Abstract
We assessed whether multiplex real-time PCR plus conventional microbiological testing is safe and more effective than conventional microbiological testing alone for reducing antibiotic use in community-acquired pneumonia (CAP). In this randomised trial, we recruited adults hospitalised with CAP at four Spanish hospitals. Patients were randomly assigned (1:1) to undergo either multiplex real-time PCR in non-invasive respiratory samples plus conventional microbiological testing or conventional microbiological testing alone. The primary endpoint was antibiotic use measured by days of antibiotic therapy (DOT). Between February 20, 2020, and April 24, 2023, 242 patients were enrolled; 119 were randomly assigned to multiplex real-time PCR plus conventional microbiological testing and 123 to conventional microbiological testing alone. All but one of the patients allocated to multiplex real-time PCR plus conventional microbiological testing underwent PCR, which was performed in sputum samples in 77 patients (65.2%) and in nasopharyngeal swabs in 41 (34.7%). The median DOT was 10.04 (IQR 7.98, 12.94) in the multiplex PCR plus conventional microbiological testing group and 11.33 (IQR 8.15, 16.16) in the conventional microbiological testing alone group (difference −1.04; 95% CI, −2.42 to 0.17; p = 0.093). No differences were observed in adverse events and 30-day mortality. Our findings do not support the routine implementation of multiplex real-time PCR in the initial microbiological testing in hospitalised patients with CAP. Clinicaltrials.gov registration: NCT04158492.
Similar content being viewed by others
Introduction
Community-acquired pneumonia (CAP) is a major cause of morbidity and mortality worldwide and one of the leading drivers of antibiotic use in hospitalised patients1,2. However, in many cases, the causative agent is not identified and patients are overtreated with antibiotics3,4. Overuse of antibiotics is a major cause of antimicrobial resistance and increases the risk of Clostridioides difficile infection and other antibiotic-related adverse events4,5. Antibacterial resistance is accelerating at an alarming pace and is raising morbidity and mortality rates worldwide6. In this scenario, antimicrobial stewardship is recognised as a crucial component in strategies to deal with the threat of antibiotic resistance7,8.
The development of multiplex real-time polymerase chain reaction (PCR) in automated platforms currently allows rapid screening of non-invasive respiratory specimens, such as sputum samples and nasopharyngeal swabs, for a wide array of respiratory pathogens9. Several observational studies have found that comprehensive molecular testing significantly improved pathogen detection in CAP, particularly in antimicrobial-exposed patients10,11. Two recent studies investigating the efficacy of multiplex real-time PCR in non-invasive respiratory samples for antimicrobial stewardship in CAP have yielded conflicting findings12,13.
Current guidelines for CAP do not incorporate multiplex PCR pneumonia panels into their recommendations for initial microbiological diagnostic testing2,14,15. Furthermore, the guidance regarding conventional microbiological testing methods like sputum culture, blood cultures, and urinary antigen tests lacks consistency and is predominantly grounded in low or very low-quality evidence.
We conducted a randomised controlled trial to test the hypothesis that multiplex real-time PCR in non-invasive respiratory samples plus conventional microbiological testing is safe and more effective than conventional microbiological testing alone for reducing antibiotic use in hospitalised patients with CAP.
Results
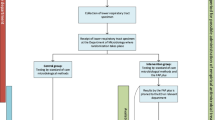
Between February 20, 2020, and April 24, 2023, we assessed 315 patients with CAP for eligibility. After excluding 73 patients who were considered ineligible, the remaining 242 were enrolled and then randomly assigned to undergo multiplex real-time PCR plus conventional microbiological testing (n = 119; 49%) or conventional microbiological testing alone (n = 123; 51%). The primary endpoint was analysed by intention-to-treat in all 242 patients and per-protocol in 230. The trial profile is shown in Fig. 1. During the study period, 11,938 patients with suspected or confirmed COVID-19 were admitted in specific emergency areas or buildings of the participating hospitals to which the trial investigators did not have access.
