Thus, scarb2a mutants feature defective CNS angiogenesis, accompanied by a dysfunctional and leaky BBB and sporadic intracranial hemorrhage.
scarb2a is expressed in RGCs, and its absence results in excessive neurogenesis
Notably, the presence of Scarb2 in ECs has not been described17. Analysis of single-cell RNA-sequencing (scRNA-Seq) data18 derived from mouse developing brain and spinal cord, highlighted microglia, newly-formed OLs (NFOLs), and oligodendrocyte precursor cells (OPCs) among the top 10 cell types expressing Scarb2 (Supplementary Fig. 2a). Similarly, Scarb2 is primarily expressed by OPCs19,20 and microglia21 in humans. In zebrafish embryos, the presence of scarb2a mRNA was reported in the notochord and throughout the developing CNS at 24 hpf22. However, the specific cell type in the brain expressing scarb2a was not described. To accurately map scarb2a expression, we generated a transgenic reporter-TgBAC(scarb2a:KalTA4)- expressing the KalTA4 driver downstream of the scarb2a promoter, following removal of the scarb2a coding sequence (Supplementary Fig. 2b). This strategy enabled visualization of scarb2a-expressing cells both in wt and mutant embryos. Confocal images of TgBAC(scarb2a:KalTA4;UAS:mkate2;kdrl:EGFP) at 22, 48 and 60 hpf confirmed the absence of scarb2a expression in kdrl+ ECs (Fig. 2a, a”,b and Supplementary Fig. 2c-d”). Instead, the scarb2a reporter highlighted cells in the ventricular zone (vz, Fig. 2a’-a”, b) highly reminiscent of radial glial cells (RGCs), a population of multipotent progenitor cells that serve as the primary source of neurons and glia in the developing embryo23. RGCs display a unique elongated morphology (Fig. 2a”’) and express specific markers, such as glial fibrillary acidic protein (GFAP)24 and SOX2, a transcription factor crucial for maintaining them in a progenitor state, preventing their premature neuronal differentiation25,26. Confocal imaging of TgBAC(scarb2a:KalTA4;UAS:EGFP;GFAP:dTomato) embryos revealed that at 24 hpf, a subset of scarb2a+ cells is also labeled by the GFAP transgene (Fig. 2c–d’) and by Sox2 antibody27 (Fig. 2e–f’) in both wt and mutant embryos. To further discern the identity of the scarb2a expressing cells, we immunostained TgBAC(scarb2a:KalTA4;UAS:EGFP) embryos at 36 and 48 hpf, with Fabp7a, previously shown to label RGCs in the zebrafish hindbrain28,29. As seen in Supplementary Fig. 2e-f‘, we found scarb2a (green) and Fabp7a (red) co-expressed (yellow) in a restricted subset of RGCs in the ventricular zone. Thus, based on their morphology, gene expression, and temporal distribution, we identified scarb2a+ cells as RGCs and concluded that they are unaffected by the mutation up to this developmental stage.
a-a‘, Confocal images of Tg(kdrl:TagBFP;scarb2a:KalTA4;UAS-mkate2) showing no colocalization of scarb2a (magenta) and the kdrl (blue) signals at 48 hpf (dashed square in a marks the region displayed in a”). a”, Transverse optical section at the level shown in a demonstrates the presence of scarb2a+ cells in the ventricular zone (vz; dashed square depicts the region enlarged in a”’). a”’, scarb2a+ cells (dashed lines) display morphology typical of RGCs (Nexperiments=3, for each nwt = 9, nscarb2a−/−=9). b Schematic illustration depicting the anatomical organization of RGCs and blood vessels in the zebrafish hindbrain at 48 hpf. c–d‘, Dorsal views of wt and mutant hindbrains showing co-localization of Tg(scarb2a:KalTA4;UAS-EGFP) and Tg(GFAP:dTomato) signal in a subpopulation of RGCs (yellow channel denotes co-localization, dashed squares in c, d mark the level of the optical sections in c’, d’, Nexperiments = 3, for each nwt = 6, nscarb2a−/−=6). e–f’ Dorsal views of wt and mutant hindbrains showing co-localization of Tg(scarb2a:KalTA4;UAS-EGFP) and SOX2 immunostaining in RGCs at the vz (co-localization depicted in white, dashed squares in e–f mark the level of the optical sections in e', f', Nexperiments = 3, for each nwt = 4, nscarb2a−/−=4). g–l Transverse optical sections from a time-lapse series of Tg(scarb2a:KalTA4;UAS-mkate2) in wt (g, i, k) and mutant (h, j, l) embryos. Images show gradual restriction of scarb2a expression to the vz at 28 (g, h), 32 (i, j), and 48 (k, l) hpf in wt fish. In mutant, scarb2a-labeled cells invade the ventricle space starting at 32 hpf (j–l, Nexperiments = 3, for each nwt = 1, nscarb2a−/−=1). m Schematic representation of the zebrafish hindbrain depicting organization of different cell types at 48 hpf. n, o Immunofluorescence staining of Tg(scarb2a:KalTA4;UAS-EGFP) at 48 hpf showing SOX2 expression in scarb2a+ RGCs in the vz of wt embryos (n, magenta), that is utterly absent in the vz of scarb2a mutants (o, Nexperiments = 3, for each nwt = 4, nscarb2a−/−=4). p, q, HuC immunostaining on Tg(scarb2a:KalTA4;UAS-EGFP) embryos at 48 hpf showing HuC+ cells in the mantle zone of wt embryos (p, yellow) and scarb2a+ RGCs in the vz (p, blue). In scarb2a mutants, scarb2a-labeled cells co-express HuC in the vz, mantle zone, and while invading the ventricular space (q, arrow), indicating a neuronal fate (white depicts co-localization channel, Nexperiments = 3, for each nwt = 3, nscarb2a−/−=3). Scale bars, a-a'=50 μm; a"=25 μm; a"'=5 μm; c–f = 50 μm; c'–f', g–l, n–q = 30 μm. vz ventricular zone, v ventricle space.
Since the vascular phenotypes typically appear around 30 hpf, we carried out time-lapse imaging of wt and mutant TgBAC(scarb2a:KalTA4;UAS:mkate2) embryos between 24–40 hpf to identify potential defects in scarb2a+ cells that could lead to the observed vascular abnormalities (Supplementary movie 2). Transverse optical sections at the hindbrain level showed no apparent defects at the initial stages of secondary neurogenesis (~28 hpf) in scarb2a mutants (Fig. 2g, h). However, starting at ~32 hpf, scarb2a−/−-labeled cells from the ventricular zone begin invading the ventricular space (v) in mutant hindbrains (Fig. 2i, j, arrows; Supplementary Fig. 3a, b). These phenotypes become more pronounced as development progresses, leading to disrupted morphology and loss of the midline and bilateral cell organization by ~40 hpf (Fig. 2k, l; Supplementary Fig. 3c, d). At 48 hpf, we observed two well-defined domains in the hindbrains of wt embryos: the vz containing undifferentiated RGCs co-expressing scarb2a:EGFP and Sox2 (Fig. 2m, n and Supplementary Fig. 3e) and the sub-ventricular and mantle zones, with cells labeled by the pan-neuronal marker HuC+ that were largely devoid of scarb2a expression (Fig. 2p and Supplementary Fig. 3g). In contrast, there were no scarb2a/Sox2 double-positive cells in the vz of mutant embryos (Fig. 2o and Supplementary Fig. 3f); rather most cells highlighted by the scarb2a:EGFP reporter were also labeled by the HuC+ antibody (Fig. 2q and Supplementary Fig. 3h), suggesting that they have acquired a neuronal fate. Furthermore, the neuronal domain had significantly expanded, with newly differentiated neurons overtaking the vz and invading the ventricular space (Fig. 2q, arrow). Thus, our findings indicate that scarb2a is expressed in RGCs during early neurogenesis, and its depletion results in the loss of SOX2+ undifferentiated RGCs in the vz accompanied by excessive neurogenesis and overall abnormal hindbrain morphogenesis. Moreover, our data define a concerted temporal onset of both neural and vascular phenotypes.
scarb2a mutants display defective gliogenesis
After neurogenesis is complete, the remaining undifferentiated RGCs generate OLs and astrocytes through asymmetric cell division8,24,30. The marked exhaustion of the RGC pool in scarb2a mutants raises the question of how this impacts gliogenesis. In the zebrafish hindbrain, Olig2-labeled OPCs emerge around 30 hpf from the pMN clusters located at the midline in rhombomeres r5 and r631. To investigate whether Scarb2a depletion affects the OPC and/or OL lineages, we generated scarb2a mutants under olig2:EGFP and mbp:EGFP transgenic backgrounds. Confocal images at 30 hpf showed no differences in the pMNs of wt and scarb2a−/− embryos (Supplementary Fig. 3i, j). By 60 hpf, when the newly generated OPCs begin to migrate to colonize the entire CNS31, wt olig2+ cells were found in the pMNs as well as spread radially throughout the hindbrain (Fig. 3a, a’, c, d). In contrast, scarb2a mutant hindbrains exhibited significantly reduced numbers of olig2+ OPCs, both within and outside the pMNs (Fig. 3b–d), suggesting that the mutation affects the differentiation of a subset of OPCs. Mature OLs produce myelin and express myelin binding protein (MBP), a major constituent of the myelin sheath32. In zebrafish, myelin production begins at 3 dpf, when OPCs start maturing to become myelinating OLs33. Importantly, Tg(mbp:EGFP);scarb2a−/− embryos display a significant decrease in the number of myelinating OLs at 5 dpf (Fig. 3e–g), indicating that the early oligodendrogenesis defects do not recover at later developmental stages. Interestingly, analysis of publicly available scRNA-Seq data derived from 310 cells isolated from Tg(olig1:memEYFP), a zebrafish reporter that specifically labels OPCs and OLs34, revealed the presence of scarb2a in the oligodendrocyte lineage (Supplementary Fig. 3k), suggesting that Scarb2a may potentially play additional roles during later stages of oligodendrogenesis.
