Abstract
Despite recent advances in systemic therapy for hepatocellular carcinoma (HCC), the prognosis of hepatitis B virus (HBV)-induced HCC patients remains poor. By screening a sgRNA library targeting human deubiquitinases, we find that ubiquitin-specific peptidase 26 (USP26) deficiency impairs HBV-positive HCC cell proliferation. Genetically engineered murine models with Usp26 knockout confirm that Usp26 drives HCC tumorigenesis. Mechanistically, we find that the HBV-encoded protein HBx binds to the promoter and induces the production of USP26, which is an X-linked gene exclusively expressed in the testis. HBx consequently promotes the association of USP26 with SIRT1 to synergistically stabilize SIRT1 by deubiquitination, which promotes cell proliferation and impedes cell apoptosis to accelerate HCC tumorigenesis. In patients with HBV-positive HCC, USP26 is robustly induced, and its levels correlate with SIRT1 levels and poor prognosis. Collectively, our study highlights a causative link between HBV infection, deubiquitinase induction and development of HCC, identifying a druggable target, USP26.
Similar content being viewed by others
Introduction
Liver cancer, also known as hepatic cancer, is the sixth most common cancer worldwide and the fourth leading cause of cancer-related deaths. Hepatocellular carcinoma is the most common type, accounting for about 75-85% of all liver cancer cases1,2. Several risk factors have been documented for HCC; among them, chronic hepatitis B is a leading pathogenic factor causing 50–80% of HCC cases3. HBV DNA sequences integrated into the host genome include the X, C, enhancer, and S genes, among which the X and C genes are the most common integration genes in the genomes of patients with HCC4. The integration of HBV DNA into the host genome can result in gene mutations, chromosome deletions, and rearrangement of gene sequences5,6. Several known tumor suppressors or oncogenes are significantly mutated in HBV-associated HCC, including TP53 (58%), Axin1 (18%), RB1 (6%), CTNNB1 (19%) and Keap1 (7%)7. Among them, most are tumor suppressor genes that are not druggable targets. In addition, by inactivating CD8+ T cells8 and recruiting Foxp3+ Treg cells9, the tumor microenvironment of HBV-associated HCC also shows severe immunosuppression and exhaustion, which contributes to the poor prognosis of patients with HBV3.
Treatment for liver cancer depends on the stage of the cancer and may include surgery, chemotherapy, radiation therapy, targeted therapy, immunotherapy, or a combination of these approaches10. Although recent advances in systematic therapy for HCC, such as the invention of sorafenib11, lenvatinib12, and regorafenib13, the overall survival of advanced HCC patients remains extremely poor due to lower response rates14. More seriously, despite recent breakthroughs in immunotherapy trials for the treatment of advanced HCC using immune checkpoint blockade, such as anti-PD-L115,16 and anti-PD-117, overall response rates remain very low at around 20%, suggesting that complexities remain in understanding the HCC immune landscape18. It is necessary to reconsider disease-causing druggable targets at the cancer cell-autonomous level.
Sirtuin 1 (SIRT1) is a nicotinamide adenine dinucleotide-dependent deacetylase that plays a key role in regulating a variety of cellular processes, including metabolism, DNA repair, aging and cancer progression through epigenetic reprogramming or non-histone modulation19,20,21. Although studies have suggested that SIRT1 may function as a tumor suppressor by preventing the accumulation of DNA damage22, more studies have demonstrated that SIRT1 is overexpressed in cancer cells such as liver cancer cells and actually promotes cancer growth by regulating the expression of genes involved in cell proliferation and survival23. In addition, SIRT1 may help cancer cells evade the immune system and resist chemotherapy11. Moreover, some studies have suggested that SIRT1 may promote the development of liver cancer in patients with chronic HBV infection by promoting cell proliferation and inhibiting apoptosis24. However, the relationship between SIRT1 and HBV and how HBV infection regulates SIRT1 in HCC progression need to be fully understood.
Ubiquitination is a post-translational modification process in which a small protein called ubiquitin is attached to a target protein, thereby marking it for degradation by the proteasome or altering its function25. SIRT1 can be regulated by either autophagosome-lysosome26 or ubiquitin-proteasome through polyubiquitination27,28. In contrast, SIRT1, like many other key proteins associated with cancer, such as CHK1 and EZH2, as we previously found29,30, can also be regulated by deubiquitination. Although USP22 has been reported to deubiquitinate SIRT1 and promote deacetylation of p53, it is difficult to use as a drug target considering that USP22 is widely expressed in normal tissues and that loss of USP22 leads to embryonic lethality in mice31. Further research is needed to identify the oncogenic and druggable deubiquitinase responsible for SIRT1 overexpression and hyperactivation in hepatocellular carcinoma.
In this study, we conduct unbiased screening to investigate whether loss of deubiquitinase causes lethality in HCC cells. By using all individual DUB (sgRNA library covering 87 human deubiquitinases (DUBs)) knockout stable cell lines, we identify that USP26 is important for the survival and proliferation of HCC cells. Furthermore, we find that USP26 is transcriptionally activated by HBx and highly expressed in HBV-positive HCC samples. Aberrantly expressed USP26 consequently binds and stabilizes SIRT1 by deubiquitination, which drives HCC development both in vitro and in genetic or xenograft murine tumor models. Taken together, our study reveals a causal role of USP26–SIRT1 signaling in HBV-induced liver pathogenesis, uncovers a mechanism of aberrant SIRT1 regulation in HCC and identifies a potential therapeutic target, a cancer/testis deubiquitinase.
Results
Identification of USP26 as a potential driver of hepatocellular carcinogenesis
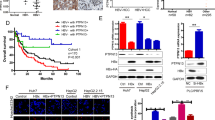
The deubiquitinating enzyme (DUB) library screening has been used to identify novel dependencies in drug resistance by us and other groups32,33. To investigate the druggable DUBs driving HBV-associated liver cancer, we performed CRISPR/Cas9 screening using a DUB knockout library that contains 348 sgRNAs against 87 human DUBs (pooled 4 sgRNA targeting one DUB), which represent most human DUBs (Supplementary Data 1). Here, a highly metastatic HBV-positive HCC cell line, HCCLM3, was used to generate all individual DUB knockout stable cell lines for initial screening (Fig. 1a). Accordingly, a colony formation assay and EdU analysis were cooperatively carried out to select the key DUBs responsible for HCCLM3 cell survival and proliferation. The results showed that the loss of some candidate DUBs inhibited the survival and proliferation ability of HCCLM3 cells compared to the sg-Control group (Fig. 1b, c). We considered DUBs to be significant if they had a p < 0.01 and a log2 fold change < −0.58 (Supplementary Data 2, 3). Based on these criteria, we identified 8 DUBs that were significant according to colony formation (Fig. 1d, red pie graph) and 10 DUBs that were significant according to EdU analysis (Fig. 1d, blue pie graph); among these DUBs, 3 DUBs (USP26, BAP1 and COPS6) were significant in two screening assays (Fig. 1d, dark brown oval graph). Further experiments showed that USP26 deletion led to more apoptosis than did the other two DUBs (Supplementary Fig. 1a). Thus, we focused on USP26 and its potential as a drug target.
a Schematic strategy for CRISPR-based DUB library screening. A total of 348 sgRNAs targeting 87 DUBs (4 sgRNAs/DUB) were transfected into the HCCLM3 cell line, which was subsequently screened. Left: Dot plots showing the rank of DUBs from 1 (lowest) to 87 (highest) based on colony formation assay (b) and EdU staining assay (c) of sg-DUBs HCCLM3 cells, which were measured and calculated by log2fold change. The top DUBs with inhibitory effects on cell growth were listed as indicated. Right: The percentage of significantly changed DUBs in each group (p < 0.01, log2 fold change < −0.58, down group; p < 0.01, log2fold change > 0.26, up group; p > 0.05, −0.58 <log2 fold change < 0.26, no difference group (No-diff)). d Venn diagram showing the overlapping DUBs that inhibited cell survival in the two screening assays described in b and c. e, f Western blot analysis and colony formation assay in HCCLM3 cells with or without CRISPR/Cas9-mediated USP26 knockout. g Flow cytometry analysis showing the percentages of cells in S phase in the sg-Control and USP26 knockout groups. h. Representative immunoblots showing USP26 protein levels in tumors and paired normal tissues from HBV-positive (HBV+) HCC patients. N, normal tissue; T, tumor tissue. Quantification of relative USP26 protein levels in tumors and paired normal tissues from HBV+ HCC patients (i) and HBV-negative (HBV−) HCC patients (j). k Kaplan-Meier survival analysis of HCC patients with high or low USP26 expression. The data were obtained from the Kaplan-Meier plotter database. Each graph is presented as the mean ± SEM. b, c n = 3 biologically independent experiments; (e–h) n = 3 biologically independent experiments; n = 42 (i) and n = 8 (j) paired tumor and normal samples. Statistical significance was calculated by (b–d) one-way ANOVA; (e, g) two-tailed unpaired t-test; (i, j) two-tailed paired t-test; (k) log-rank test. Source data are provided as a Source Data file.