The baseline characteristics in the intention-to-treat population were well-balanced between groups (Table 1). Median age, the percentage of patients older than 75 years, and the frequency of chronic pulmonary and heart diseases were slightly higher in the group undergoing multiplex real-time PCR plus conventional microbiological testing. Charlson comorbidity index score, pneumonia severity index score, and CURB-65 were similar in both groups. The baseline characteristics of the per-protocol population were similar and are provided in Supplementary Table 1. The initial antibiotic therapy was also similar in the two study groups (Table 2). Most patients received conventional antibiotics used in CAP. Only 12 patients in the multiplex real-time PCR plus conventional microbiological group and 17 in the conventional microbiological testing alone group received anti-pseudomonal β-lactams. No patient received vancomycin or linezolid.
Table 3 shows the microbiological studies performed in each study group in the intention-to-treat population. All but one of the patients allocated to multiplex real-time PCR plus conventional microbiological testing underwent PCR, which was performed in sputum samples in 77 patients (65.2%) and in nasopharyngeal swabs in 41 (34.7%). Twenty-four (31.2%) of the 77 sputum samples were obtained from induced sputum. The proportion of patients who underwent the different types of conventional microbiological examinations was similar in the two study groups. The diagnostic yield based on the sample used for PCR testing is detailed in Supplementary Table 2. Sputum samples and induced sputum samples had a higher yield than nasopharyngeal swabs. The time to positivity for each diagnostic test is presented in Supplementary Table 3. Multiplex real-time PCR results were available more quickly than those from non-PCR-based diagnostic tests. An aetiological diagnosis was established in 76 (63.9%) of 119 patients in the multiplex real-time PCR plus conventional microbiological testing group and in 32 (26.02%) of 123 patients in the conventional microbiological testing alone group (difference 37.85; 95% CI, 25.42–50.28; p < 0.0001). The CAP causative organisms identified in each group are shown in Table 4; the most frequent in both groups were Streptococcus pneumoniae, Legionella pneumophila, and Haemophilus influenzae. Polymicrobial infections and those caused by respiratory viruses were more frequent in the multiplex real-time PCR plus conventional microbiological testing group. Gram-negative bacilli, including Pseudomonas aeruginosa, were uncommon in both study groups. There were only three cases of CAP due to Staphylococcus aureus, all in the PCR group, and none of the strains were resistant to methicillin (MRSA).
The results for primary and secondary endpoints in the intention-to-treat population are shown in Table 5. Median DOT was 10.04 (IQR 7.98, 12.94) in the 119 patients in the multiplex real-time PCR plus conventional microbiological testing group and 11.33 (IQR 8.15, 16.16) in the 123 patients in the conventional microbiological testing alone group (difference −1.04; 95% CI, −2.42 to 0.17; p = 0.093). The results of the primary endpoint are also shown in Fig. 2. Results for the primary endpoint were confirmed by adjusted analysis (Supplementary Table 4). No significant differences in length of antibiotic therapy (LOT) were found between the groups: the median LOT was 9.00 days (7.42, 11.0) in the experimental group and 8.76 days (6.92, 12.73) in the control group (difference 0.12, 95% CI −0.79 to 0.96; p = 0.775).
The figure presents a box-plot analysis of the primary endpoint, days of antibiotic therapy (DOT), in both study groups; multiplex real-time PCR plus conventional microbiological testing (left, red box-plot) and conventional microbiological testing alone (right, teal box-plot). The median DOT in the multiplex real-time PCR plus conventional microbiological testing is 10.04, with an interquartile range spanning from 7.98 to 12.94. The median DOT in the conventional microbiological testing alone is 11.33, with an interquartile range spanning from 8.15 to 16.16. Outliers were distributed similarly between bout groups with extended DOT up to 60 days.
Time to switch from intravenous to oral antibiotic therapy and time to reach an aetiological diagnosis was significantly shorter in patients undergoing multiplex real-time PCR plus conventional microbiological testing. More patients in the conventional microbiological testing alone group were admitted to the ICU. There were no significant differences in other secondary endpoints, including de-escalation to narrowed spectrum antibiotic treatment, time to clinical stability, days of mechanical ventilation, antibiotic-related side effects, length of hospital stay, hospital readmission (≤30 days), and death from any cause at 48 h and at 30 days after randomisation. Per-protocol analyses of primary and secondary endpoints produced similar results to those of the intention-to-treat population (Supplementary Tables 5 and 6).