a, b’, Dorsal views (a, b) and transverse optical sections (a’, b’) of Tg(olig2:EGFP) wt (a, a’) and scarb2a−/− (b, b’) at 60 hpf showing significant reduction of olig2+ migrating cells and in the pMN domain (dashed lines) in scarb2a mutants. (dashed squares in a, b mark the level of the optical sections showed in a’, b’). c Quantification of olig2+ migrating OPCs (n = 14 embryos, two-tailed Student’s t-test, P < 0.0001) and d olig2+ OPCs in the pMN domain (embryos n = 14 embryos, two-tailed Student’s t-test, P < 0.0001). e–f’, Dorsal views (e, f) and transverse optical sections (e’,f’) of Tg(mbp:EGFP) embryos at 5 dpf depicting reduced numbers of myelinating OLs in scarb2a−/−, quantified in (g) (n = 14 embryos, two-tailed Student’s t-test, P < 0.0001). h–i’ Dorsal views (h, i) and transverse optical sections (h’, i’) of Tg(slc1a3b:MYRGFP-2A-H2AmCherry) x wt (h, h’) and scarb2a−/− (i, i’) showing significantly fewer astrocyte nuclei (red) in the vz of mutant hindbrains at 60 hpf (dashed squares in h, i mark the level of the optical sections shown in h’, i’), quantified in j (n = 11 embryos, two-tailed Student’s t-test, P < 0.0001). k–l', Dorsal views (k, l) and transverse optical sections (k', l') of Tg(slc1a3b:MYRGFP-2A-H2AmCherry) embryos at 6 dpf showing reduced numbers of astrocytes, quantified in m (n = 10 embryos, two-tailed Student’s t-test, P < 0.0001). n–p Dorsal views of Tg(olig2:EGFP;GFAP:Gal4FF) hindbrain in wt (n), scarb2a−/− (o), and scarb2a−/− mutant following injection of UAS-scarb2a-P2A-RFP (p). q Quantification of migrating olig2+ cells (n = 13 embryos; One-way ANOVA, multiple comparisons with Tukey posthoc test). r–t Dorsal views of Tg(slc1a3b:MYRGFP-2A-H2AmCherry;GFAP:Gal4FF) hindbrains in wt (r), scarb2a−/− (s) and scarb2a−/− mutant following injection of UAS-scarb2a,cmcl2:EGFP (t). u Quantification of slc1a3b+ red nuclei (n = 12 embryos; One-way ANOVA, multiple comparisons with Tukey posthoc test). Error bars are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not statistically significant. Scale bars, a, b, e, f, h, i, k, l, n–p, r–t = 50 μm; a', b', e', f’, h', i', k', l' = 30 μm. Source data are provided as a Source Data file.
Astrocytes also arise from RGCs35. To evaluate whether astrogenesis is also impaired in scarb2a mutants, we imaged Tg(slc1a3b:MYRGFP-2A-H2AmCherry) embryos that express the membrane-bound myristoyl-GFP (myrGFP) and the nuclear marker H2AmCherry under the control of the astrocyte marker slc1a3b/Glast36. Confocal images at 60 hpf showed the presence of slc1a3b+ cells throughout the hindbrain, particularly at the midline, midbrain-hindbrain junction (MHB), and rhombic lips (rl) (Fig. 3h). Transverse optical sections indicated that the nuclei of these cells (red) are situated in the vz and the rhombic lips, from where long GFP+ processes extend ventrally into the neural tube (Fig. 3h’, j). Moreover, images of double transgenic Tg(slc1a3b:MYRGFP-2A-H2AmCherry;scarb2a:KalTA4:UAS:EGFP) embryos depicted scarb2a-EGFP+ and slc1a3b-mCherry+ colocalization in the vz nuclei, indicating that scarb2a is also expressed in this cell population at 60 hpf (Supplementary Fig. 3l, l’). When compared to wt siblings, scarb2a−/− embryos exhibited significantly fewer slc1a3b+ nuclei at all locations (Fig. 3i, i’, j), especially in the vz. These defects persisted through 6 dpf (Fig. 3k–m) when astrocytes become functional37. Finally, we confirmed that these defects are not secondary to the morphological malformations derived from hemorrhagic events. As seen in Supplementary Fig. 3m, n the reduction in the number of oligodendrocytes and astrocytes at 60 dpf was not changed by the presence or absence of hemorrhage.
To understand the genetic programs linked to the observed phenotypes, we performed bulk RNAseq on scarb2a+ cells isolated from wt and mutant heads at 60 hpf. Analysis of transcription factor (TF) expression revealed a significant reduction of factors involved in OPC differentiation38,39 (Supplementary Fig. 4a), accompanied by a concomitant increase in TFs driving neuronal fate40 (Supplementary Fig. 4b) in scarb2a mutants. These results were further validated by examining Tg(isl1a:GFP) embryos, which revealed a marked increase in the number of differentiated motoneurons41 in the hindbrain of scarb2a mutants as compared to their wt siblings, both at 60 hpf and at 6 dpf (Supplementary Fig. 4c–h). Taken together, our data indicate that Scarb2a depletion results in precocious neuronal differentiation of RGCs and exhaustion of the progenitor pool, ultimately leading to impaired gliogenesis.
To functionally confirm these results, we conducted rescue experiments by expressing the wt scarb2a coding sequence in GFAP+ cells. Injection of a UAS:scarb2a-P2A-RFP or UAS:scarb2a;cmcl2:EGFP construct into Tg(GFAP:Gal4FF);scarb2a−/− embryos, fully rescued the number and distribution of OPCs (Fig. 3n–q; Supplementary Fig. 4i–k) and astrocytes (Fig. 3r–u; Supplementary Fig. 4l–n) at 60 hpf. Similar results were obtained upon expression of wt scarb2a in Tg(scarb2a:KalTA4);scarb2a−/− embryos (Supplementary Fig. 5a–j). Interestingly, ~40% of the olig2+ cells co-expressed the injected wt scarb2a construct (Supplementary Fig. 5d, d’, f, n, n’, o), roughly corresponding to the number of OPCs lost in scarb2a mutants (Fig. 3c, d).
Scarb2 depletion results in impaired acidification of the endolysosomal compartment in RGCs, thereby affecting Notch3 S3 cleavage and release of N3ICD
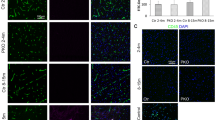
The pronounced neurogenic phenotype observed in scarb2a mutants resembled the one resulting from Notch deficiency42,43,44, raising the possibility that scarb2a depletion hinders Notch signaling, thereby leading to excessive neurogenesis and impaired glia differentiation. To test this hypothesis, we time-lapse imaged Notch signaling during different phases of neurogenesis using the Tg(EPV.Tp1-Mmu.Hbb:EGFP) (a.k.a. 12NRE:EGFP45) Notch reporter under wt and scarb2a mutant backgrounds (Supplementary movie 3). Analysis of transverse optical sections revealed comparable levels of Notch activity in the rhombic lips of both wt and mutant embryos during the initial phase of secondary neurogenesis (28 hpf) (Fig. 4a–b’; Supplementary Fig. 6a, b). However, while at 32 hpf, vz progenitors display evident Notch activity in wt embryos (Fig. 4c, c’; Supplementary Fig. 6c), the EGFP signal was barely detected in these cells in mutant siblings (Fig. 4d, d’; Supplementary Fig. 6d). By 40 hpf, only residual levels of Notch-derived EGFP signal were observed in the rl and midline of scarb2a mutants (Fig. 4e–f’; Supplementary Fig. 6e, f). These results were further validated by qRT-PCR analyses carried out on Tg(scarb2a:KalTA4;UAS:mKate2) cells isolated from the heads of wt and scarb2a−/− embryos. The expression of the Notch downstream targets her4.2 and her6 was significantly reduced (Fig. 4g), whereas no differences were observed in the levels of notch1a and notch3 (Fig. 4h), indicating that Scarb2a depletion impairs Notch signaling without affecting transcription of the receptors. Finally, injection of UAS:scarb2a-P2A-RFP into Tg(GFAP:Gal4FF;12NRE:EGFP);scarb2a−/− embryos significantly rescued Notch activation in RGCs, with ~80% of the cells expressing the exogenous Scarb2, displaying Notch activity (Supplementary Fig. 6g–l).