USP26 is a member of the deubiquitinating enzyme family that cleaves ubiquitin from protein substrates. It has been reported to be an X-linked gene exclusively expressed in the testis and plays important roles in spermatogenesis. Although our results indicated that USP26 deficiency reduced the proliferative ability of HCCLM3 cells (Fig. 1e–g), whether and how USP26 correlates with HCC remain unclear. To further clarify the oncogenic role of USP26 in HCC progression, we detected the expression of USP26 in 50 paired HCC tissues and adjacent tissues, including 42 paired HBV-positive HCC tissues and 8 paired HBV-negative HCC tissues, and found that USP26 was significantly upregulated in tumors from HBV-positive HCC patients (Fig. 1h and i, and Supplementary Fig. 1b) but not in tumors from HBV-negative HCC patients (Fig. 1j). Kaplan-Meier survival analysis of HCC patients revealed that higher expression of USP26 was associated with poorer prognosis (Fig. 1k). Collectively, these results suggested that USP26 is a cancer/testis deubiquitinase responsible for HCC cell survival and proliferation, and its expression may correlate with HBV infection status and patient survival status.
USP26 deficiency inhibits HCC tumorigenesis
To validate these findings, we established stable cell lines by restoring USP26 in USP26-depleted HCCLM3, MHCC97H and Huh7 HCC cell lines. Depletion of USP26 had a significant inhibitory effect on HCC cell growth, while restoration of USP26 rescued the phenotype induced by USP26 deficiency (Fig. 2a–j, and Supplementary Fig. 2a-i), indicating that USP26 has a significant effect on promoting cell proliferation in HCC cell lines. To further clarify whether the effect of USP26 deletion on HCC cells is related to HBV status, we assessed the function of USP26 in three other HBV-negative cell lines, HepG2, HLE and SMMC7721. USP26 knockdown had no effect on the proliferation of HepG2 or HLE cells but slightly inhibited that of SMMC7721 cells (Supplementary Fig. 2j–l), suggesting that USP26 plays an important role in HBV-related HCC cell survival and proliferation. In addition, knockdown of USP26 in MHCC97H cells increased total apoptosis, including early apoptosis and late apoptosis (cell death), by 3-fold, which could be completely reversed by restoration of USP26 (Fig. 2k–m). Consistently, the expression of cleaved PARP, the hallmark of apoptosis, was dramatically induced by USP26 deficiency, and re-expression of USP26 reversed this phenotype (Fig. 2n).
Western blot analysis (a), MTT (b), colony formation (c) and EdU staining assays (d, e) of USP26-knockdown HCCLM3 cells with or without USP26 (shRNA-resistant USP26) restoration. Scale bar: 100 μm. Western blot analysis (f), MTT (g), colony formation (h) and EdU staining assays (i, j) of USP26-knockdown MHCC97H cells with or without USP26 (shRNA-resistant USP26) restoration. Scale bar: 100 μm. k–m. The apoptosis rate of MHCC97H cells as described in f was measured by flow cytometry after Annexin/V and PI dual staining. n Cleaved PARP (C-PARP) expression in MHCC97H cells as described in f was measured by western blotting. The red stars (*) in a, f and n indicate the positions of USP26. Each graph is presented as the mean ± SEM. Each blot data is representative of three independent experiments. (b, g) n = 4 independent experiments; (c, d, h, i, k) n = 3 independent experiments. Statistical significance was calculated by (b, g) two-way ANOVA; (c, e, h, j, l, m) one-way ANOVA. Source data are provided as a Source Data file.
We then asked whether USP26 is a functional regulator in hepatocellular carcinoma tumorigenesis. To investigate the physiological and pathologic role of USP26, Usp26 knockout mice were generated via CRISPR/Cas9 technology, and then the expression of Usp26 in the mouse testis was detected to verify the knockout efficiency because the expression of Usp26 is very low in the liver and other normal tissues34 (Supplementary Fig. 3a, b). Previous studies have established that the cooperation of ΔN90-β-catenin and YAP-S127A could induce the hepatoblastoma model in mice35. Accordingly, we used hydrodynamic tail vein injection (HDI) and the Sleeping Beauty (SB) transposon system to deliver the plasmids stably into the male mouse liver, which would consistently express integrated genes36. Seven weeks later, the livers of male mice were collected for subsequent analysis (Fig. 3a). As expected, deletion of Usp26 in mice resulted in a decreased tumor burden in comparison to that in Usp26+/Y mice, as shown by morphological analysis (Fig. 3b). Similarly, the Usp26 knockout group exhibited 70%, 70% and 80% reductions in liver weight, the liver-to-body weight ratio, and tumor nodules, respectively, relative to the Usp26+/Y group (Fig. 3c–f). In addition, Ki67 staining and TUNEL staining showed that Usp26 deficiency resulted in growth inhibition and cell apoptosis, which collaboratively dampened liver tumorigenesis (Fig. 3g–j). Kaplan–Meier survival analysis showed that mice with Usp26-depleted tumors had prolonged survival compared with Usp26+/Y mice (Fig. 3k). It should be noted that the expression of Usp26 was robustly induced in genetically engineered murine liver tumors compared to normal hepatocytes or the adjacent tissues (Supplementary Fig. 3c, d), indicating the causal role of USP26 in liver tumorigenesis. Consistently, Usp26 deficiency significantly inhibited female mouse liver tumorigenesis (Supplementary Fig. 3e–g). To corroborate the results, we used the same HDI and SB transposon system to co-deliver the β-catenin-S33/37 A and MYC plasmids to generate another HCC murine model37,38. In parallel, we constructed a luciferase reporter gene co-expressed with MYC to monitor tumors (Fig. 3l). Eight weeks after injection, we confirmed tumor establishment by bioluminescence imaging (Fig. 3m, n). Similarly, loss of Usp26 markedly suppressed oncogene-induced HCC progression (Fig. 3o–t and Supplementary Fig. 3h), confirming that Usp26 deficiency inhibits liver tumor progression.
a Schematic illustration of hepatoblastoma model by hydrodynamic injection (HDI) of ΔN90-β-catenin and YAP-S127A plasmids. IR/DR refers to inverted repeats and direct repeats. Images of liver tumors (b), liver weight (c) and the ratio of liver weight to body weight (d) from Usp26+/Y and Usp26−/Y mice generated in a. Scale bar: 10 mm. Representative images and quantitative results of H&E staining (e, f), Ki67 staining (g, h) and TUNEL-positive cells (i, j) in tumor nodules from Usp26+/Y and Usp26−/Y mouse liver sections. The areas circled by black dashed lines in e point to tumor nodules. N normal tissue, T tumor. Scale bar: 200 μm (e), 50 μm (g, i). k Kaplan‒Meier curves of the HDI hepatoblastoma model. l Schematic illustration of the HCC model with HDI of oncogenic β-catenin-S33/37 A and MYC-luciferase plasmids. IRES indicates the internal ribosome entry site. m, n Representative images and quantitative bioluminescence intensity of liver tumors in l. Representative liver images (o), liver-to-body ratios (p), H&E staining (q, r) and Ki67 staining (s, t) of the tumor nodules in Usp26+/Y and Usp26−/Y mouse liver sections. Scale bar: 10 mm (o), 200 μm (q), 50 μm (s). u. DNA sequencing was used to validate the gene editing of USP26 in single cell-derived knockout clones. Schematic representation with various insertions (red) and guide sequences (blue) are marked. v–x Representative images, volumes, and weights of xenograft tumors derived from USP26 knockout (KO) HCCLM3 cells with or without USP26 restoration. Scale bar: 10 mm. Each graph is presented as the mean ± SEM. c, d n = 10 in Usp26+/Y group, 9 in Usp26−/Y group; (f, h) n = 6 per group; (k) n = 11 in Usp26+/Y group, 7 in Usp26−/Y group; (n) n = 9 in Usp26+/Y group, 6 in Usp26−/Y group; (j, p, r, t, w, x) n = 5 per group. Statistical significance was calculated by (c, d, f, h, j, n, p, r, t, x) two-tailed unpaired t-test; (k) log-rank test; (w) two-way ANOVA. Source data are provided as a Source Data file.
Next, we established a xenograft model to validate the effects of USP26 on human HCC tumorigenesis. The USP26-KO HCCLM3 cells (Fig. 3u) with or without USP26 restoration were injected into male nude mice, and tumor volume was then recorded. Notably, the tumor volume and weight were markedly reduced in the USP26-KO group compared with the control group, and these effects were fully reversed by restoration of USP26 expression (Fig. 3v–x). Thus, our findings from three different mouse models indicate that USP26 is a cancer driver for the occurrence and progression of HCC.