All patients who received at least one dose of antibiotic treatment were included in the safety analysis. The proportion of patients with any adverse events (17.65% [21/119] vs. 21.14% [26/123]; risk difference −3.49; 95% CI, −14.27 to 7.28; p = 0.600), serious adverse events (14.29% [17/119] vs. 20.33% [25/123]; risk difference −6.04; 95% CI, −16.36 to 4.28; p = 0.284), and antibiotic-related events (5.04% [6/119] vs. 4.07% [5/123]; risk difference 0.98; 95% CI, −5.11 to 7.06; p = 0.955) were similar in the two study groups. A description of all adverse events according to system organ class reported in both study groups is provided in Supplementary Table 7.
Discussion
In this randomised, controlled, open-label, multicentre trial, we aimed to evaluate whether multiplex real-time PCR in non-invasive respiratory samples plus conventional microbiological testing is safe and more effective than conventional microbiological testing alone for reducing antibiotic use in hospitalised patients with CAP. The primary endpoint was DOT, chosen as a measure of antibiotic consumption on the basis of the guidelines published by the IDSA and the Society for Healthcare Epidemiology of America for the implementation of antibiotic stewardship programmes16. Specifically, this metric takes into account the use of more than one antibiotic per day by summing up the total days on which any antibiotic is administered.
The main result of our stuy is that we found a modest reduction in the median number of DOT in patients undergoing PCR testing that was not statistically significant. Furthermore, we did not find significant differences in the LOT between the study groups. The results for most secondary endpoints, including adverse events and 30-day all-cause mortality, were similar. As expected, and in line with the findings of several observational studies, the use of multiplex real-time PCR was associated with an increased microbial yield11,17. We also found that the time to reach an aetiological diagnosis and the time to switch from intravenous to oral antibiotics was shorter in patients undergoing multiplex real-time PCR plus conventional microbiological testing. Importantly, timely switching from intravenous to oral antibiotic therapy can enhance patient outcomes by reducing the risk of catheter-related complications and shortening hospital stays, which can result in cost savings for healthcare facilities18.
The results of our study are in line with the findings of a randomised trial showing that the routine implementation of urine antigen detection tests did not bring any substantial outcome-related benefit to hospitalised patients with CAP in terms of pneumonia-related complications, length of hospitalisation, or mortality19. Of concern, narrowing the antibiotic treatment according to the urine antigen test results was associated with a higher risk of relapse.
On the other hand, a randomised trial found that, compared to conventional microbiology, a multiplex bacterial PCR examination of bronchoalveolar lavage shortened the duration of inappropriate antibiotic therapy by 38.6 h in patients admitted to hospital with pneumonia and at risk of Gram-negative rod infection20. However, this result did not translate into a significant difference in terms of time to reach clinical stability, antibiotic-adverse events, length of hospital stay, or in-hospital mortality. An important proviso regarding the applicability of the results of that study in clinical practice is the fact that most institutions do not perform invasive techniques such as bronchoscopy and bronchoalveolar lavage to identify the cause of pneumonia in non-intubated, clinically stable patients. In contrast, our trial included the overall population of non-severely immunosuppressed patients hospitalised with CAP (without focusing on any subgroup of patients at risk for specific pathogens) and used non-invasive respiratory samples that were easy to collect, thus avoiding the risk of complications associated with invasive procedures and patient discomfort and increasing the likelihood of widespread implementation.
Two recent randomised trials have explored the efficacy of multiplex real-time PCR in non-invasive respiratory samples to reduce antibiotic use in CAP, yielding divergent results12,13. Our main finding aligns with a trial conducted across three Danish medical emergency departments, wherein the integration of point-of-care PCR into the diagnostic regimen did not increase the number of CAP patients with a more targeted and appropriate use of antibiotics12. Conversely, a single-centre randomised trial in Norway revealed that implementing a PCR-based panel for rapid testing in the emergency department facilitated swifter and more tailored antibiotic therapy for individuals with suspected CAP13. However, the difference in the time taken to administer pathogen-directed treatment between patients undergoing PCR and those undergoing standard microbiological diagnostic tests alone was modest. Furthermore, this difference was not correlated with significant variations in length of hospital stay, mortality rates, and hospital readmissions13.