a–f' Optical transverse sections from a time-lapse series of Tg(12NRE:EGFP;scarb2a:KalTA4;UAS-mkate2) wt and mutant embryos at 28 (a–b'), 32 (c–d') and 40 hpf (e–f') showing reduced Notch signaling in scarb2a mutant vz starting from 32 hpf (d, d’; f, f’, Nexperiments = 3, for each nwt = 1, nscarb2a−/−=1). g-h qRT-PCR showing mRNA levels of her4.2, her6 (g) and notch1a, notch3 (h) in mKate2+ cells sorted from Tg(scarb2a:KalTA4;UAS-mkate2) heads (N = 5, n = pull of 20 embryos, two-tailed Student’s t-test, Pher4.2 = 0.0017; Pher6 = 0.0012,; Pnotch1a = 0.6586, Pnotch3 = 0.9277). i–j’ Transverse optical sections of Tg(12NRE:EGFP;scarb2a:KalTA4;UAS-mkate2) wt embryos untreated (i, i’) or treated with the v-ATPase inhibitor Bafilomycin A1 (BafA1, j, j') showing intraventricular RGC invasion (j, arrow) and reduced Notch signaling in treated embryos (j’, Nexperiments = 3, for each nwt = 5, nBafA1-=5). k–l” Lysotracker Deep Red staining of Tg(hsp70l:lamp1-RFP) wt (k–k”) and scarb2a mutant (l–l”) embryos showing reduction of Lamp1+ acidic punctae in mutant hindbrains; quantified in o. m–n’, Transverse optical sections of Tg(hsp70l:lamp1-RFP) wt embryos treated with BafA1 (m–m”) or with the γ-secretase inhibitor (LY-411575) (n, n’, Nexperiments = 3, for each group n = 5 embryos). o Quantification of double-positive Lamp1/Lysotracker punctae (Nexperiments=3, n = 5 embryos/group, One-way ANOVA, multiple comparisons with Tukey posthoc test). p Quantification of Lamp1+ vesicle size in wt, scarb2a mutants, BafA1, and LY-411575 wt-treated embryos (Nexperiments=3,for each group n = 5 embryos, One-way ANOVA, multiple comparisons with Tukey posthoc test, PANOVA < 0.0001). q–r’ Lysotracker Deep Red staining of Tg(h2afx:EGFP-RAB7) wt (q–q”) and scarb2a mutant (r–r”) embryos showing reduction of double positive Rab7/Lysotracker punctae in mutant hindbrains; quantified in (u, Nexperiments = 3, for each group n = 5 embryos, One-way ANOVA, multiple comparisons with Tukey posthoc test, PANOVA < 0.0001). s–t’ Transverse optical sections of Tg(h2afx:EGFP-RAB7) wt embryos treated with BafA1 (s–s”, u) or LY-411575 (t, t’, Nexperiments = 3, n = 5 embryos/group). v Quantification of Rab7 vesicle size in wt, scarb2a mutants, BafA1, and LY -411575 wt-treated embryos (Nexperiments=3, n = 5 embryos/group, One-way ANOVA, multiple comparisons with Tukey posthoc test, PANOVA < 0.0001). g, h, o, u = Error bars are mean ± s.e.m.; p, v = bars are median. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not statistically significant. Scale bars, a–f', i–j’,k, l, m, n, q, r, s, t = 30 μm; k’, l’, m’, n’, q’, r’, s’, t’ = 5 μm. Source data are provided as a Source Data file.
Notch1a signaling is active during early neurogenesis (18–24 hpf), whereas Notch3 is the predominant isoform acting at the stages when the scarb2a phenotypes become evident (30–55 hpf)27,46,47. Accordingly, notch3fh332/fh332 mutants display defective oligodendrogenesis46 (Supplementary Fig. 6m–o) as well as premature neuronal differentiation27, highly reminiscent of the scarb2a−/− phenotypes. Similarly, the Tg(slc1a3b:MYRGFP-2A-H2AmCherry) astrocyte population was also significantly reduced in notch3fh332/fh332 embryos (Supplementary Fig. 6p–r), further reinforcing the link between Scarb2 absence, Notch3 signaling deficiency and impaired gliogenesis.
In both mice and humans, Scarb2 is found in the membrane of late endosomes and lysosomes, where it was shown to fulfill several functions12. Therefore, we speculated that alterations to Scarb2a might disrupt the trafficking and/or processing of Notch3 within the endolysosomal compartment. Previous research in drosophila, zebrafish, and human cells has established a connection between endolysosomal activity and Notch signaling48,49. In particular, it has been shown that the acidity of the endolysosomal compartment, via the V-ATPase proton pump activity, can significantly affect the efficiency of the γ-secretase-dependent cleavage of the NICD in the receiving cell, thereby altering proper Notch signaling50,51,52,53.
As a first step toward testing our hypothesis, we exposed 32 hpf Tg(12NRE:EGFP, scarb2a:KalTA4;UAS:mKate2) wt embryos to Bafylomycin A1 (BafA1), a well-established V-ATPase inhibitor. As seen in Fig. 4i–j’, the treatment led to a reduction of Notch signaling48 and increased numbers of scarb2a+ cells invading the ventricular space, strongly resembling the phenotypes observed in scarb2a mutants. Next, we asked whether the acidity of the endolysosomal compartment is indeed affected in scarb2a mutants. To answer this question, we took advantage of Tg(hsp70l:lamp1-RFP) fish, expressing an RFP-tagged form of the Lysosomal Associated Membrane Protein 1 (Lamp1) and Tg(h2afx:EGFP-rab7) embryos, ubiquitously expressing an EGFP-fused form of the late endosome marker Rab754 under the heat-shock promoter55. We combined these transgenic reporters with scarb2a mutant embryos and stained them with far red-coupled Lysotracker, which labels acidic organelles. High-magnification images of the vz demonstrated that scarb2a−/− embryos exhibited fewer numbers of both Lamp1/Lysotracker (Fig. 4k–l”, o) and Rab7/Lysotracker (Fig. 4q–r”, u) double-positive punctae, as compared to wt siblings, indicating impaired acidification of these intracellular compartments. Similar, albeit more pronounced results, were obtained upon exposure of Tg(hsp70l:lamp1-RFP) and Tg(h2afx:EGFP-rab7) wt embryos to BafA1- in this case, no punctate Lysotracker staining was detected (Fig. 4m–m”, o, s–s”, u) further supporting the notion that scarb2a mutants display impaired acidification of Rab7 and Lamp1 organelles. Interestingly, we also noticed that both Rab7 and Lamp1 vesicles were significantly enlarged in the mutants (Fig. 4p, v), possibly due to the accumulation of undegraded materials as has been shown in the context of several Lysosomal Storage Diseases (LSDs)56, or as a result of defective fusion.
Two lines of evidence confirmed that impaired S3 cleavage of Notch3 and generation of the NICD are among the main drivers of scarb2a mutant phenotypes. First, inhibition of γ-secretase via LY-411575 resulted in a marked reduction in the numbers of OPCs (Supplementary Fig. 7a–c’, d) and astrocytes (Supplementary Fig. 7e–g’, h), as well as in enlarged Lamp1 (Fig. 4n, n’, p) and Rab7 (Fig. 4t, t’, v) organelles, fully phenocopying the scarb2a−/− phenotypes. Second, we designed rescue experiments based on overexpression of the Notch3 intracellular (N3ICD) or extracellular (N3ECD) domains. To this end, we utilized stable transgenic reporters- Tg(hsp70I:N3ICD-EGFP) and Tg(hsp70I:N3ECD-EGFP)57 expressing constitutively active forms of N3ICD and N3ECD under the control of the heat shock promoter hsp70I, and induced their expression at ~28–30 hpf, mimicking the endogenous expression of Notch327. As seen in Fig. 5a–c’, e, N3ECD-overexpression in olig2:GFP;scarb2a−/− embryos did not prevent defective oligodendrogenesis, as opposed to N3ICD induction, which exerted a significant recovery of the number of olig2+ migrating cells (Fig. 5d, e). Likewise, overexpression of N3ICD, but not of N3ECD, led to a significant increase in the number of slc1a3b:MYRGFP-2A-H2AmCherry+ cells in the hindbrains of scarb2a−/− embryos (Fig. 5f–j). Finally, we could confirm that these phenotypes are primarily attributed to defective Notch3 signaling, as overexpression of the N1aICD fragment using Tg(hsp70l:myc-notch1a-intra;cryaa:Cerulean)58 was not sufficient to restore neither astrocyte (Supplementary Fig. 7i–l) or OPC (Supplementary Fig. 7m–p) populations in scarb2a mutants.
a–d’ Dorsal views and transverse optical sections of Tg(olig2:EGFP) hindbrain in wt (a, a’), scarb2a−/− (b, b’) and scarb2a−/− following heat-shock induced overexpression of Notch3 extracellular (N3ECD) (c–c‘) and Notch3 intracellular (N3ICD) domains (d–d') (dashed square in a–d, marks the region shown in a’–d’). e Quantification of migrating OPCs in a–d (n = 9 embryos, One-way ANOVA, multiple comparisons with Tukey post-hoc test, PANOVA < 0.0001). f–i’ Dorsal views and transverse optical sections of Tg(slc1a3b:MYRGFP-2A-H2AmCherry) hindbrains in wt (f), scarb2a−/− (g) and scarb2a−/− mutant following N3ECD (h-h’) or N3ICD (i–i’), heat-shock induction (dashed square in f–i, marks the region shown in f’–i’) j, Quantification of slc1a3b+ astrocytes in f–i (n = 9 embryos, One-way ANOVA, multiple comparisons with Tukey posthoc test, PANOVA < 0.0001). Error bars are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not statistically significant. Scale bars, a’, b’, c’, d’, f’, g’, h’, i’ = 30 μm; a, b, c, d, f, g, h, i = 60 μm.Source data are provided as a Source Data file.