USP26 directly interacts with SIRT1
To elucidate the mechanism by which USP26 dictates HCC tumorigenesis, we attempted to identify the protein associated with USP26 using a triple-epitope (S-protein, FLAG tag and streptavidin binding peptide)-tagged version of USP26 (SFB-USP26) (Fig. 4a and Supplementary Table 1). Tandem affinity purification followed by mass spectrometric analysis identified several strong interactors of USP26, including reported MDM239, as well as a USP26 interactor, SIRT1. As a deacetylase, SIRT1 serves as a crucial regulator in the development of HCC. We next confirmed the exogenous interaction between Myc-SIRT1 and SFB-USP26 in HEK293T cells (Fig. 4b and Supplementary Fig. 4a), as well as the endogenous interaction of USP26 and SIRT1 by Co-immunoprecipitation assay in HCCLM3 and Huh7 cells (Fig. 4c and Supplementary Fig. 4b). Furthermore, purified recombinant GST-tagged SIRT1 could be pulled down by His-tagged USP26; reciprocally, purified His-USP26 could be pulled down by GST-SIRT1 under cell-free conditions, verifying that USP26 directly interacted with SIRT1 (Fig. 4d, e, and Supplementary Fig. 4c). In addition, immunofluorescence assays indicated that USP26 (green) colocalized with endogenous SIRT1 (red) in the nucleus (Fig. 4f).
a A Co-IP assay combined with mass spectrometry analysis was performed to screen for USP26-interacting proteins. A partial list of accession numbers and the matched peptides of candidate proteins is shown. b HEK293T cells were co-transfected with SFB-USP26 and Myc-SIRT1, and then Co-IP was performed using S-protein beads. c The interaction between endogenous USP26 and SIRT1 was measured by Co-IP. Lysates of HCCLM3 cells were immunoprecipitated with an anti-USP26 antibody or nonspecific IgG, and the associated SIRT1 was detected by immunoblotting. d. A GST pull-down assay was carried out to detect the interaction between USP26-His and GST-SIRT1. e His pull-down assay was carried out to detect the interaction between USP26-His and GST-SIRT1. f U2OS cells were transfected with the GFP-USP26 plasmid. The intracellular localization of USP26 (green) and endogenous SIRT1 (red) was visualized by using a laser confocal microscope. Scale bar: 10 μm. g Mapping the domain structure of SIRT1 that interacts with USP26. Full-length SIRT1 and its various deletion mutants together with USP26 were co-expressed in HEK293T cells. The binding regions between USP26 and SIRT1 were detected by Co-IP. A schematic diagram is shown above the panel. h The domain structure of USP26, which interacts with SIRT1, was measured by Co-IP. The binding regions were detected by Co-IP. A schematic diagram is shown above the panel. The red star (*) in c indicates the position of SIRT1. Data in (b–h) are representative from three independent experiments. Source data are provided as a Source Data file.
To further explore the regions involved in the interaction between USP26 and SIRT1, we constructed truncated variants of these two proteins accordingly (Fig. 4g, h). Co-immunoprecipitation assays showed that the H domain (residues 256-464) and the N-terminus (residues 1-256) of SIRT1 contributed to the interaction with USP26 (Fig. 4g and Supplementary Fig. 4d, e). Reciprocally, deleting the USP catalytic domain of USP26 markedly dampens the association between USP26 and SIRT1 (Fig. 4h). Taken together, our findings indicate that USP26 directly interacts with SIRT1 and that the interaction regions are the H domain and N domain of SIRT1 and the USP domain of USP26.
USP26 is a SIRT1 deubiquitinase
USP26 is a typical deubiquitinase, so we assumed that USP26 might stabilize SIRT1 by deubiquitination. To investigate whether USP26 regulates the stability of the SIRT1 protein, we first assessed the correlation between the expression of SIRT1 and the expression of USP26 in multiple HCC cell lines and normal hepatocytes and found a positive correlation between USP26 and SIRT1 (Fig. 5a). Consistently, USP26 overexpression indeed upregulated the endogenous protein level of SIRT1, whereas the mRNA level was unchanged (Fig. 5b, c, and Supplementary Fig. 5a), indicating posttranslational regulation of SIRT1 by USP26. Conversely, knockout of USP26 by using the CRISPR-Cas9 strategy (Fig. 3u) decreased the protein level of SIRT1 in HCCLM3 cells (Fig. 5d). Similarly, the ablation of Usp26 in mouse liver tumor tissues (Fig. 5e) and MHCC97H showed a reduction in SIRT1 expression, and the reduction could be rescued when USP26 was restored (Fig. 5f).
a The relative protein levels of SIRT1 and USP26 in different HCC cell lines. b The relative protein levels of SIRT1 in Huh7 cells transfected with the indicated doses of USP26. c. U2OS cells stably expressing GFP-USP26 were immunostained for endogenous SIRT1. Scale bar: 10 μm. The relative fluorescence intensity of SIRT1 in cells with or without GFP-USP26 expression was quantified in the same visual fields. d The protein levels of SIRT1 in USP26 knockout (KO) HCCLM3 cells. e The SIRT1 protein levels in liver tumors from Usp26+/Y and Usp26−/Y mice generated through the strategy described in Fig. 3a. f The protein levels of SIRT1 in USP26-knockdown and re-expressed shRNA-resistant USP26 MHCC97H cells. g The half-life of SIRT1 was measured by adding CHX to HEK293T cells transfected with wild-type (WT) USP26 or mutant USP26 (C304S). h The half-life of SIRT1 in USP26-knockdown and re-expressed shRNA-resistant USP26 MHCC97H cells was measured after treatment with CHX. i. HCCLM3 cells stably expressing SIRT1 were transfected with USP26 and then treated with MG132 (10 μM). Endogenous polyubiquitinated SIRT1 was detected by Western blotting. j, k USP26 KO HEK293T cells were co-transfected with HA-Ub, Myc-SIRT1, USP26 or mutant USP26 as indicated and then treated with MG132. Finally, the cells were lysed and subjected to a ubiquitination assay. l, m Ubiquitinated Myc-SIRT1 was purified from HEK293T cells, followed by incubation with bacteria-purified WT or mutant USP26 for an exogenous ubiquitination assay. The reaction buffer was subjected to immunoblotting or stained with Coomassie blue. n–q Similar to j, K29 and K48 ubiquitination assays were carried out. The red stars (*) in f indicate the positions of USP26. Each graph presents the mean ± SEM. Each blot data is representative of three independent experiments. Data in (c) n = 42 or 48 cells for WT or USP26 overexpressing group from three independent experiments. Data in (e) n = 7 mice per group. Statistical significance was calculated by (b, d, k, m, o, q) one-way ANOVA; (c) two-tailed unpaired t-test; (g, h) two-way ANOVA. Source data are provided as a Source Data file.
Further studies showed that overexpression of WT USP26 but not the catalytically dead mutant USP26C304S in HEK293T cells led to a pronounced increase in SIRT1 stability in the presence of cycloheximide (CHX), an inhibitor of protein translation (Fig. 5g). Conversely, depletion of USP26 in MHCC97H cells reduced SIRT1 stability, which could be reversed by restoration of USP26 (Fig. 5h). Since SIRT1 degradation occurs mainly through the 26S proteasome system (Supplementary Fig. 5b), we reasoned that USP26 may regulate SIRT1 protein levels through ubiquitin-dependent mechanisms. Consistent with the stabilization of SIRT1, overexpression of WT USP26, but not its catalytically inactive variant USP26C304S, markedly reduced the polyubiquitination of SIRT1 (Fig. 5i–k). To directly examine the deubiquitinating activity of USP26 toward SIRT1, we purified His-USP26 and its dead mutant from bacteria and ubiquitinated SIRT1 from HEK293T cells as indicated and then incubated them in a cell-free system. As expected, USP26 purified from bacteria decreased SIRT1 polyubiquitination by 75% in vitro; however, inactive USP26 had little effect on SIRT1 polyubiquitination (Fig. 5l, m). These results suggested that the enzymatic activity of USP26 is essential for protecting SIRT1 from degradation by deubiquitination.
Ubiquitin is known to be ubiquitinated on its seven lysine (K) residues, K6, K11, K27, K29, K33, K48 and K6340. To identify which ubiquitination type of SIRT1 was regulated by USP26, we generated all seven lysine-specific mutants of ubiquitin (e.g., the K48 mutant contains only a single lysine, K48, with all other six lysines mutated to arginine) and found that USP26 mainly cleaved K11-, K29-, K48- and K63-linked polyubiquitin chains of SIRT1 (Supplementary Fig. 5c–g). Further validation assays showed that only the deubiquitination effect of USP26 on K29- and K48-linked polyubiquitin chains of SIRT1 was significantly inhibited when the catalytic site was mutated (Fig. 5n–q and Supplementary Fig. 5h, i). Taken together, these results demonstrated that USP26 is a deubiquitinase of SIRT1 and may play an oncogenic role by deubiquitinating SIRT1.