In our trial, although the multiplex real-time PCR plus conventional microbiological testing group achieved an aetiological diagnosis more frequently and more rapidly than the control group, this did not correlate with a significant reduction in antibiotic consumption. Among the factors that may have contributed to this finding is physician behaviour. Physicians may retain reservations regarding the reliability of PCR results, often placing more confidence in their clinical judgement than in microbiological data when determining antibiotic treatment, particularly in cases where patients show signs of improvement even when a virus is detected. Additionally, concerns may arise regarding the true aetiological significance of certain microorganisms identified through molecular techniques21. We should stress that, in our trial, attending physicians received clinical interpretations of the PCR results, but the research team did not provide specific recommendations regarding antibiotic use based on the microbiological findings. Interestingly, a recent cross-sectional, stepped-wedge, cluster-randomised, non-inferiority trial demonstrated that in patients hospitalised with CAP, a multifaceted antibiotic stewardship intervention might reduce broad-spectrum antibiotic use without improving diagnostic yield22.
Our study has several limitations. The first is its open-label design, which may have introduced a bias in the evaluation of the primary endpoint. The lack of blinding may have influenced researcher behaviours, responses, and assessments. However, DOT is an objective metric of antibiotic consumption, which was assessed by a DSMB blinded to microbiological testing allocation. Second, the multiplex real-time PCR was performed in nasopharyngeal swabs in around one-third of cases. Although concerns have been voiced regarding the value of nasopharyngeal swabs for PCR testing, a growing body of evidence shows the reliability and utility of these easy-to-obtain samples, especially in non-immunocompromised patients and in those for whom sputum samples are difficult to collect23,24. Third, in a planned interim analysis when half the sample size required had been achieved, the DSMB committee proposed to stop recruitment owing to a concern with futility. Therefore, and also in view of the slow recruitment rate due to the impact of the COVID-19 pandemic the steering committee decided to discontinue the trial. It should be noted, that the premature discontinuation of the study might have limited the robustness and generalisability of its results. Additionally, mortality was low in both treatment groups, and the trial was not powered to detect survival differences. Notably, the pandemic may have contributed to the low frequency of viral infections and invasive pneumococcal disease observed in our trial. This effect was likely due to the implementation of universal masking and other non-pharmaceutical interventions, such as social distancing, hand hygiene, and lockdowns25,26,27. Finally, since the number of ICU patients was low, and the trial did not include severely immunocompromised patients, our conclusions do not apply to these populations, that may have risk factors for unusual pathogens and might benefit from comprehensive microbiological testing.
In conclusion, our study does not support the routine implementation of multiplex real-time PCR in non-invasive respiratory samples in the initial microbiological testing in CAP patients not admitted to the ICU and without severe immunocompromise. Our trial not only broadens the understanding of the difficulties of antibiotic optimisation in CAP management but also highlights the practical implications and considerations for implementing advanced microbiological diagnostic approaches in real-world clinical settings. To thoroughly evaluate the safety and effectiveness of multiplex real-time PCR testing in improving antibiotic use and enhancing relevant clinical outcomes, further studies are needed. These studies should ideally be conducted on adaptive platforms with electronic data capture. They should incorporate prescribers’ qualitative behavioural analysis, cluster-randomised interventions using individual patient data, and include education and audit tools. Such studies are necessary before recommending the integration of this testing method into the initial microbiological assessment of all hospitalised patients with CAP.
Methods
Study design
We performed a randomised, controlled, open-label trial with two parallel groups of patients hospitalised for CAP at four Spanish teaching hospitals (the RADICAP trial). Participants were recruited from February 20, 2020, to April 24, 2023. The study was authorised by the Spanish Medicines and Healthcare Products Regulatory Agency (AEMPS; 19-0388) and by the Bellvitge University Hospital Ethics Committee (PR214/18). The protocol has been published elsewhere and followed the SPIRIT initiative28. Patients’ personal and clinical information was managed in line with European Regulation (2016/679). The results are presented in accordance with the Consolidated Standards of Reporting Trials (CONSORT) statement. The trial is registered in ClinicalTrials.gov (NCT04158492) and EudraCT (2018-004880-29).