Defective RGC differentiation in scarb2a mutants impacts the production of Vegfaa and Wnt7a, thereby affecting the normal establishment of the CNS vasculature and functionality of the BBB
Having identified the specific cell populations affected by the absence of Scarb2, we aimed to investigate whether the exhaustion of RGCs and consequent glia reduction is responsible for the observed vascular phenotypes, and examine the underlying molecular mechanisms. The active crosstalk between neuronal, glial, and vascular compartments is fundamental for the correct development and functionality of the CNS and the BBB. Similar to mammals59,60,61,62,63, CNS angiogenesis and barriergenesis in zebrafish are tightly coordinated by the VEGF13,64,65 and Wnt signaling pathways66,67,68,69,70,71. Chemical inhibition of VEGF signaling during specific stages of hindbrain development led to faulty EC migration and survival, resulting in malformed CtAs72. During the same angiogenic phases, Wnt signaling is highly active in the sprouting CtA ECs, and its inhibition prompted diminished glut1 expression72 and defective CtA anastomosis68, causing intracerebral hemorrhages at later developmental stages.
First, we verified that reducing neurogenesis and restoring proper gliogenesis via expression of UAS:scarb2a-P2A-RFP or UAS:scarb2a;cmcl2:EGFP in Tg(GFAP:Gal4FF);scarb2a−/− (Fig. 3n–u) or Tg(scarb2a:KalTA4);scarb2a−/− embryos (Supplementary Fig. 5a–j) resulted in significant recovery of CtA morphology, reduced numbers of ectopic interconnections (Fig. 6a–d; Supplementary Fig. 8a–d), and rescued glut1b expression to wt levels (Fig. 6e–g’; Supplementary Fig. 8e–g’), confirming that scarb2a depletion in RGCs prevents proper glia differentiation and maturation, leading to subsequent defects in the establishment of the hindbrain vasculature and the BBB.
a–c Dorsal views of the hindbrain vasculature in Tg(kdrl:TagBFP;GFAP:Gal4FF) embryos wt (a), scarb2a−/− (b) and scarb2a−/− mutant following injection of UAS-scarb2a-P2A-RFP (c). d Quantification of CtA interconnections (n = 13 embryos; One-way ANOVA, multiple comparisons with Tukey posthoc test, PANOVA < 0.0001). e–g’ Dorsal views of Tg(kdrl:EGFP;glut1b:mCherry;GFAP:Gal4FF) depicting full restoration of glut1b expression (white) following injection of UAS-scarb2a,cmcl2:EGFP in mutant embryos (g, g’, Nexperiments = 3, for each nwt = 6, nscarb2a−/−=6). h–i' Transverse optical sections of kdrl:TagBFP;vegfaa:EGFP 60 hpf embryos showing increased Vegfa signal in scarb2a−/− (i, i', Nexperiments = 3, for each nwt = 5, nscarb2a−/−=5). j–l Dorsal views of Tg(kdrl:TagBFP) wt (j), scarb2a−/− (k) and scarb2a−/− after treatment with the Vegfr inhibitor SU5416 (l). m Quantification of CtA anastomoses following SU5416 treatment (n = 6 embryos; One-way ANOVA, multiple comparisons with Tukey posthoc test, PANOVA < 0.0001). Error bars are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001; ***P < 0.001 ns, not statistically significant. Scale bars, a–c; e–g' = 60 μm; h-i’ = 30 μm. Source data are provided as a Source Data file.
Subsequently, we assessed potential alterations in the VEGF and Wnt signaling pathways. Previous works in zebrafish highlighted the crucial role of Vascular Endothelial Growth Factor aa (Vegfaa) in the development of the hindbrain vascular network64. Analysis of the TgBAC(vegfaa:EGFP)pd26073 reporter revealed that during the initial stages of CtA sprouting and migration (~32 dpf), similar levels of vegfaa expression are detected in scarb2a + RGCs both in wt and mutant embryos (Supplementary Fig. 8h–k’). These results are in accordance with the normal development of the BA and initial sprouting of the CtAs from the PHBCs observed in scarb2a−/− (Fig. 1d). In contrast, by 60 hpf, higher levels of vegfaa were observed in the hindbrains of scarb2a−/− embryos (Fig. 6h–i’; Supplementary Fig. 8l–o), likely produced by RGCs that adopted a neuronal fate, which results in aberrant angiogenesis, as previously described64,65,66,72. To functionally confirm these results, we treated wt and mutant embryos with the VEGFR inhibitor SU5416 at ~32 hpf (right before the initial appearance of the vascular phenotypes). As seen in Fig. 6j–m, we observed a strong decrease in the number of CtA anastomoses, confirming that at least part of the altered angiogenesis phenotypes seen in scarb2a−/− is mediated by the excess of vegfaa production due to the excessive neuronal differentiation.
We further investigated whether Wnt signaling is also altered in the hindbrains of scarb2a−/− embryos. Wnt7a has been previously shown to be important for BBB formation in mammals70 and zebrafish68,69. Accordingly, we assessed the expression of Wnt7Aa in both GFAP+ and scarb2a-labeled cells at 2 dpf and confirmed a consistent decrease in the mutant brains (Supplementary Fig. 9a, b). Moreover, Wnt activation, as depicted by the Tg(7xTCF-Xla.Sia:GFP)74 reporter, was reduced in CtA-ECs of mutant embryos (Fig. 7a–b’; Supplementary Fig. 9c–d’), confirming that indeed, Wnt signaling is not properly activated in the hindbrains of scarb2a mutants.
a–b', Optical transverse sections of Tg(kdrl:mcherry;7xTCF-Xla.Sia:GFP) in wt (a, a') and scarb2a−/− (b, b') showing reduced Wnt activity in ECs of scarb2a−/− (b, b', Nexperiments = 3, for each nwt = 3, nscarb2a−/−=3). c–e', Dorsal views of Tg(kdrl:TagBFP;glut1b:mCherry) depicting normalized glut1b expression (white) following treatment with LiCl (e, e', Nexperiments = 3, for each nwt = 6, nscarb2a−/−=6). f–h LiCl treatment significantly reduces leakage of 2000KDa Rhodamine from scarb2a−/− hindbrain vessels (h, Nexperiments = 3, for each nwt = 4, nscarb2a−/−=4). i–k Confocal images of Tg(kdrl:TagBFP) showing complete restoration of the brain vascular network after administration of LiCL and SU5416 to scarb2a mutants (k, Nexperiments = 3, for each nwt = 4, nscarb2a−/−=4). l Graphical model depicting the series of events affecting NVU formation and function in scarb2a mutant embryos. Defective endolysosomal acidification hinders Notch3 processing in RGCs, leading to imbalanced neurogenesis vs. gliogenesis, altered levels of Vegf and Wnt activation, which in turn result in defective cerebral angiogenesis, micro-hemorrhages, and compromised BBB. Scale bars a–b' = 30 μm; c–k = 60 μm.
To functionally corroborate these results, we treated scarb2a mutants with the Wnt pathway activator LiCl. This treatment, carried out at 32 dpf, resulted in significant rescue of CtA anastomoses (Supplementary Fig. 9e), Glut1 expression (Fig. 7c–e’), and BBB leakiness (Fig. 7f–h). These data demonstrate that Wnt secreted from RGCs and their glia derivatives are responsible for inducing barriergenesis in accordance with previous reports66,67,68,69,70,71,72,75. Finally, combined treatment with SU5416 and LiCl at 32 hpf led to complete normalization of CtAs anastomosis and vascular morphology (Fig. 7i–k; Supplementary Fig. 9f). Taken together, these data indicate that the increased neurogenesis and depletion of RGCs and/or other glia populations in scarb2a mutants have a direct impact on the production of VEGF and Wnt7Aa, thereby affecting the normal establishment of the CNS vasculature and its barrier function.
Overall, our results demonstrate that dysfunction of the endolysosomal compartment caused by Scarb2 depletion hinders Notch3 signaling in RGCs, thereby affecting the maintenance of the undifferentiated RGC progenitor pool and modulating the balance between neurogenesis and gliogenesis. We further demonstrate that alterations to this balance result in severe vascular defects, hemorrhages, and a leaky BBB (Fig. 7l). In a broader sense, our data illustrate the exquisite interactions between different cell types contributing to the establishment and functionality of the NVU, and reveal a relationship between endolysosomal dysfunction in non-vascular cells of the brain parenchyma (i.e., RGCs and glia) and cerebrovascular health.
Discussion
The study of zebrafish scarb2a mutants has revealed insights into the mechanisms controlling neuronal vs. glial differentiation of RGC progenitors that, when disrupted, lead to phenotypes associated with cerebrovascular diseases and demyelinating neuropathies. By generating a scarb2a transgenic reporter, we revealed the specific expression of scarb2a initially in RGCs, and subsequently in glial cells, including a subset of OPCs and OLs, as well as astrocytes. The absence of Scarb2 results in impaired acidification of the endolysosomal compartment in RGCs, which in turn affects the cleavage of Notch3 and the release of N3ICD. Consequently, scarb2a mutants display excessive differentiation towards neuronal lineage at the expense of glial cells. This disruption affects the normal activation of the VEGF and Wnt signaling pathways, thereby impairing the normal development of the CNS vasculature, including abnormal interconnections of the CtAs, micro-hemorrhages, and a compromised BBB.
Notably, a complete rescue of the glial and vascular abnormalities associated with Scarb2a deficiency was achieved through the expression of the wt form of scarb2a, exclusively in RGCs, underscoring the specificity of the observed phenotypes and the significant connection between proper RGC differentiation, gliogenesis, and CNS angiogenesis.
At the molecular level, we show significant downregulation of Notch signaling in scarb2a mutant hindbrains starting from 30 hpf and demonstrate that this reduction is specifically attributed to impaired Notch3 activity. Through a series of genetic manipulations, we demonstrate that restoring the Notch3-intracellular, but not the extracellular domain, leads to a recovery of OPC and astrocyte populations. These results confirm earlier findings supporting the crucial role of Notch3 in regulating RGC fate determination11,27,76 and highlight Scarb2a and the proper function of the endolysosomal compartment as key players in the process.