USP26 promotes HCC tumorigenesis by stabilizing SIRT1
Whereas SIRT1 has been shown to dictate HCC tumorigenesis41,42, the role and mechanism of USP26 in HCC progression remain unclear. We wondered whether SIRT1 is a functional mediator of USP26 in HCC initiation. MTT and colony formation assays showed that silencing USP26 reduced the proliferative capacity of MHCC97H and HCCLM3 cells, which could be reversed by restoration of either USP26 or SIRT1 (Fig. 6a–f). Consistent with this finding, flow cytometry analysis also showed that SIRT1 overexpression significantly rescued the low proliferative capacity of HCC cells induced by USP26 knockdown (Fig. 6g). In addition, SIRT1 overexpression markedly blocked the apoptosis of MHCC97H cells induced by USP26 deficiency, which is consistent with the expression of cleaved PARP (Fig. 6h, i). Furthermore, in SIRT1 knockdown and USP26 knockout cells, overexpressing USP26 did not promote HCC cell proliferation, suggesting that USP26 promotes HCC tumorigenesis mainly through SIRT1 (Supplementary Fig. 6a–d). In addition, we found that ablation of USP26 robustly enriches the SIRT1-dependent epigenetic marks H3K9ac and H4K16ac43, which could be completely reversed by restoration of either USP26 or SIRT1 expression (Fig. 6j). Consistently, the SIRT1 target gene p66Shc44 was also regulated by USP26 (Fig. 6k). Finally, we generated a subcutaneous xenograft model to verify whether SIRT1 is the functional effector of USP26 in vivo. As expected, USP26 depletion dramatically shrunk the tumors, which could be rescued by overexpression of SIRT1 (Fig. 6l–n). Taken together, these results indicate that USP26 plays an oncogenic role in HCC by regulating SIRT1 and the corresponding epigenetic remodeling.
Western blot analysis (a), MTT (b) and colony formation (c) assays of USP26-knockdown MHCC97H cells with or without shRNA-resistant USP26 or SIRT1 overexpression. Western blot analysis (d), MTT (e) and colony formation (f) assays of USP26-knockdown HCCLM3 cells with or without USP26 (shRNA-resistant USP26) or SIRT1 overexpression. g. An EdU staining assay was carried out on USP26-knockdown and SIRT1 restored Huh7 and HCCLM3 cells. Flow cytometry was carried out to screen the active DNA replicated cells. h The apoptosis rates of USP26-knockdown and SIRT1 restored MHCC97H cells were measured by flow cytometry after Annexin/V/PI dual staining. i Immunoblotting of cleaved PARP in USP26-knockdown SIRT1restored MHCC97H cells. j Immunoblotting of the indicated proteins in USP26-knockdown MHCC97H cells with or without shRNA-resistant USP26 or SIRT1 overexpression. k The relative p66Shc mRNA levels in USP26-knockdown MHCC97H cells with or without shRNA-resistant USP26 or SIRT1 were detected by RT‒qPCR. l Representative images of xenograft tumors derived from USP26-knockout (KO) HCCLM3 cells with or without USP26 or SIRT1 overexpression. Scale bar: 10 mm. m The volumes of tumors burdened in nude mice receiving the HCCLM3 cells were measured on different days after implantation. n Weights of the mouse xenograft tumors. Each graph is presented as the mean ± SEM. Each blot data is representative of three independent experiments. b, e n = 4 independent experiments; c, f, g, h, k n = 3 independent experiments; (m, n) n = 4 mice per group. Statistical significance was calculated by (b, e, m) two-way ANOVA; (c, f, g, h, k) one-way ANOVA; (n) two-tailed unpaired t-test. Source data are provided as a Source Data file.
HBV transcriptionally activates USP26 expression and enhances the association between USP26 and SIRT1 to stabilize the SIRT1 protein
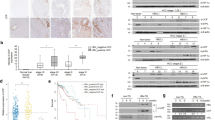
As mentioned, 1) USP26 was robustly expressed in HBV-positive HCC tissues (Fig. 1h–j), and 2) USP26 and SIRT1 were highly expressed and tightly correlated in HBV-positive HCC cells (Figs. 5a and 7b). Moreover, USP26 was highly co-expressed with SIRT1 in human HBV-positive tumor tissues and was significantly related to HBV infection status (Fig. 7a and Supplementary Fig. 7a–c). To further characterize the relationship between HBV and USP26 and SIRT1, Huh7, HepG2 and L02 cells (non-HBV integrated) were transfected with HBx, one of the core proteins encoded by HBV. Interestingly, we found that USP26 and SIRT1 protein levels were upregulated in the presence of HBx (Fig. 7c). Further analysis showed that HBx could promote USP26 but not SIRT1 at the transcriptional level, indicating that HBV regulates SIRT1 by activating USP26 (Fig. 7d, e). Indeed, in USP26-depleted Huh7 cells, HBx overexpression no longer increased SIRT1 expression compared to that in the control group (Fig. 7f). To verify whether HBx transactivates USP26, we identified the USP26 promoter region (−2000 to +100 base pair (bp) nucleotides) according to bioinformatics analysis and constructed it into a luciferase reporter vector and then cotransfected the cells with HBx and another two proteins encoded by HBV, HBc and HBs. Interestingly, HBx exhibited the predominant effects and increased the luciferase activity of the USP26 promoter in a dose-dependent manner (Fig. 7g, h), suggesting that HBx may be a transcriptional regulator of USP26.
a Left: Representative immunoblots of USP26 and SIRT1 protein levels in tumor and paired normal tissues of HBV-positive (HBV+) HCC patients. N normal tissue; T tumor. Right: Spearman correlation analysis between USP26 and SIRT1 protein expression in HBV+ HCC samples. b Quantification of relative USP26 and SIRT1 expression in HBV-negative (HBV−) and HBV+ HCC cell lines as indicated in Fig. 5a. c The endogenous protein levels of USP26 in Huh7 and HepG2 cells transfected with HBx. The relative mRNA levels of SIRT1 and USP26 in Huh7 (d) and L02 (e) cells transfected with HBx. f The protein levels of USP26 and SIRT1 in USP26-knockdown Huh7 cells transfected with HBx. g USP26 promoter activity was measured in Huh7 cells transfected with HBx, HBc or HBs. h USP26 promoter activity was measured in Huh7 cells transfected with HBx. i, j A series of USP26 promoter truncations were constructed to identify the regulatory region mediated by HBx. k, l In vivo and in vitro interaction between biotinylated USP26 promoter and HBx were detected by DNA pull-down assay. m EMSA assay was used to examine binding between HBx and the −450 to +100 region of the USP26 promoter. When the band of the free probe became weaker, it denoted strong binding. n The interaction between Myc-USP26 and HBx-Flag was detected in HEK293T cells. o The interaction between SFB-USP26 and Myc-SIRT1 was detected in the presence of HBx-Flag in HEK293T cells. p USP26-KO HEK293T cells were co-transfected with HA-Ub and Myc-SIRT1 with or without SFB-USP26 or SFB-HBx. The ubiquitination of SIRT1 was detected by Western blotting. Each graph presents the mean ± SEM. Data in (a) n = 41 human HBV+ HCC samples. Each blot data is representative of three independent experiments. Data in (d, e, g–j) n = 3 independent experiments. Statistical significance was calculated by (a) Spearman’s rank correlation coefficient, analysis of two-tailed t-test; (d, e, h) two-tailed unpaired t-test; (f, g, p) one-way ANOVA; (i, j) two-way ANOVA. Source data are provided as a Source Data file.
To delineate the specific regulatory region of the USP26 promoter activated by HBx, we generated serial truncations of the USP26 promoter as indicated (Fig. 7i). Luciferase reporter assays showed that the 4th promoter (Pro#4) (−450 to +100 bp) of USP26 is the regulatory region activated by HBx (Fig. 7i). We further identified that the region (−450 to −318 bp) is the core element for the transactivation of USP26 (Fig. 7j). As HBx could localize in the nucleus (Supplementary Fig. 7d, e), we hypothesized that HBx directly binds to the promoter of USP26. Through a DNA pull-down assay, we found that the purified HBx was obviously enriched by the biotinylated USP26 promoter (Fig. 7k, l). EMSA assays further confirmed that the Pro4# (−450 to +100 bp) region is responsible for direct binding to HBx (Fig. 7m and Supplementary Fig. 7f). These results prompted us to determine whether HBx serves as a transcription factor and thus regulates USP26 transcription. The GAL4 system was introduced to verify this hypothesis. We constructed a GAL4-DBD vector fused with HBx and then cotransfected it with a 5 x TATA luciferase reporter to detect luciferase activity in HEK293T cells. Although positive controls such as GAL4-DBD-P53 and GAL4-DBD-TCF4 could activate the reporter, GAL4-DBD-HBx had no effect on the reporter, indicating that HBx could not function as a transcriptional activator (Supplementary Fig. 7g), which is consistent with a previous study45. Since HBx can activate multiple transcription factors and thereby promote transcription, we speculate that HBx may enhance the transcription of USP26 as a transcriptional coactivator. Interestingly, when we analyzed the mRNA levels of Usp26 in mouse tumors and adjacent normal liver tissues, we found that either the β-catenin + YAP oncogenic combination or the β-catenin + MYC oncogenic combination could induce the expression of Usp26 (Supplementary Fig. 7h, i), suggesting that β-catenin-mediated transactivation plays a key role in USP26 transcription. To further validate this hypothesis, we transfected β-catenin into Huh7 cells and found that β-catenin indeed significantly induced USP26 mRNA expression (Supplementary Fig. 7j). Conversely, loss of β-catenin dramatically decreased the expression of USP26 in MHCC97H cells (Supplementary Fig. 7k). In addition, we found that HBx could interact with β-catenin and that HBx regulates the expression of USP26 in a manner dependent on β-catenin expression (Supplementary Fig. 7l–o), suggesting that HBx promotes the activation of USP26 by β-catenin. Considering that HBx localizes in the nucleus (Supplementary Fig. 7d) and colocalizes with USP26 (Supplementary Fig. 7e), we wondered whether HBx interacts with USP26. Co-immunoprecipitation assays showed that HBx indeed binds to USP26; more importantly, overexpression of HBx markedly enhances the interaction between USP26 and SIRT1 in a dose-dependent manner (Fig. 7n, o, and Supplementary Fig. 7p). Consequently, deubiquitination assays showed that HBx could synergize with USP26 to robustly remove the ubiquitination of SIRT1 (Fig. 7p).