Participants
All patients aged ≥18 years diagnosed with CAP in the emergency department were screened for eligibility within the first 24 h of admission. CAP was defined as the presence of an infiltrate on the chest radiograph plus one or more of the following: fever (temperature, ≥38.0 °C) or hypothermia (<35.0 °C), new cough with or without sputum production, pleuritic chest pain, dyspnoea, and altered breath sounds on auscultation. Exclusion criteria were pregnancy or lactation; severe immunocompromise (i.e., patients receiving antineoplastic chemotherapy or radiotherapy in the previous 90 days, use of immunosuppressive drugs, use of corticosteroids at a minimum dose 15 mg/day in the previous 2 weeks, haematopoetic progenitor transplant, solid organ transplant, patients with HIV infection and CD4 count ≤200 cells/mm3); pleural empyema; imminent death (life expectancy ≤ 24 h); and participation in another clinical trial. Sex was recorded from the official documentation of each participant. Acute SARS-CoV2 infection and COVID-19 in the previous 90 days were added as exclusion criteria by a protocol amendment after the start of the pandemic. The amendment was approved by the Ethics Committee and by AEMPS. Before inclusion in the trial, all participants or legal representatives provided written informed consent.
Randomisation and masking
Patients were randomly assigned (1:1) to multiplex real-time PCR in non-invasive respiratory samples plus conventional microbiological testing or to conventional microbiological testing alone. A centralised electronic computer randomisation schedule was developed by the Biostatistics Unit at the Bellvitge Biomedical Research Institute (IDIBELL). The randomisation was performed in computed-generated blocks of 10 patients stratified by hospital site so as to conceal the sequence until the intervention was assigned. The code numbers for eligible patients were assigned in ascending sequential order. The allocation list was stored at IDIBELL and was not available to any member of the research team. At each participating hospital, patients who provided written informed consent and met the study criteria were randomised by investigators, who obtained the microbiological testing assigned and code number from a computer-assisted website.
Procedures
We randomly allocated participants to undergo either multiplex real-time PCR (Biofire® Filmarray® Pneumonia Plus panel, Biofire Diagnostics, LLC, Salt Lake City, Utah, US) plus conventional microbiological testing or conventional microbiological testing alone. In participants assigned to undergo multiplex real-time PCR, sputum samples (either spontaneous or induced) were obtained when available. If sputum samples could not be obtained, nasopharyngeal swabs were collected instead. All samples for PCR testing were obtained within 24 h of randomisation. All participants in both study groups underwent conventional microbiological testing at the discretion of the attending physician, which usually included two sets of blood cultures, sputum for Gram stain and culture when available, and urine for detection of antigens of Streptococcus pneumoniae and Legionella pneumophila serogroup 1. Testing for respiratory viruses (e.g., influenza, respiratory syncytial virus, and human metapneumovirus) was indicated at the discretion of the attending physician. All participants underwent SARS-COV-2 PCR testing. The results of the multiplex real-time PCR were communicated to the attending physicians immediately upon availability. This information, along with the clinical interpretation by the investigators, was shared both via telephone and through the electronic medical record system28. Additionally, the results of conventional microbiological tests were provided to the attending physicians through the electronic medical record system.
Outcomes
The primary endpoint was antibiotic use measured by days of antibiotic therapy (DOT). DOT refers to the number of days that a patient receives an antimicrobial agent, regardless of the dose, route or frequency of administration16. The secondary endpoints were de-escalation to narrower spectrum antibiotic treatment, time to switch from intravenous to oral antibiotics, time to reach an aetiological diagnosis, days to clinical stability after randomisation, need for intensive-care unit admission, days of mechanical ventilation, antibiotic-related side effects, any adverse event, length of hospital stay, need for hospital readmission within 30 days of randomisation, death from any cause within 48 h and within 30 days of randomisation.
Antibiotic therapy, follow-up, and outcomes assessment
Initial empirical antibiotic therapy was administered in the emergency department in accordance with participating hospitals’ guidelines, which recommend the administration of a β-lactam agent with or without a macrolide or fluoroquinolone. Initial empirical combination antimicrobial therapy was recommended for patients with severe CAP and/or those without any positive microbiological test. Levofloxacin monotherapy was indicated for Legionella pneumonia and for selected patients such as those with β-lactam allergy. Carbapenems, piperacillin-tazobactam and cefipime were considered broad-spectrum antibiotics. Narrow-spectrum antibiotics were generally considered when penicillin or ceftriaxone was used. All decisions regarding empirical and definitive antibiotic therapy, de-escalation, switch from intravenous to oral antibiotic therapy, and duration of treatment were made by the attending clinicians. The investigators were not involved in any decisions regarding antibiotic treatment.