Analysis of N3ECD and N3ICD overexpression experiments showed that only the intracellular fragment is able to rescue the scarb2a mutant phenotype, suggesting that lack of Scarb2a might potentially affect the cleavage operated by the γ-secretase complex. The activation of the Notch signaling pathway involves two sequential proteolytic cleavages of the Notch receptor that generate first a Notch extra-cellular domain (NECD) and, subsequently, a Notch intracellular domain (NICD) fragment. The NECD is internalized within the signaling cell, whereas the product of the second (S2) cleavage, consisting of the transmembrane and intracellular domains (also referred to as Notch Extracellular Truncation- NEXT), undergoes an additional cleavage by γ-secretase at the S3 site, releasing the NICD, which can then translocate into the nucleus and activate transcription of downstream targets77. Experiments carried out in Drosophila50,78 as well as in mammalian cells52,79 have demonstrated that the acidity of the endolysosomal compartment can significantly affect the efficiency of the S3 cleavage (following NEXT endocytosis)51. Notably, inhibition of vacuolar H+-ATPase (V-ATPase) (a protein involved in the acidification of endosomes and lysosomes) by bafilomycin-A1 (BafA1)50,78 was shown to hinder the processing and activation of Notch receptors69, further supporting the notion that Notch S3 cleavage by γ-Secretase requires an acidified intracellular compartment and that this step is crucial for Notch activation. Given that SCARB2/LIMP2 is predominantly located in the membrane of late endosomes and lysosomes, we speculated that impaired production of N3ICD observed in scarb2a mutants could be linked to alterations in these organelles. In this context, we show that Scarb2a depletion results in reduced acidity and increased size of both Lamp1 and Rab7 compartments, features associated with several LSDs. These data, along with the strong similarity observed between scarb2a mutant phenotypes and wt embryos treated with the V-ATPase inhibitor BafA1 or with the γ-Secretase inhibitor LY-411575, reinforce the notion that Scarb2-dependent malfunction of the endolysosomal machinery stands at the basis of the Notch3 signaling defects observed in scarb2a−/− mutants. Accordingly, Scarb2a depletion could lead to changes in the endolysosomal machinery and γ-secretase activity, resulting in altered N3ICD cleavage52.
Previous studies have demonstrated that RGCs and their derived lineages of neurons, oligodendrocytes, and astrocytes control different aspects of embryonic and postnatal CNS angiogenesis1. During the development of the embryonic mouse cortex, for instance, Wnt ligands secreted by RGCs control Glut-1 and Gpr124 expression, and their depletion results in defective CNS angiogenesis and compromised BBB integrity67,69,71,72,80,81,82. Additionally, recent research has demonstrated that RGCs secrete TGF-β1, which promotes both EC migration and tube formation in vitro83. Finally, VEGFa, secreted by neural progenitors and their progenies, is known to control CNS angiogenesis in a dose-dependent manner59,60. Our results, showing defective sprouting and anastomosis of the CtAs in scarb2a mutant embryos, accompanied by a compromised BBB and clear downregulation of Glut1 expression, align with these previous reports and highlight the coordinated and tightly regulated nature of cell interactions within the developing brain, which is essential for the proper functioning of the CNS vasculature.
Brain vascular abnormalities can lead to various, sometimes life-threatening forms of cerebrovascular disease6, including ischemic and hemorrhagic stroke, vascular malformations, vascular cognitive impairment, and dementia. Within this category, cerebral small vessel diseases (CSVDs) encompass a range of sporadic or hereditary pathological alterations of cerebral arterioles, capillaries, and venules, occurring primarily within the white matter. These changes contribute to demyelination, BBB dysfunction, glial activation, and axonal loss7. Notably, CADASIL, the most common inherited form of CSVD, is caused by mutations in the NOTCH3 gene84, which have been shown to primarily affect vascular smooth muscle cells and pericytes. One of the outstanding open questions is how the vascular phenotype is linked to the white matter changes observed in CADASIL (and in SVDs in general). While the current prevalent idea is that the initial vascular defects lead to white matter damage, the possibility exists that impaired Notch3 processing and/or signaling within glia, cell-autonomously account for the observed changes. This hypothesis could also explain the fact that despite the monogenic nature of the disease, CADASIL patients display a large phenotypic variability and an unclear genotype-phenotype correlation. Our findings thus bring about a fresh perspective on cerebrovascular diseases, suggesting that defects in cellular populations within the vascular microenvironment, beyond the commonly studied vascular cells, could also play a crucial role.
Methods
Zebrafish husbandry, transgenic and mutant lines
Zebrafish were raised by standard methods and handled according to the guidelines of the Weizmann Institute Animal Care and Use Committee85. For all imaging, in situ hybridization, and immunofluorescence experiments, embryos were treated with 0.003% phenylthiourea (PTU, Sigma-Aldrich) from 8 hpf to inhibit pigment formation. Zebrafish lines used in this study were: Tg(fli1:EGFP)yl86; Tg(gata1a:DsRed)sd286; Tg(kdrl:EGFP)s84386; Tg(kdrl:mCherry)y20687; Tg1(kdrl:NLS-mCherry)y17313; Tg(7xTCF-Xla.Sia:GFP)ia474; TgBAC(vegfaa:EGFP)pd26073; Tg(gfap:Tomato)nns1788; Tg(gfap:GAL4FF)wa3289; Tg(olig2:EGFP)vu1231; Tg(olig2:dsRed)vu1931; Tg(isl1a:EGFP)rw031; Tg(slc1a3b:MYRGFP-2A-H2AmCherry)37; Tg(EPV.Tp1-Mmu.Hbb:EGFP)ia1245; Tg(kdrl:TagBFP)mu29390; Tg(glut1b:mCherry)sj191; Tg(mbp:EGFP)ck133; Tg(hsp70I:N3ECD-EGFP)co1557; Tg(hsp70I:N3ICD-EGFP)co1757; Tg(hsp70l:MYC-notch1a,cryaa:Cerulean)fb1258; notch3fh332/fh33257, Tg(hsp70l:lamp1b-RFP)pd106455, Tg(h2ax:EGFP-rab7a)mw754.
The scarb2a mutant was generated using CRISPR/CAS technology. The CRISPR guide was designed with CHOPCHOP, and potential off-target sequences were assessed using the MIT CRISPR Design site. The guide sequence GGCTGAGAACAGCAGAGTAT was cloned into the BsmBI sites of the pT7-gRNA plasmid (Addgene plasmid 46759). Cas9 mRNA was generated from pCS2-nCas9n92 using the mMACHINE T7 ULTRA kit (Ambion, AM1345). Cas9 mRNA (250 ng/μL) and guide RNA (100 ng/μL) were co-injected into 1-cell-stage embryos. For genotyping, genomic DNA was extracted and amplified with 5′-TGTCTGTGTTTGGTTACAGGAG-3‘ and 5′-TTCCCCGCCAAGAACTCA-3‘ (138 bp) primers and analyzed on a 2% agarose gel.
The TgBAC(scar2ba:KalTA4) line was generated using the DKEY-177l10 BAC (BioScience, HUKGB735J035Q) containing the full scarb2a sequence. The KalTA4 fragment was cloned downstream to the ATG following the removal of the scarb2a CDS. The resulting BAC(scar2ba:KalTA4) construct was injected at the 1-cell stage into AB zebrafish and integrated through transposon-mediated BAC transgenesis to generate a stable reporter line that was then crossed with scarb2a mutant fish93.
The Tg(UAS-scarb2a-P2A-RFP) and Tg(UAS-scarb2a;cmcl2:EGFP) lines were generated by amplifying the coding sequences of scarb2a with the following primers: F 5‘- ATGACTAGAAGATCTTGTACTATTTACGCC-3‘; R 5‘- TCAACACTTTTGTGCCTCCACT-3‘. The resulting fragment was then cloned in pDONR221 plasmid and combined with p5E-UAS, p3E-P2A-mKate2, and pDestTol2pA2 using the Gateway system as previously described94.
For rescue experiments Tg(UAS-scarb2a-P2A-RFP) and Tg(UAS-scarb2a;cmcl2:EGFP) constructs were injected at 1 cell stage (800 pg per embryos) into Tg(GFAP:Gal4FF);scarb2a−/− and Tg(scarb2a:KalTA4);scarb2a−/− transgenic lines, respectively.
Angiography
Tetramethylrhodamine Dextran (molecular mass, 2,000,000 Da; Thermo Fisher Scientific, D7139) was injected intravascularly on anesthetized larvae as previously described95. Imaging was initiated within 5 min of the injection.
In situ hybridization and immunostaining
In situ hybridization was performed as described85. The following primers were used to generate riboprobes:
glut1b:
5′- ATTGGCATCCTCATGGCACA-3‘;
5‘- ATGAAAACGTATGGGCCGGT-3‘
scarb2a:
5′-CTGAGCATCCGAGAAATACTCCCG-3′;
5′-CACCAATTCATCTGACTGATGCCG-3′22
For immunostaining, 60 hpf embryos were fixed in 4% PFA overnight, permeabilized in acetone for 30 min in ice, blocked in PBS containing 2% BSA, 10% goat serum, and 0.1% Tween20 for 3 hrs. at RT, and incubated overnight at 4 °C with rabbit anti-Sox2 (1:100; Abcam ab97959)), anti-fabp7a (1:100, Millipore ABN14) or mouse anti-HuC (1:400; Thermo Fisher Scientific A-21271) antibodies. After extensive washing with PBST, embryos were incubated overnight at 4 °C with secondary antibodies conjugated with Alexa Fluor 633 (1:500; Goat anti-Rabbit Invitrogen A21070, Goat anti-Mouse Invitrogen A21053).