Taken together, these results suggest that HBx transactivates USP26 expression by recruiting β-catenin and promotes the induced-USP26 interaction with SIRT1 to stabilize SIRT1 and drive hepatocellular carcinoma tumorigenesis by deubiquitination (Fig. 8).
Discussion
It has been reported that deubiquitinases are involved in regulating the immune response to viral infections, including hepatitis B virus46,47. Specifically, USP18 is known to inhibit the expression of interferon-stimulated genes (ISGs), and inactivation of USP18 has been shown to improve the immune response to HBV infection in vitro. In addition, USP15 can increase HBx protein expression by deubiquitination and promote subsequent HBV infection48. These studies suggest that DUBs play a critical role in regulating the immune response to HBV. However, whether and how deubiquitinases are involved in HBV-induced HCC is largely unclear. In this study, we identified USP26 as an oncogenic driver of HCC and the mechanism underlying the association between HBV infection and HCC occurrence (Fig. 8): in response to HBV infection, USP26 is transcriptionally activated and consequently recruits the HBV-encoded X protein HBx to interact with SIRT1, which synergistically deubiquitinates and stabilizes SIRT1.
Deubiquitinases have been established to control multiple cellular processes that are dysregulated in cancer, including cell cycle progression, DNA damage response, and apoptosis49. To date, several DUBs have been implicated in liver cancer progression. USP9X is a ubiquitin-specific protease that is overexpressed in liver cancer and regulates the stability of several proteins, such as PTENα/β, to promote cell proliferation50. However, studies have shown that complete knockout of the Usp9x gene is lethal in mice, indicating that this gene is essential for normal development and survival. Additionally, conditional knockout of Usp9x in mice has been shown to result in pancreatic tumorigenesis, suggesting that USP9X is a major tumor suppressor gene51. As a member of the SAGA complex, USP22 has been shown to regulate the expression of several genes involved in lipogenesis and immune evasion, including PPARγ and SPI1, which collaboratively drive liver cancer progression52,53. Unfortunately, USP22 expression has also been shown to be important in the normal development and homeostasis of various tissues, including the heart, brain, and immune system, and loss of Usp22 leads to embryonic lethality in mice54. These studies indicate that DUBs play an important role in the development and progression of liver cancer, but whether DUBs could be potential targets for the development of novel therapies for the treatment of liver cancer needs to be systematically assessed. Interestingly, we identified that USP26, which has been previously linked to spermatogenesis and male infertility due to its restricted expression in normal tissues except the testis, plays an important role in HCC tumorigenesis. Moreover, USP26 expression is robustly induced by HBV infection and plays a causal role in HBV-related HCC tumorigenesis.
Whether HBx could be a transcription factor is controversial, studies have shown that HBx could be a transcription factor and recruit histone acetyltransferases or histone deacetylases onto the HBV minichromosome and host chromatin, modifying its epigenetic status and function55,56. However, another study showed that HBx may act as a coactivator but not a transcription factor45. Our study supports that HBx directly binds DNA and promotes the transcription of USP26 by β-catenin.
Currently, SIRT1 overexpression has been known to be associated with advanced HCC bearing higher α-fetoprotein levels and the absence of β-catenin signaling mutations, which occur in 30% of HCC patients57. Numerous studies have demonstrated that the expression and activity of SIRT1 can be regulated by multiple cellular stresses, including inflammation58. One study indicated that HBV status may correlate with the expression of SIRT1, but the mechanism was not elucidated59. Our study uncovers that HBx plays dual roles in stabilizing the SIRT1 protein through the deubiquitinase USP26, which further clarifies how HBV infection induces HCC progression. As a promising drug target, SIRT1 has continuously attracted more scientists to identify specific inhibitors. It is now known that the most selective SIRT1 inhibitor is selisistat, also known as EX527. Although EX527 was tolerated in patients with Huntington’s disease in phase I clinical trials, it may reduce effector T-cell proliferation and differentiation while enhancing the function of T regulatory cells in mice. For example, EX527 has been shown to play opposite roles in treating pancreatic cancer cells in vitro and in vivo, suggesting the complexity of the tumor microenvironment. Moreover, EX527 may target SIRT2 or other unknown targets, which increases the unpredictable outcome for the treatment of cancer patients60. Intriguingly, our data strongly supported that USP26 is highly expressed and is tightly correlated with SIRT1 expression in HCC tissues compared to normal adjacent tissues. In addition, our and other results have shown that Usp26 knockout mice are viable and do not exhibit any overt developmental defects61, suggesting that Usp26 is not essential for normal development and survival in mice. Taken together, our study suggests that the HBx-USP26-SIRT1 axis plays a causal role in HBV-positive HCC occurrence and may represent an attractive therapeutic option for HBV-related HCC. Further development of a clinical USP26 enzymatic inhibitor may be promising for treating patients with HBV-associated HCC.
Methods
Ethics statement
All animal studies were approved by the Animal Experimentation Ethics Committee of Huazhong University of Science and Technology (IACUC Number: 2918), and conducted according to the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) guidelines.
This paper describes studies involving human tissue samples that were approved by the Ethics Committee of Tongji Hospital, Huazhong University of Science and Technology, China. All tissue samples were collected and used in compliance with informed consent policy.
CRISPR-Cas9 library screening
A human deubiquitinase CRISPR lentiviral library (EdiGene, ARR60307), which contains sgRNAs targeting 87 human DUBs (348 sgRNAs targeting 87 DUBs, 4 sgRNAs/DUB), was used to determine the effects of the indicated DUBs on HCC cell growth. For the screening flow, we first transduced HBV-positive HCCLM3 cells with Cas9-mCherry lentivirus and selected the positive cells by flow cytometry. Cas9-positive cells were cultured with medium containing 15 μg/mL blasticidin for 7 days to generate a stable cell pool. Then, HCCLM3 cells stably expressing Cas9 were infected with each DUB or control lentivirus containing pooled 4 sgRNAs for 2 days. After selection with 1 μg/mL puromycin for another 7 days, 87 designated pooled DUB-knockout and control cell populations were obtained for subsequent experiments. DUBs were considered to be significant if they had a p < 0.01 and a log2-fold change < −0.58. Based on these criteria, we identified DUBs that were significant according to colony formation and DUBs that were significant according to EdU analysis. Finally, we identified DUBs that were significant in two screening assays as potential candidates.
Reagents
MTT Cell Proliferation and Cytotoxicity Assay Kit (Solarbio, M1020); FITC Annexin V Apoptosis Detection Kit (BD, 556547); BeyoClick™ EdU-594 or 488 Cell Proliferation Kit (Beyotime, C0078 or C0071). MG132 (Santa Cruz Biotechnology, SC-201270); CHX (Sigma, C7698); S-protein agarose (Millipore, 69704); anti-Myc affinity gel (GNI, 4510-MC); and anti-Flag affinity gel (GNI, 4510-FG). The following antibodies were used in the study: anti-USP26 (homemade); anti-mouse-Usp26 (Dr. ZJ Chen lab34); anti-USP26 (ABclonal, A7999); anti-SIRT1 (CST, 1:1000, 8469S); anti-HA (Santa Cruz, 1:2000, SC-7392); anti-FLAG (Sigma, 1:3000, F3165), anti-Myc (GNI, 1:3000, 4110-MC); anti-cleaved PARP (CST, 1:1000, 5625 T); anti-β-actin (Santa Cruz, 1:3000, SC-47778) and anti-GAPDH (Proteintech, 1:40000, 60004-1-Ig); anti-H3K9ac (ABclonal, 1:5000, A7255); anti-histone H3 (CST, 1:3000, 9715); anti-H4K16ac (ABclonal, 1:3000, A23091); anti-histone H4 (ABclonal, 1:5000, A19815); Anti-Ub (Santa Cruz, 1:1000, SC-8017); Anti-GFP (CST, 1:1000, 2956); Anti-GST (Proteintech, 1:20000, 66001-1-Ig); Anti-Lamin B1 (Proteintech, 1:5000, 12987-1-AP); Anti-β-tublin (Proteintech, 1:20000, 66240-1-Ig); Anti-β-catenin (CST, 1:1000, 8480); goat anti-rabbit IgG (Boster, 1:5000, BA1005); and goat anti-mouse IgG (Boster, 1:5000, BA1038).
Tissue samples and cell culture
Human HCC tissue samples were obtained from patients who received surgical resection at Tongji Hospital of Huazhong University of Science and Technology (China). Tissue samples were then collected for different experiments.