All participants were seen daily during their hospital stay by their attending physicians and by at least one of the study investigators. The investigators recorded all outcome measures. DOTs were calculated as the days elapsed from the initial dose of antimicrobial until the last dose of antimicrobial therapy for the CAP episode. The DOT for a given patient on multiple antibiotics was the sum of DOT for each antibiotic that the patient received. All antibiotics administered to patients for an episode of CAP and its related complications were included in the primary endpoint calculation. A new treatment for CAP was considered if there was an interruption in antibiotic therapy lasting more than 48 h. Antimicrobial de-escalation was considered when a broad-spectrum antimicrobial treatment regimen was replaced with narrower-spectrum antimicrobials or when one or more initial combination empiric antimicrobials were discontinued. Participants attended an outpatient visit 30 days after hospital discharge. The investigators recorded readmissions for any reason or death from any cause in the 30 days after randomisation. The information was obtained from specific searches of hospital databases and was checked by asking patients at the outpatient visit 30 days after hospital discharge. For patients who did not attend this outpatient visit, a structured telephone interview was used to assess outcomes. Adverse events were recorded in all patients who received at least one dose of antibiotic therapy. All adverse events were recorded according to the Common Terminology Criteria for Adverse Events. The study was monitored by the IDIBELL Clinical Research and Clinical Trials Unit. All data were recorded on a secure web application for building and managing databases (REDCap). The study endpoints were assessed by a Data and Safety Monitoring Board (DSMB), which was blinded to study group and patient identity.
Microbiological testing
Sputum samples or nasopharyngeal swabs were processed immediately after reception at the Microbiology Laboratory. The multiplex real-time PCR used was the Biofire® Filmarray® Pneumonia Plus panel (Biofire Diagnostics, LLC, Salt Lake City, Utah, US). This panel is an automated multiplex PCR test for the rapid detection of 15 typical bacteria (four Gram-positive, 11 Gram-negative) with a semiquantification result, three atypical bacteria, and nine respiratory viruses (https://www.biomerieux-diagnostics.com/biofire-filmarray-pneumonia-panel). Bacterial load detections were categorised as positive when ≥106 CFU/mL were detected. In cases of Streptococcus pneumoniae detection, the cut-off point for considering the test as positive was ≥105 CFU/mL. The results for atypical bacteria (Legionella pneumophila, Mycoplasma pneumoniae, Chlamydiophila pneumoniae) and viruses were reported as detected or not detected. The multiplex real-time PCR results were provided to the attending physicians via the electronic medical record. Conventional microbiological studies in both study groups were carried out by standard methods and usually included Gram stain and culture of good quality sputum samples (<10 squamous cells and >25 leucocytes by low-power field [100X] in the Gram stain examination) when available, two sets of blood cultures (BACTEC® FX, BD, Madrid, Spain), and culture of pleural fluid when present. Furthermore, S. pneumoniae antigen in urine was detected using a rapid immunochromatographic assay (Binax®, Abbott, Chicago, Illinois, U.S.) and L. pneumophila serogroup 1 antigen was detected using an immunoenzymatic method (Bartels®, Trinity Biotech Plc., Bray, Ireland).
Statistical analysis
Based on our experience using conventional microbiological testing, the expected DOT is about 8 days when the aetiology of CAP is known and 11 days when it is not known. The primary endpoint (DOT) was expected to be non-normally distributed. If the true difference between the two microbiological testing study groups is two DOT, we estimated that we needed 220 participants undergoing multiplex real-time PCR plus conventional microbiological testing and 220 participants undergoing conventional microbiological testing alone to be able to reject the null hypothesis with a probability above 0.8. The type I error probability associated with this test of the null hypothesis is 0.05, assuming an expected dropout rate of 10%. The planned interim analysis was performed on March 27, 2023, when half of the sample required had been recruited, in order to evaluate the safety and to ensure sufficient statistical power. The DSMB, which was blinded to the microbiological testing allocation, raised no concerns regarding safety; however, it mentioned that the difference in DOT specified in the sample size calculation was 2 days and that the difference observed in the interim analysis was around one DOT. The estimated conditional power was 40%, and various simulations of the expected difference at the end of the study yielded a difference of around one DOT, an effect far removed from the expected clinical significance; as a result, the DSMB committee proposed to stop recruitment owing to concerns regarding futility. In view of the DSMB’s recommendation and the slow recruitment rate due to the impact of the COVID-19 pandemic, the steering committee decided to halt the trial on April 24, 2023.