Lysotracker staining
Lysotracker™ Deep Red (Thermo Fisher L12492) staining was previously described96. Briefly, 25–30 embryos were transferred into 6-well plates containing LysoTracker™ Deep Red dissolved to a final concentration of 10 μM in PTU water and incubated at 28.5 °C for 45 min in a dark environment. Embryos were then rinsed 3 times with ~1 ml of fresh fish water and imaged immediately using a 633 nm laser for far red staining.
Heat shock induction
Heat shock was performed at 37 °C for 45 minutes. For Tg(hsp70I:N3ECD-EGFP)co15, Tg(hsp70I:N3ICD-EGFP)co17 and Tg(hsp70l:MYC-notch1a,cryaa:Cerulean)fb12 experiments, the heat shock was repeated every 6-7 h on embryos from 28 hpf until 60 hpf.
Drug treatments
To inhibit γ‐secretase, 28 hpf manually dechorionated embryos were incubated at 28.5 °C in the dark into a 2 ml petri dish containing 0.4% DMSO in embryo water with LY-411575 (Sigma, SML0506) at a final concentration of 50 μM. To specifically block V‐ATPase proton pump activity, we incubated 28 hpf zebrafish embryos with Bafylomicin A1 inhibitor (Sigma, B1793) that was added directly to the fish medium at a final concentration of 100 nM. 0.4% DMSO was used as control. LiCl97 (Merck) 50 mM and SU541698 (EMD Millipore) 5 μM were added to dechorionated embryos at 30 hpf and incubated at 28.5 °C in the dark into a 2 ml petri dish containing 0.4% DMSO in embryo water. For all treatments, embryos were left in the incubation medium at 28.5 °C until 60 hpf.
Cell sorting
For each experimental condition, a pool of 50 heads (3 biological replicates) was dissected from euthanized 48 hpf scarb2a mutants and wt siblings and dissociated into a single-cell suspension. After a brief enzymatic treatment with a cocktail of Liberase Blendzyme 3 (Roche), trypsin B (BI), and DNAseI (Roche), the cell suspension was strained through a 70 µm filter and stained with SYTOXTM blue (ThermoFisher) for live/dead discrimination. 5000 live scarb2a+ single cells were sorted into 40 µl of lysis/binding buffer solution (ThermoFisher) containing RNase inhibitor RNasinTM (Promega). FACS analysis and sorting were performed on a (BD FACS Aria III) using a 70 µm nozzle.
For qRT-PCR analysis of wnt7aa, 50 heads/sample were dissected at 48 hpf and 50000 cells were sorted from Tg(gfap:Tomato)nns17 and TgBAC(scar2ba:KalTA4; UAS-mKate2) embryos under wt and mutant backgrounds.
Bulk MARS-Seq
RNA was captured using Dynabeads™ mRNA DIRECT™ Purification Kit (ThermoFisher) prior to library preparation. A bulk adaptation of the MARS-seq protocol99 was used to generate RNA libraries for the expression profile of scarb2+ mutant and wt cells. The RNA was further fragmented and transformed into a sequencing-ready library by tagging the samples with Illumina sequences during ligation, RT, and PCR. The final library concentration was measured by Qubit, TapeStation, and qPCR for zebrafish actin as previously described99. Sequencing was performed on a Nextseq500/550 High Output Kit v2.5 75 cycles (Illumina; paired-end sequencing), and each sample was sequenced for 6 M reads. The Initial quality control report was done using the User-friendly Transcriptomic Analysis Pipeline100. Upregulated and downregulated transcription factors were identified in the sequenced dataset using gene ontology from Uniprot.org.
Total RNA isolation and quantitative reverse transcription PCR
A pool of 10–20 heads/sample was dissected from 48 hpf wt and mutant embryos. The samples were homogenized in TRIzol (Invitrogen, Thermo Fisher Scientific, 15596026) and processed for RNA extraction following standard procedures95. A total of 1 μg of RNA per reaction was reverse-transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific, 4368814).
The following primers were used:
notch1a 5′-GTGGCACAAGGGCAAAGATG -3′, 5′-CAGGGGTTCGGGAATTGACA-3′;
notch3 5′- GCATTGACCGACCTAATGGA -3′, 5′-TGCTCTCACACAGTCTTCCTTC-3′;
her4.2 5‘-GGCTCAATCAGCAGCAGAGA-3‘; 5‘-GCACTGCTTTTCTGAGAGCG-3‘;
her6 5‘-AATGACCGCTGCCCTAAACA-3‘; 5‘-TCACATGTGGACAGGAACCG-3‘.
vegfaa 5′-AGAAAGAAAACCACTGTGAG-3′;5‘-AGGAATGTTCTTCCTTAGGT-3′;
vegfab 5′-TGCTGAACACAGTGAA-TGCCAG-3′; 5′- ACATCCATCTCCAACCACTTCAC-3′;
wnt7aa 5‘-CACGGGAGATCAAGCAGAAC-3‘; 5‘- TGGTGCAGGATCCAGATACG-3‘
Expression levels were standardized to the primer set specific for β-actin
5′-TGACAGGATGCAGAAGGAGA-3′ and 5′-GCCTCCGATCCAGACAGAGT-3′.
Microscopy and imaging
Confocal imaging was performed using a Zeiss LSM780 upright confocal microscope or LSM880 upright confocal microscope equipped with an Airyscan module 32-channel GaAsP-PMT area detector, water-immersed ×20/1.0 NA or ×10/0.5 NA objective lens. Embryos were mounted on 1.5% (w/v) low-melting agarose.
Image processing
Confocal images were processed offline using the Fiji101 version of ImageJ (NIH) or Imaris v.9.3 (Bitplane). The images shown in this study are single-views, 2D-reconstructions of collected z-series stacks. The co-localization channel was created using the Imaris Colocalization Module. Co-localization thresholds were set manually. Transverse sections were prepared using Imaris 3D software from z-series stacks. Detection, colocalization, and quantification of Lysotracker, Lamp1, and Rab7 positive puncta were performed using ComDet v.0.5.3 plugin for ImageJ, available on Github [https://github.com/UU-cellbiology/ComDet] and Zenodo [https://zenodo.org/record/4281064].
In silico analyses
For Supplementary Fig. 1a, the amino acid sequences for zebrafish (Q8JGR8, 531aa), mouse (O35114, 478 aa), and human (Q14108, 478 aa) were obtained from Uniprot.org. Percentage identity were calculated using Uniprot.org.
For Supplementary Fig. 2a, previously published single-cell RNA sequencing data from developing mouse brain and spinal cord were used102. Bar graphs were made using R (version 4.3.1) with the ggplot2 package103.
For Supplementary Fig. 3k, single-cell transcriptomic data from 310 cells isolated from Tg(olig1:memEYFP) zebrafish at 5 dpf were obtained from34. Plots were obtained from a searchable database made publicly available at https://castelobranco.shinyapps.io/zebrafish_OPCs/34.
Statistics and reproducibility
Statistical significance between two samples was calculated using unpaired two-tailed Student’s t-tests assuming unequal variance from at least three independent experiments unless stated otherwise. Ordinary one-way ANOVA with Tukey’s multiple comparisons was used when comparing more than two groups to test mean differences. In all cases, normality was assumed, and variance was comparable between groups. Sample size was selected empirically according to previous experience in assessing experimental variability. Each experiment was repeated at least 3 times independently, unless stated otherwise.The investigators were blind to allocation during experiments and outcome assessment. Numerical data are the mean ± s.e.m. unless stated otherwise. Statistical calculations and the graphs for the numerical data were performed using Prism 8 software (GraphPad Software).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All cartoons and schemes in the paper were manually created using Adobe Illustrator 2024 and are original, not copied from any sources. The datasets generated and/or analysed during the current study are available in the GEO repository, GSE270309 and GSE110823. Source data are provided with this paper.
References
Paredes, I., Himmels, P. & Ruiz de Almodóvar, C. Neurovascular Communication during CNS Development. Dev. Cell 45, 10–32 (2018).
Segarra, M., Aburto, M. R., Hefendehl, J. & Acker-Palmer, A. Neurovascular Interactions in the Nervous System. Annu. Rev. Cell Dev. Biol. 35, 615–635 (2019).
Langen, U. H., Ayloo, S. & Gu, C. Development and Cell Biology of the Blood-Brain Barrier. Annu. Rev. Cell Dev. Biol. 35, 591–613 (2019).
Schaeffer, S. & Iadecola, C. Revisiting the neurovascular unit. Nat. Neurosci. 24, 1198–1209 (2021).
Kalucka, J. et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764–779.e20 (2020).
Andjelkovic, A. V., Keep, R. F. & Wang, M. M. Molecular Mechanisms of Cerebrovascular Diseases. Int. J. Mol. Sci. 23, 7161 (2022).
Wardlaw, J. M., Benveniste, H. & Williams, A. Cerebral Vascular Dysfunctions Detected in Human Small Vessel Disease and Implications for Preclinical Studies. Annu. Rev. Physiol. 84, 409–434 (2022).
Beattie, R. & Hippenmeyer, S. Mechanisms of radial glia progenitor cell lineage progression. Febs Lett 591, 3993–4008 (2017).
Kim, H. et al. Notch-regulated oligodendrocyte specification from radial glia in the spinal cord of zebrafish embryos. Dev. Dyn. 237, 2081–2089 (2008).