HEK293T (CRL-11268), HepG2 (HB-8065) and U2OS (HTB-96) cells were obtained from American Type Culture Collection (ATCC). Huh7 (TCHu182), HCCLM3 (SCSP-5093), MHCC97H (SCSP-5092), and the human normal liver cell line L02 were purchased from National Collection of Authenticated Cell Cultures. HLE(CBP60200) and SMMC7721(CBP60210) were purchased from CoBioer Inc. (Nanjing, China). Cells were cultured in DMEM supplemented with 10% FBS, penicillin (100 units/mL) and streptomycin (100 μg/mL) at 37 °C in an incubator with 5% CO2.
RNA extraction and real-time PCR
Total RNA was extracted after cells were treated as indicated by using TRIzol (Invitrogen, 15596026). Then, cDNA synthesis was obtained by using a Synthesis Kit (BIO-RAD, 1708891), and qRT-PCR was performed with SYBR Green qPCR Master Mix (BIO-RAD, 1708880). The qRT-PCR data were collected by a CFX Connect Real-Time PCR Detection System (BIO-RAD) and calculated by the 2−ΔΔCt method. GAPDH or β-Actin was used as an internal control. Primer sequences are listed in the Supplementary Table 2.
DNA constructs
pcDNA-3.1-HBx, pcDNA-3.1-HBc and pcDNA-3.1-HBs were constructed as described previously62. pMH-Myc-SIRT1 was amplified by PCR and subcloned and inserted into the pLenti-SFB vector or pET-N-GST vector. Full-length USP26 and its truncated variants were subcloned and inserted into a pLenti-SFB or Myc-tagged expression vector or along with an N-terminal GFP tag in the pLVX-puro vector. Full-length β-catenin was cloned and inserted into the pMH-Myc vector (Addgene, 101765). pT2HB-EF1a-YAP-S127A and pT2HB-EF1a-ΔN90-β-catenin plasmids along with sleeping beauty transposase (SB) were provided by Dr. Xinjun Zhang’s laboratory. For the pT2HB-EF1a-β-catenin-S33/37 A and pT2HB-EF1a-MYC-luciferase plasmid, oncogenic β-catenin-S33/37 A or MYC and internal ribosome entry site (IRES) followed by a luciferase linkage were cloned and inserted into the pT2HB plasmid. For the luciferase plasmids, the USP26 promoter or its truncated variants were subcloned and inserted into the pGL4.11 (Promega, E6661) vector or GAL-DBD-basic vector (Addgene, 145245). The USP26 shRNA or SIRT1 shRNA was subcloned and inserted into the pLKO.1 vector. The USP26 shRNA-resistance plasmid was generated by site-directed mutagenesis. Primer sequences are listed in the Supplementary Table 2.
Generation of CRISPR/Cas9 knockout (KO) cell lines
Human USP26 guide sequences and β-catenin guide sequence were designed through the Benchling platform (https://benchling.com/) and ligated into the LentiCRISPR v2 plasmid (Addgene, 52961) according to the manufacturer’s instructions. Then, the plasmid containing the corresponding sgRNA was cotransfected with the lentiviral packaging vectors psPAX2 (Addgene, 12260) and pMD2. G (Addgene, 12259) into HEK293T cells at a ratio of 3:2:1. The lentiviral particles were collected 48 h after transfection, and the indicated cell lines were infected with the collected lentiviral particles for 24 h, followed by selection using 1 μg/mL puromycin for 72 h. Finally, the monoclonal knockout cell lines were obtained by limited dilution.
Tandem-affinity purification and mass spectrometry
HEK293T cells were first transfected with SFB-USP26 and then homogenized in lysis buffer (10 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40, 1 × protease cocktail and 1 × phosphatase cocktail, pH 7.4) after transfection for approximately 48 h. Next, cell lysates were centrifuged, and the supernatant was extracted for pulldown of SFB-tagged USP26 by preincubation with Streptavidin Sepharose High Performance (Cytiva, 17511301) for 3 hr. After washing with lysis buffer 5 times, the beads were incubated with 2 mg/mL biotin solution to elute proteins conjugated on the beads. Then, the supernatant was collected and incubated with S-protein beads (Millipore, 69704) at 4 °C overnight. Finally, the beads were washed 5 times and then analyzed by SDS-PAGE. The gel was excised and the proteins were subjected to mass spectrometry analysis by Beijing Protein Innovation Co., Ltd. (China). The resulting MS/MS data were processed using Proteome Discoverer 1.3. Tandem mass spectra were searched against 2021_uni_human human (194,324 sequences; 58,129,772 residues) database. Trypsin was specified as cleavage enzyme allowing up to 2 missing cleavages. Mass error was set to 10 ppm for precursor ions and 0.02 Da for fragment ions. FDR <0.05, Peptide ion score was set > 20. See details in the Supplementary Table 1.
Immunoprecipitation and Western blot
Cell pellets or tissue samples were lysed in NETN lysis buffer (10 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40, 1 × protease cocktail and 1 × phosphatase cocktail, pH 7.4) on ice for 30 min and then disrupted by ultrasonication for 30 s (6 pulses, 5 s each). After centrifugation at 12,000 × g for 10 min, cell lysates were incubated with the indicated antibody-conjugated beads at 4 °C overnight. Subsequently, the beads were washed approximately 4-5 times, and then western blotting was performed as described below. First, proteins were separated by SDS-PAGE and then transferred onto PVDF membranes (Millipore, IPVH00010). Then, the membrane loaded with proteins was blocked in 5% milk and incubated with the indicated primary antibody at 4 °C overnight. Finally, the membrane was incubated with a secondary antibody conjugated with horseradish peroxidase (HRP). Images of the Western blots were obtained by ChemiDoc XRS+ (BIO-RAD). Statistical analyses of protein bands were performed using ImageJ/Fiji.
Ubiquitination assays
For the ubiquitination assay, HEK293T cells were first transfected with the HA-Ub and Myc-SIRT1 plasmids with or without SFB-USP26 or SFB-USP26-C304S. Next, the cells were treated with 10 μM MG132 for 6 h and lysed with 1% SDS buffer. After heating at 95 °C for 10 min, the cell lysate was diluted with 900 μL of lysis buffer and ultrasonicated for 5 min (10 pulses, 60 s each). The subsequent steps were the same as those for immunoprecipitation described above. For the ubiquitination assay in vitro, HEK293T cells were first transfected with the HA-Ub and Myc-SIRT1 plasmids and then treated with MG132. Next, SIRT1 was purified with anti-Myc magnetic beads and then incubated with purified recombinant USP26 or USP26-C304S in deubiquitination buffer (5 mM MgCl2, 20 mM Tris-Cl, 2 mM DTT, 10 mM ATP, pH 7.5) for 3 h at 37 °C.
GST pull-down assay
For the GST affinity isolation assay, SIRT1 was first cloned and inserted into the pET-N-GST vector, and then the pET-N-GST-SIRT1 plasmid was transformed into E. coli BL21 (DE3). The bacteria were cultured at 37 °C for approximately 6 h until the OD600 of the bacterial solution reached 0.6–1.0. Then, IPTG was added to the bacteria to a final concentration of 100 μM and incubated at 25 °C for 12 h. Next, the bacteria were lysed by ultrasonication for 10 min (10 pulses, 60 s each) in PBS containing protease inhibitor cocktail and 1 mg/mL lysozyme. To purify the target proteins, the supernatant was incubated with glutathione agarose at 4 °C overnight. Subsequently, glutathione Sepharose was eluted with glutathione buffer after washing with PBS 3 times. Finally, western blotting was carried out to verify the proteins.
Immunofluorescence and immunohistochemistry staining
For immunofluorescence staining, the indicated cells were plated on glass coverslips (NEST Biotechnology, 801010) overnight and then fixed with 4% paraformaldehyde (PFA) for 20 min. After permeabilization with 0.1% Triton X-100 for 10 min, the cells were blocked in PBS containing 1% BSA for 1 h. Subsequently, the cells were incubated with the indicated primary antibody at 4 °C overnight and with the fluorophore-conjugated secondary antibody for 1 h at room temperature. Finally, the cells were visualized and photographed using a laser confocal microscope (Olympus, FV3000).
For immunohistochemistry (IHC) staining, tissues were fixed with 4% PFA at 4 °C after washing with PBS 3 times. The formalin-fixed tumor tissues were embedded in paraffin and then routinely stained with hematoxylin and eosin (H&E). Paraffin-embedded tissue sections were subjected to deparaffinization, antigen retrieval and antibody conjugation for IHC. Then, the images were scanned by a panoramic slice scanner (3DHISTECH, Pannoramic MIDI).
Cell viability assay and colony formation assay
MTT and colony formation assays were adopted to assess cell proliferation as described below. Briefly, cells were seeded in 96-well plates at approximately 2 × 103 cells per well and then cultured with MTT for 4 h at the indicated times. Then, the stained cells were washed with PBS and dissolved in DMSO. The absorbance values were detected at 490 nm using Biotek Synergy H1 (Agilent). For the colony formation assays, cells were seeded into 6-well plates at a density of 1000 cells per well. After incubation for 12 days, clones were stained with 0.1% crystal violet within 1 h for further analysis.
EdU analysis
The BeyoClick™ EdU-594 or 488 Cell Proliferation Kit was adopted to detect cell proliferation ability according to the manufacturer’s protocol. In brief, cells were plated in 12-well plates at approximately 60% concentration. Next, an equal volume of 2× EdU working solution (20 μM), preheated at 37 °C, was added to the 12-well plate to make the final 1x concentration of EdU in the 12-well plate and then cultured for 1.5 h at 37 °C. After fixation with 4% formaldehyde for 10 min, the cells were permeabilized with 0.1% Triton X-100 for 20 min. For flow cytometry assays, adherent cells were trypsin-digested and resuspended in culture medium before fixation. Then, the Click Reaction Mixture was added to the culture for 30 min, and the cells were incubated with Hoechst 33342 for 10 min. The cells were washed 3 times with washing buffer after each staining. Finally, the results were visualized or analyzed by fluorescence microscopy or flow cytometry. The flow cytometric gating strategies are indicated in the supplementary information (Supplementary Fig 8a).
Mice and treatments
Mice were housed in individually ventilated cages (IVC) with controlled temperature (22–25 °C) and relative humidity (40–60%) on a constant light dark period of 12/12 h cycle (07:00–19:00 light, 19:00–07:00 dark). The density of mice housing should not exceed 5 mice per cage. To generate Usp26-/- C57BL/6 J mice, 2664 bp on the coding region of Usp26 was deleted by Cyagen Biosciences (Guangzhou, China). In this study, 6- to 8-week-old mice were used to establish a liver cancer model.
For the xenograft model, HCCLM3 cells (7 × 106 cells) were mixed with Matrigel (Corning, 356234) at a ratio of 1:1 and then injected subcutaneously into 5-week-old male BALB/c nude mice. Tumor volume was measured every 2 or 3 days after cell implantation, and tumors were weighed at the fixed time for further experiments. Tumor volumes (mm3) were estimated by V = 0.5 × length (mm) × width (mm2). According to the Committee of the Use of Live Animals in Teaching and Research (CULTAR) in Huazhong University of Science and Technology guidelines, the tumor diameter should not exceed 20 mm. At the endpoint, mice were euthanized using cervical dislocation under anesthesia as approved by CULTAR.
Hydrodynamically injected oncogene-induced liver cancer model
pT2HB-EF1a-YAP-S127A and pT2HB-EF1a-ΔN90-β-catenin plasmids or pT2HB-EF1a-MYC-luciferase and pT2HB-EF1a-β-catenin-S33/37 A plasmids along with sleeping beauty transposase (SB) at a ratio of 10:1 (10 µg per oncogene plasmid, 1 µg per SB plasmid) were diluted into 0.9% sodium chloride solution at a volume equivalent to 10% of body weight and injected into the lateral tail vein of 6- to 8-week-old mice at 5–7 s as described35,36,63. Mice (male and female) were euthanized at 6-7 weeks after hydrodynamic tail vein injection, and livers were collected to examine various parameters. Within 3 months of the injection, mice were followed up for overall survival.
For the pT2HB-EF1a-MYC-luciferase and pT2HB-EF1a-β-catenin-S33/37 A plasmids along with the sleeping beauty transposase-induced HCC model, after injection for 8 weeks, mice (male) were intraperitoneally injected with luciferin (150 mg/kg body weight; Promega, P1042), and then the bioluminescence intensity was detected by BLI using the SPECTRAL Lago X Imaging System after 15 min.
Cell apoptosis assay
The Annexin V-FITC/PI double staining kit was used to examine cell apoptosis according to the manufacturer’s protocol. Cells were harvested with trypsin-free EDTA and then resuspended in 1× binding buffer after washing twice in cold PBS. Next, 100 µl of cell suspension (1 × 105 cells) was transferred to a PI and Annexin V-FITC mixed solution and stained for 30 min at RT in the dark. Finally, 1X binding buffer was added to each tube up to 500 µl, and the cells were gently vortexed. The data were collected by a CytoFLEX LX flow cytometer (Beckman Coulter) and analyzed using Cytoexpert 2.40. The flow cytometric gating strategies are indicated in the supplementary information (Supplementary Fig 8b).
DNA pull-down assay
Genomic DNA was extracted from Huh7 cells, and the promoter region of USP26 was amplified with biotin-labeled primers. Then, 1 μg of biotin-labeled DNA was incubated with streptavidin Sepharose beads for 6 h at 4 °C. Next, bacteria-purified HBx or overexpressed HBx in Huh7 cells was incubated with USP26 promoter-bound beads at 4 °C overnight. Another USP26 promoter fragment without biotin labeling but with the same sequence was used as a competitor at a 20-fold concentration. Finally, western blotting was carried out to verify the pulled down proteins.
Electrophoretic mobility shift assay (EMSA)
The USP26 promoter or its truncated variants were first amplified, and bacteria-purified HBx was also purified. Then, 0–800 ng of bacteria-purified HBx and 50 ng of USP26 promoter or its truncated variants were incubated at RT (25 °C) for 20 min in a protein binding buffer containing 10 mM Tris-HCl (pH 8.3), 0.05 M KCl, 1 mM MgCl2, 0.05 mg/mL bovine serum albumin (BSA), 0.5 mM DTT, 0.5 mM EDTA and 0.2% glycerol. Boiled inactive HBx was used as a negative control. Next, the reacted DNA-protein complexes were subjected to 5% native polyacrylamide gel electrophoresis in cold 0.5×TBE buffer for 90 min. Finally, the gel was stained with EB for 10 min and then exposed to UV light. The DNA-protein complex migrates more slowly than unbound DNA double-stranded oligonucleotides on native polyacrylamide gels.
Dual-luciferase reporter assay and GAL4 assay
The USP26 promoter region or its truncated variants were cloned and inserted into the pGL4 luciferase reporter vector (Promega, USA) and then transfected into Huh7 cells together with the Renilla luciferase vector at a ratio of 5:1. After 48 h of transfection, cells were collected and lysed at 25 °C in the dark for 15 min. The luciferase activity was measured using a dual-luciferase reporter assay system (Promega, E2920), and then the luminescence ratio of firefly to Renilla was calculated.
For the GAL4 assay, HBx, TCF4 or P53 was first cloned and inserted into the GAL-DBD-basic vector (Addgene, 145245) and then transfected into HEK293T cells together with 5× TATA- firefly luciferase plasmid (Addgene, 46756) as described. Luciferase activity was detected by Biotek Synergy H1 (Agilent). Luciferase reporter activity represented by a ratio of firefly: Renilla luminescence.
Statistical analyses
All experiments were independently repeated at least three times and were analyzed as the mean ± SEM. Two-tailed unpaired Student’s t-tests were used to determine significant differences between two independent groups and two-sided paired Student’s t-test was used for paired data. One- or two-way analysis of variance (ANOVA) was used to compare three or more group means, depending on the situation. Survival curves were plotted by Kaplan–Meier and analyzed by log-rank test. Spearman’s analysis was used for correlation of protein expression in tumor tissues. Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software, La Jolla, USA). p < 0.05 was defined as statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org/ui) under accession code PXD051958. The association between USP26 and the prognosis of HCC patients are publicly available and downloaded from Kaplan–Meier plotter database (http://kmplot.com/analysis/). The FACS sequential gating strategies are indicated in the supplementary information (Supplementary Fig 8). The remaining data are available within the Article, Supplementary Information or Source Data file. Correspondence and requests for materials should be addressed to P. Z. Source data are provided with this paper.
References
Islami, F. et al. Disparities in liver cancer occurrence in the United States by race/ethnicity and state. CA Cancer J. Clin. 67, 273–289 (2017).
Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 380, 1450–1462 (2019).
Jiang, Y., Han, Q., Zhao, H. & Zhang, J. The Mechanisms of HBV-Induced Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 8, 435–450 (2021).
Lee, W. Y., Bachtiar, M., Choo, C. C. S. & Lee, C. G. Comprehensive review of Hepatitis B Virus-associated hepatocellular carcinoma research through text mining and big data analytics. Biol. Rev. Camb. Philos. Soc. 94, 353–367 (2019).
Sung, W. K. et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet 44, 765–769 (2012).
Fujimoto, A. et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet 44, 760–764 (2012).
Gao, Q. et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell 179, 1240 (2019).
Fisicaro, P. et al. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B. Nat. Med. 23, 327–336 (2017).
Yang, P. et al. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell 22, 291–303 (2012).
Couri, T. & Pillai, A. Goals and targets for personalized therapy for HCC. Hepatol. Int 13, 125–137 (2019).
Llovet, J. M. et al. Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 19, 151–172 (2022).
Kudo, M. et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 391, 1163–1173 (2018).
Bruix, J. et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 389, 56–66 (2017).
Pan, J. et al. Genome-Scale CRISPR screen identifies LAPTM5 driving lenvatinib resistance in hepatocellular carcinoma. Autophagy 19, 1184–1198 (2023).
Lee, M. S. et al. Atezolizumab with or without bevacizumab in unresectable hepatocellular carcinoma (GO30140): an open-label, multicentre, phase 1b study. Lancet Oncol. 21, 808–820 (2020).
Finn, R. S. et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 382, 1894–1905 (2020).