Ge, W. et al. Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J. Neurosci. Res. 69, 848–860 (2002).
Tanigaki, K. et al. Notch1 and Notch3 Instructively Restrict bFGF-Responsive Multipotent Neural Progenitor Cells to an Astroglial Fate. Neuron 29, 45–55 (2001).
Gonzalez, A., Valeiras, M., Sidransky, E. & Tayebi, N. Lysosomal Integral Membrane Protein-2: A New Player in Lysosome-Related Pathology. Mol. Genet. Metab. 111, 84–91 (2014).
Fujita, M. et al. Assembly and patterning of the vascular network of the vertebrate hindbrain. Development 138, 1705–1715 (2011).
Quiñonez-Silvero, C., Hübner, K. & Herzog, W. Development of the brain vasculature and the blood-brain barrier in zebrafish. Dev. Biol. 457, 181–190 (2020).
Yaniv, K. et al. Live imaging of lymphatic development in the zebrafish. Nat Med 12, 711–716 (2006).
Zheng, P.-P. et al. Glut1/Slc2a1 Is Crucial for the Development of the Blood-Brain Barrier in Vivo. Ann. Neurol. 68, 835–844 (2010).
Gamp, A.-C. et al. LIMP-2/LGP85 deficiency causes ureteric pelvic junction obstruction, deafness and peripheral neuropathy in mice. Hum. Mol. Genet. 12, 631–646 (2003).
Rosenberg, A. B. et al. SPLiT-seq reveals cell types and lineages in the developing brain and spinal cord. Science 360, 176–182 (2018).
Velmeshev, D. et al. Single-cell genomics identifies cell type–specific molecular changes in autism. Science 364, 685–689 (2019).
Chaerkady, R. et al. Quantitative temporal proteomic analysis of human embryonic stem cell differentiation into oligodendrocyte progenitor cells. PROTEOMICS 11, 4007–4020 (2011).
van der Poel, M. et al. Transcriptional profiling of human microglia reveals grey–white matter heterogeneity and multiple sclerosis-associated changes. Nat. Commun. 10, 1139 (2019).
Diaz-Tellez, A., Zampedri, C., Ramos-Balderas, J. L., García-Hernández, F. & Maldonado, E. Zebrafish scarb2a insertional mutant reveals a novel function for the Scarb2/Limp2 receptor in notochord development. Dev. Dyn. 245, 508–519 (2016).
Jurisch-Yaksi, N., Yaksi, E. & Kizil, C. Radial glia in the zebrafish brain: Functional, structural, and physiological comparison with the mammalian glia. Glia 68, 2451–2470 (2020).
Johnson, K. et al. Gfap-positive radial glial cells are an essential progenitor population for later-born neurons and glia in the zebrafish spinal cord. GLIA. https://doi.org/10.1002/glia.22990 (2016).
Graham, V., Khudyakov, J., Ellis, P. & Pevny, L. SOX2 Functions to Maintain Neural Progenitor Identity. Neuron 39, 749–765 (2003).
Peretz, Y. et al. A new role of hindbrain boundaries as pools of neural stem/progenitor cells regulated by Sox2. BMC Biol 14, 57 (2016).
Hevia, C. F., Engel-Pizcueta, C., Udina, F. & Pujades, C. The neurogenic fate of the hindbrain boundaries relies on Notch3-dependent asymmetric cell divisions. Cell Rep 39, 110915 (2022).
Belmonte-Mateos, C., Meister, L. & Pujades, C. Hindbrain rhombomere centers harbor a heterogenous population of dividing progenitors which rely on Notch signaling. Front. Cell Dev. Biol. 11, 1170–1189 (2023).
Than-Trong, E. & Bally-Cuif, L. Radial glia and neural progenitors in the adult zebrafish central nervous system. Glia 63, 1406–1428 (2015).
Lyons, D. A. & Talbot, W. S. Glial Cell Development and Function in Zebrafish. Cold Spring Harb. Perspect. Biol. 7, a020586 (2015).
Zannino, D. A. & Appel, B. Olig2+ Precursors Produce Abducens Motor Neurons and Oligodendrocytes in the Zebrafish Hindbrain. J. Neurosci. 29, 2322–2333 (2009).
Emery, B. Regulation of Oligodendrocyte Differentiation and Myelination. Science 330, 779–782 (2010).
Jung, S.-H. et al. Visualization of myelination in GFP-transgenic zebrafish. Dev. Dyn. 239, 592–597 (2010).
Marisca, R. et al. Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat. Neurosci. 23, 363–374 (2020).
Zheng, K., Huang, H., Yang, J. & Qiu, M. Origin, molecular specification, and stemness of astrocytes. Dev. Neurobiol. 82, 149–159 (2022).
Shibata, T. et al. Glutamate Transporter GLAST Is Expressed in the Radial Glia–Astrocyte Lineage of Developing Mouse Spinal Cord. J. Neurosci. 17, 9212–9219 (1997).
Chen, J., Poskanzer, K. E., Freeman, M. R. & Monk, K. R. Live-imaging of astrocyte morphogenesis and function in zebrafish neural circuits. https://doi.org/10.1038/s41593-020-0703-x.
Wang, J. et al. Paired Related Homeobox Protein 1 Regulates Quiescence in Human Oligodendrocyte Progenitors. Cell Rep 25, 3435–3450.e6 (2018).
Naruse, M., Shibasaki, K. & Ishizaki, Y. Temporal Changes in Transcription Factor Expression Associated with the Differentiation State of Cerebellar Neural Stem/Progenitor Cells During Development. Neurochem. Res. 43, 205–211 (2018).
Sagner, A. & Briscoe, J. Establishing neuronal diversity in the spinal cord: a time and a place. Development 146, dev182154 (2019).
Hutchinson, S. A. & Eisen, J. S. Islet1 and Islet2 have equivalent abilities to promote motoneuron formation and to specify motoneuron subtype identity. Development 133, 2137–2147 (2006).
Bingham, S. et al. Neurogenic phenotype ofmind bomb mutants leads to severe patterning defects in the zebrafish hindbrain. Dev. Dyn. 228, 451–463 (2003).
Gaiano, N. & Fishell, G. The Role of Notch in Promoting Glial and Neural Stem Cell Fates. Annu. Rev. Neurosci. 25, 471–490 (2002).
Engler, A., Zhang, R. & Taylor, V. Notch and Neurogenesis. in Molecular Mechanisms of Notch Signaling (Borggrefe, T. & Giaimo, B. D. eds.) 223–234 (Springer International Publishing, Cham, 2018).
Templehof, H., Moshe, N., Avraham-Davidi, I. & Yaniv, K. Zebrafish mutants provide insights into Apolipoprotein B functions during embryonic development and pathological conditions. JCI Insight 6, e130399 (2021).
Zaucker, A., Mercurio, S., Sternheim, N., Talbot, W. S. & Marlow, F. L. Notch3 Is Essential for Oligodendrocyte Development and Vascular Integrity in Zebrafish. Dis. Model. Mech. 6, 1246–1259 (2013).
Qiu, X., Lim, C.-H., Ho, S. H.-K., Lee, K.-H. & Jiang, Y.-J. Temporal Notch activation through Notch1a and Notch3 is required for maintaining zebrafish rhombomere boundaries. Dev. Genes Evol. 219, 339–351 (2009).
Kobia, F., Duchi, S., Deflorian, G. & Vaccari, T. Pharmacologic inhibition of vacuolar H+ ATPase reduces physiologic and oncogenic Notch signaling. Mol. Oncol. 8, 207–220 (2014).
Tognon, E. et al. Control of lysosomal biogenesis and Notch-dependent tissue patterning by components of the TFEB-V-ATPase axis in Drosophila melanogaster. Autophagy 12, 499–514 (2016).
Vaccari, T., Duchi, S., Cortese, K., Tacchetti, C. & Bilder, D. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Dev. Camb. Engl. 137, 1825–1832 (2010).
Yan, Y., Denef, N. & Schüpbach, T. The Vacuolar Proton Pump, V-ATPase, Is Required for Notch Signaling and Endosomal Trafficking in Drosophila. Dev. Cell 17, 387–402 (2009).
Valapala, M. et al. Impaired endolysosomal function disrupts Notch signaling in optic nerve astrocytes. Nat. Commun. 4, 1629 (2013).
Pasternak, S. H. et al. Presenilin-1, Nicastrin, Amyloid Precursor Protein, and γ-Secretase Activity Are Co-localized in the Lysosomal Membrane *. J. Biol. Chem. 278, 26687–26694 (2003).
Clark, B. S., Winter, M., Cohen, A. R. & Link, B. A. Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev. Dyn. 240, 2452–2465 (2011).
Ellis, K., Bagwell, J. & Bagnat, M. Notochord vacuoles are lysosome-related organelles that function in axis and spine morphogenesis. J. Cell Biol. 200, 667–679 (2013).
Parenti, G., Andria, G. & Ballabio, A. Lysosomal Storage Diseases: From Pathophysiology to Therapy. Annu. Rev. Med. 66, 471–486 (2015).
Wang, Y., Pan, L., Moens, C. B. & Appel, B. Notch3 establishes brain vascular integrity by regulating pericyte number. Development 141, 307–317 (2014).
Zhao, L. et al. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl. Acad. Sci. 111, 1403–1408 (2014).
Mackenzie, F. & Ruhrberg, C. Diverse roles for VEGF-A in the nervous system. Development 139, 1371–1380 (2012).
Tata, M. & Ruhrberg, C. Cross-talk between blood vessels and neural progenitors in the developing brain. Neuronal Signal 2, NS20170139 (2018).
Daneman, R. & Prat, A. The Blood–Brain Barrier. Cold Spring Harb. Perspect. Biol. 7, a020412 (2015).
Daneman, R. et al. Wnt/β-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc. Natl. Acad. Sci. USA 106, 641–646 (2009).
O’Brown, N. M. et al. The secreted neuronal signal Spock1 promotes blood-brain barrier development. Dev. Cell 58, 1534–1547.e6 (2023).
Lange, M. et al. Zebrafish mutants in vegfab can affect endothelial cell proliferation without altering ERK phosphorylation and are phenocopied by loss of PI3K signaling. Dev. Biol. 486, 26–43 (2022).
Rossi, A. et al. Regulation of Vegf signaling by natural and synthetic ligands. Blood 128, 2359–2366 (2016).
Fetsko, A. R., Sebo, D. J. & Taylor, M. R. Brain endothelial cells acquire blood-brain barrier properties in the absence of Vegf-dependent CNS angiogenesis. Dev. Biol. 494, 46–59 (2023).
Cho, C., Smallwood, P. M. & Nathans, J. Reck and Gpr124 Are Essential Receptor Cofactors for Wnt7a/Wnt7b-Specific Signaling in Mammalian CNS Angiogenesis and Blood-Brain Barrier Regulation. Neuron https://doi.org/10.1016/j.neuron.2017.07.031 (2017).
Hübner, K. et al. Wnt/β-catenin signaling regulates VE-cadherin-mediated anastomosis of brain capillaries by counteracting S1pr1 signaling. Nat. Commun. https://doi.org/10.1038/s41467-018-07302-x (2018).
Vanhollebeke, B. et al. Tip cell-specific requirement for an atypical Gpr124- and Reck-dependent Wnt/β-catenin pathway during brain angiogenesis. eLife 4, e06489 (2015).
Martin, M. et al. Engineered Wnt ligands enable blood-brain barrier repair in neurological disorders. Science 375, eabm4459 (2022).
Schevenels, G. et al. A brain-specific angiogenic mechanism enabled by tip cell specialization. Nature 628, 863–871 (2024).
Ulrich, F. et al. Reck enables cerebrovascular development by promoting canonical Wnt signaling. Development 143, 1055–1055 (2016).
Karra, R. et al. Vegfaa instructs cardiac muscle hyperplasia in adult zebrafish. Proc Natl Acad Sci U A 115, 8805–8810 (2018).
Nicenboim, J. et al. Lymphatic vessels arise from specialized angioblasts within a venous niche. Nature 522, 56–61 (2015).
Posokhova, E. et al. GPR124 functions as a WNT7-specific coactivator of canonical β-catenin signaling. Cell Rep. https://doi.org/10.1016/j.celrep.2014.12.020 (2015).
Alunni, A. et al. Notch3 signaling gates cell cycle entry and limits neural stem cell amplification in the adult pallium. Dev. Camb. Engl. 140, 3335–3347 (2013).
Kopan, R., Schroeter, E. H., Weintraub, H. & Nye, J. S. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. 93, 1683–1688 (1996).
Yoshimori, T., Yamamoto, A., Moriyama, Y., Futai, M. & Tashiro, Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266, 17707–17712 (1991).
Xu, J. et al. Effects of Bafilomycin A1: An inhibitor of vacuolar H (+)-ATPases on endocytosis and apoptosis in RAW cells and RAW cell-derived osteoclasts. J. Cell. Biochem. 88, 1256–1264 (2003).
Daneman, R. et al. Wnt/β-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc. Natl. Acad. Sci. USA. https://doi.org/10.1073/pnas.0805165106 (2009).
Zhou, Y. & Nathans, J. Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical Wnt signaling. Dev. Cell. https://doi.org/10.1016/j.devcel.2014.08.018 (2014).
Hübner, K. et al. Wnt/β-catenin signaling regulates VE-cadherin-mediated anastomosis of brain capillaries by counteracting S1pr1 signaling. Nat. Commun. 9, 4860 (2018).
Siqueira, M. et al. Radial Glia Cells Control Angiogenesis in the Developing Cerebral Cortex Through TGF-β1 Signaling. https://doi.org/10.1007/s12035-017-0557-8 (2018).
Mizuno, T., Mizuta, I., Watanabe-Hosomi, A., Mukai, M. & Koizumi, T. Clinical and Genetic Aspects of CADASIL. Front. Aging Neurosci. 12, 91 (2020).
Jerafi-Vider, A. et al. VEGFC/FLT4-induced cell-cycle arrest mediates sprouting and differentiation of venous and lymphatic endothelial cells. Cell Rep 35, 109255 (2021).
Gancz, D. et al. Distinct origins and molecular mechanisms contribute to lymphatic formation during cardiac growth and regeneration. eLife 8, e44153 (2019).
Gore, A. V. et al. Rspo1/Wnt signaling promotes angiogenesis via Vegfc/Vegfr3. Development 138, 4875–4886 (2011).
Bernardos, R. L. & Raymond, P. A. GFAP transgenic zebrafish. Gene Expr. Patterns GEP 6, 1007–1013 (2006).
Shimizu, Y., Ito, Y., Tanaka, H. & Ohshima, T. Radial glial cell-specific ablation in the adult Zebrafish brain: radial Glial Cell-Specific Ablation in Adult Zebrafish. genesis 53, 431–439 (2015).
Das, R. N. et al. Generation of specialized blood vessels via lymphatic transdifferentiation. Nature 606, 570–575 (2022).
Umans, R. A. et al. CNS angiogenesis and barriergenesis occur simultaneously. Dev. Biol. 425, 101–108 (2017).
Jao, L. E., Wente, S. R. & Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA 110, 13904–13909 (2013).
Suster, M. L., Abe, G., Schouw, A. & Kawakami, K. Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6, 1998–2021 (2011).
Villefranc, J. A., Amigo, J. & Lawson, N. D. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077–3087 (2007).
Avraham-Davidi, I. et al. ApoB-containing lipoproteins regulate angiogenesis by modulating expression of VEGF receptor 1. Nat. Med. 18, 967–973 (2012).
He, C., Bartholomew, C. R., Zhou, W. & Klionsky, D. J. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy 5, 520–526 (2009).
Robertson, J. K. et al. Targeting the Wnt pathway in zebrafish as a screening method to identify novel therapeutic compounds. Exp. Biol. Med. 239, 169–176 (2014).
Shin, M. et al. Vegfa signals through ERK to promote angiogenesis, but not artery differentiation. Development 143, 3796–3805 (2016).
Keren-Shaul, H. et al. MARS-seq2.0: an experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat. Protoc. 14, 1841–1862 (2019).
Kohen, R. et al. UTAP: User-friendly Transcriptome Analysis Pipeline. BMC Bioinformatics 20, 154 (2019).
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nature Methods 9, 676–682 (2012).
Rosenberg, A. B. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. (Springer New York, New York, NY, 2009). 10.1007/978-0-387-98141-3.
Acknowledgements
The authors would like to thank H. Hasid, L. Shen, and Y. Yogev (Weizmann Institute, Israel) for technical assistance, K. Monks (Vollum Institute, US), C. Burns (Harvard Institute, US), B. Appel (University of Colorado, US), M. Bagnat (Duke Cancer Institute, US) and B. A. Link (Wisconsin Medical School, US) for providing fish lines. S. Federici (Weizmann Institute, Israel) for assistance with statistical analyses and illustrations, G. Almog, R. Hofi, A. Glozman, and R. Brihon (Weizmann Institute, Israel) for superb animal care. The authors are grateful to all the members of the Yaniv lab for discussion, technical assistance, critical reading of the manuscript, and continuous support. This work was supported in part by Minerva Foundation (714447) to KY, ERA-Net-NEURON (Tackle-CSVD) to KY, ERC StG (LIPintoECtion 335605) to KY, and research grants from The M. Judith Ruth Institute for Preclinical Brain Research (P137071), the Weizmann – Nella and Leon Benoziyo Center for Neurological Diseases and the Weizmann SABRA - Yeda-Sela - WRC Program, the Estate of Emile Mimran, and The Maurice and Vivienne Wohl Biology Endowment and the Brenden-Mann Women’s Innovation Impact Fund. K.Y. is the incumbent of the Enid Barden and Aaron J. Jade Professorial Chair. C.R.A. acknowledges support from ERA-Net-NEURON (Tackle-CSVD) and ERC CoG (OLI.VAS 864875). I.B. was supported by a postdoctoral fellowship by the Sergio Lombroso Program and a Senior Postdoc Fellowship by the Weizmann Institute. N.M. was supported by research grants from the Estate of Olga Klein Astrachan and the Estate of Mady Dukler.
Author information
Authors and Affiliations
Contributions
I.B. designed and conducted experiments, analyzed data, and co-wrote the manuscript; I.B., M.G., G.H., N.M., L.G-M., D.K. and A.S. conducted experiments and data analyses; K.A.R. and S.S. performed bioinformatics analyses; I.B. and N.M. generated transgenic lines; C.R.A contributed to interpretation of the results, provided important intellectual content and co-wrote the manuscript; K.Y. directed the study, secured funding, designed experiments, analyzed data and co-wrote the paper with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Bruce Appel, Carla Belmonte Mateos, Cristina Pujades, and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bassi, I., Grunspan, M., Hen, G. et al. Endolysosomal dysfunction in radial glia progenitor cells leads to defective cerebral angiogenesis and compromised blood-brain barrier integrity. Nat Commun 15, 8158 (2024). https://doi.org/10.1038/s41467-024-52365-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52365-8
- Springer Nature Limited