Yau, T. et al. Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): a randomised, multicentre, open-label, phase 3 trial. Lancet Oncol. 23, 77–90 (2022).
Suthen, S. et al. Hypoxia-driven immunosuppression by Treg and type-2 conventional dendritic cells in HCC. Hepatology 76, 1329–1344 (2022).
Gonzalez-Rellan, M. J. et al. Hepatic p63 regulates glucose metabolism by repressing SIRT1. Gut 72, 472–483 (2023).
Yang, L. et al. Nicotine rebalances NAD(+) homeostasis and improves aging-related symptoms in male mice by enhancing NAMPT activity. Nat. Commun. 14, 900 (2023).
Zhu, X. et al. SIRT1 deacetylates WEE1 and sensitizes cancer cells to WEE1 inhibition. Nat. Chem. Biol. 19, 585–595 (2023).
Rasti, G. et al. SIRT1 regulates DNA damage signaling through the PP4 phosphatase complex. Nucleic Acids Res. 51, 6754–6769 (2023).
Zhao, E. et al. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers (Basel) 11, https://doi.org/10.3390/cancers11121949 (2019).
Kong, F. et al. Sirtuins as Potential Therapeutic Targets for Hepatitis B Virus Infection. Front Med (Lausanne) 8, 751516 (2021).
Dikic, I. & Schulman, B. A. An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 24, 273–287 (2023).
Xu, C. et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nat. Cell Biol. 22, 1170–1179 (2020).
Yu, L. et al. Ubiquitination-mediated degradation of SIRT1 by SMURF2 suppresses CRC cell proliferation and tumorigenesis. Oncogene 39, 4450–4464 (2020).
Lee, S. H. et al. SIRT1 ubiquitination is regulated by opposing activities of APC/C-Cdh1 and AROS during stress-induced premature senescence. Exp. Mol. Med. 55, 1232–1246 (2023).
Zhang, P. et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 16, 864–875 (2014).
Zhang, P. et al. ZRANB1 Is an EZH2 Deubiquitinase and a Potential Therapeutic Target in Breast Cancer. Cell Rep. 23, 823–837 (2018).
Lin, Z. et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 46, 484–494 (2012).
Ge, F. et al. Deubiquitinating enzymes: Promising targets for drug resistance. Drug Discov. Today 27, 2603–2613 (2022).
Huang, S. et al. The deubiquitinase ZRANB1 is an E3 ubiquitin ligase for SLC7A11 and regulates ferroptotic resistance. J. Cell Biol. 222, https://doi.org/10.1083/jcb.202212072 (2023).
Liu, C. et al. Paternal USP26 mutations raise Klinefelter syndrome risk in the offspring of mice and humans. EMBO J. 40, e106864 (2021).
Chen, X. & Calvisi, D. F. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am. J. Pathol. 184, 912–923 (2014).
Tao, J. et al. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 147, 690–701 (2014).
Xue, R. et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature 612, 141–147 (2022).
Tang, M. et al. Liver cancer heterogeneity modeled by in situ genome editing of hepatocytes. Sci. Adv. 8, eabn5683 (2022).
Lahav-Baratz, S., Kravtsova-Ivantsiv, Y., Golan, S. & Ciechanover, A. The testis-specific USP26 is a deubiquitinating enzyme of the ubiquitin ligase Mdm2. Biochem Biophys. Res Commun. 482, 106–111 (2017).
Song, L. & Luo, Z. Q. Post-translational regulation of ubiquitin signaling. J. Cell Biol. 218, 1776–1786 (2019).
Liu, X. et al. SIRT1 Regulates N(6) -Methyladenosine RNA Modification in Hepatocarcinogenesis by Inducing RANBP2-Dependent FTO SUMOylation. Hepatology 72, 2029–2050 (2020).
Qiu, P. et al. Sirt1 deficiency upregulates glutathione metabolism to prevent hepatocellular carcinoma initiation in mice. Oncogene 40, 6023–6033 (2021).
Levine, D. C. et al. NADH inhibition of SIRT1 links energy state to transcription during time-restricted feeding. Nat. Metab. 3, 1621–1632 (2021).
Zhou, S. et al. Repression of P66Shc expression by SIRT1 contributes to the prevention of hyperglycemia-induced endothelial dysfunction. Circ. Res. 109, 639–648 (2011).
Lin, Y. et al. The hepatitis B virus X protein is a co-activator of activated transcription that modulates the transcription machinery and distal binding activators. J. Biol. Chem. 273, 27097–27103 (1998).
Randall, G. et al. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology 131, 1584–1591 (2006).
Ritchie, K. J. et al. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 10, 1374–1378 (2004).
Su, Z. J. et al. Deubiquitylation of hepatitis B virus X protein (HBx) by ubiquitin-specific peptidase 15 (USP15) increases HBx stability and its transactivation activity. Sci. Rep. 7, 40246 (2017).
Dewson, G., Eichhorn, P. J. A. & Komander, D. Deubiquitinases in cancer. Nat. Rev. Cancer 23, 842–862 (2023).
Shen, S. M. et al. PTENalpha and PTENbeta promote carcinogenesis through WDR5 and H3K4 trimethylation. Nat. Cell Biol. 21, 1436–1448 (2019).
Perez-Mancera, P. A. et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 486, 266–270 (2012).
Ning, Z. et al. USP22 regulates lipidome accumulation by stabilizing PPARgamma in hepatocellular carcinoma. Nat. Commun. 13, 2187 (2022).
Li, Q. et al. PRDM1/BLIMP1 induces cancer immune evasion by modulating the USP22-SPI1-PD-L1 axis in hepatocellular carcinoma cells. Nat. Commun. 13, 7677 (2022).
Wang, L. & Dent, S. Y. Functions of SAGA in development and disease. Epigenomics 6, 329–339 (2014).
Belloni, L. et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc. Natl. Acad. Sci. USA 106, 19975–19979 (2009).
Tian, Y., Yang, W., Song, J., Wu, Y. & Ni, B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol. Cell Biol. 33, 2810–2816 (2013).
Chen, H. C., Jeng, Y. M., Yuan, R. H., Hsu, H. C. & Chen, Y. L. SIRT1 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma and its expression predicts poor prognosis. Ann. Surg. Oncol. 19, 2011–2019 (2012).
Yang, Y. et al. Regulation of SIRT1 and Its Roles in Inflammation. Front Immunol. 13, 831168 (2022).
Wang, Q., Cheng, S. T. & Chen, J. HBx mediated Increase of SIRT1 Contributes to HBV-related Hepatocellular Carcinoma Tumorigenesis. Int J. Med Sci. 17, 1783–1794 (2020).
Broussy, S., Laaroussi, H. & Vidal, M. Biochemical mechanism and biological effects of the inhibition of silent information regulator 1 (SIRT1) by EX-527 (SEN0014196 or selisistat). J. Enzym. Inhib. Med Chem. 35, 1124–1136 (2020).
Tian, H. et al. Disruption of ubiquitin specific protease 26 gene causes male subfertility associated with spermatogenesis defects in mice. dagger Biol. Reprod. 100, 1118–1128 (2019).
Zhang, C. et al. Hepatitis B-Induced IL8 Promotes Hepatocellular Carcinoma Venous Metastasis and Intrahepatic Treg Accumulation. Cancer Res. 81, 2386–2398 (2021).
Carlson, C. M., Frandsen, J. L., Kirchhof, N., McIvor, R. S. & Largaespada, D. A. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc. Natl Acad. Sci. USA 102, 17059–17064 (2005).
Acknowledgements
We apologize to the colleagues whose relevant work cannot be cited here owing to space limitations. We thank the Huazhong University of Science and Technology core facility and members of the Zhang laboratory for discussion. We also thank Drs. ZJ Chen and C Liu for reagents and valuable suggestions. This research was supported by grants from the National Key Research and Development Program of China (2021YFA1201200 to P.Z.), National Natural Science Foundation of China (82172823 and 81874116 to P.Z.), MOST-Key Program for International S&T Cooperation Projects of China (2017YFE0129100 to P.Z.), the Sichuan Science and Technology Program (2024NSFJQ0047 to P.Z.) and startup fund from Huazhong University of Science and Technology to P.Z. P.Z. is a scholar in the National Young Talents Program of China.
Author information
Authors and Affiliations
Contributions
Mengru Ma: data curation, formal analysis, validation, investigation, visualization, writing—original draft; Lian Yi, Yifei Pei, Qimin Zhang, Chao Tong, Manyu Zhao, Yuanhong Chen: data curation, formal analysis, validation, methodology; Jinghan Zhu, Wanguang Zhang, Fan Yao and Pengyuan Yang: methodology, resources, review and editing; Peijing Zhang: conceptualization; resources; formal analysis; supervision; funding acquisition; visualization; writing—original draft; writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tingting Jiang, Dragana Kopanja and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ma, M., Yi, L., Pei, Y. et al. USP26 as a hepatitis B virus-induced deubiquitinase primes hepatocellular carcinogenesis by epigenetic remodeling. Nat Commun 15, 7856 (2024). https://doi.org/10.1038/s41467-024-52201-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52201-z
- Springer Nature Limited