Data for the primary and secondary outcomes were analysed by intention-to-treat and per-protocol. The intention-to-treat analysis included all randomly assigned patients, while the per-protocol analysis included all enrolled patients who completed the study without any major protocol deviations. All patients who received at least one dose of antibiotic treatment were included in the safety analysis. The primary endpoint was compared in the two study groups using the Wilcoxon rank sum test, while secondary endpoints were analysed using the chi-squared test or the Wilcoxon rank sum test depending on the type of variable. Median or risk differences were calculated and reported to quantify the observed effect with a 95% confidence interval. An adjusted analysis was performed for the primary endpoint using linear, quantile or logistic regression according to the endpoint. The adjustment variables considered were age, sex, Charlson index score, and pneumonia severity index score. All analyses and data management were performed with R software, v.4.0.529. The most relevant R packages used were dplyr, REDCapDM, rpact and compareGroups30,31,32,33.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Individual data cannot be shared because of privacy restrictions. Raw anonymised data relating to primary and secondary outcomes and safety can be shared upon request to researchers who provide a methodologically reasonable proposal. The request for data can be sent to the corresponding author (J.C.). A period of 18 months after publication of the main study results should elapse before requests are made, to allow authors to publish substudies. Interested researchers should obtain the approval of the Bellvitge University Hospital Ethics Committee.
References
GBD 2019 Diseases and Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 396, 1204–1222 (2020).
Metlay, J. P. et al. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am. J. Respir. Crit. Care Med. 200, e45–e67 (2019).
Carugati, M. et al. Microbiological testing of adults hospitalised with community-acquired pneumonia: an international study. ERJ Open Res. 4, 00096–02018 (2018).
Vaughn, V. M. et al. Excess antibiotic treatment duration and adverse events in patients hospitalized with pneumonia: a multihospital cohort study. Ann. Intern. Med. 171, 153–163 (2019).
Chalmers, J. D., Akram, A. R., Singanayagam, A., Wilcox, M. H. & Hill, A. T. Risk factors for Clostridium difficile infection in hospitalized patients with community-acquired pneumonia. J. Infect. 73, 45–53 (2016).
Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Zay Ya, K., Win, P. T. N., Bielicki, J., Lambiris, M. & Fink, G. Association between antimicrobial stewardship programs and antibiotic use globally: a systematic review and meta-analysis. JAMA Netw. Open 6, e2253806 (2023).
Viasus, D., Vecino-Moreno, M., De La Hoz, J. M. & Carratalà, J. Antibiotic stewardship in community-acquired pneumonia. Expert. Rev. Anti Infect. Ther. 15, 351–359 (2017).
Poole, S. & Clark, T. W. Rapid syndromic molecular testing in pneumonia: the current landscape and future potential. J. Infect. 80, 1–7 (2020).
Gentilotti, E. et al. Diagnostic accuracy of point-of-care tests in acute community-acquired lower respiratory tract infections. A systematic review and meta-analysis. Clin. Microbiol. Infect. 28, 13–22 (2022).
Gadsby, N. J. et al. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin. Infect. Dis. 62, 817–823 (2016).
Cartuliares, M. B. et al. Evaluation of point-of-care multiplex polymerase chain reaction in guiding antibiotic treatment of patients acutely admitted with suspected community-acquired pneumonia in Denmark: a multicentre randomised controlled trial. PLoS Med. 20, e1004314 (2023).
Markussen, D. L. et al. Diagnostic stewardship in community-acquired pneumonia with syndromic molecular testing: a randomized clinical trial. JAMA Netw. Open 7, e240830 (2024).
Lim, W. S. et al. BTS guidelines for the management of community acquired pneumonia in adults: update 2009. Thorax 64, iii1–iii55 (2009).
Woodhead, M. et al. Guidelines for the management of adult lower respiratory tract infections—full version. Clin. Microbiol. Infect. 17, E1–E59 (2011).
Barlam, T. F. et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin. Infect. Dis. 62, e51–e77 (2016).
Serigstad, S. et al. Rapid syndromic PCR testing in patients with respiratory tract infections reduces time to results and improves microbial yield. Sci. Rep. 12, 326 (2022).
Carratalà, J. et al. Effect of a 3-step critical pathway to reduce duration of intravenous antibiotic therapy and length of stay in community-acquired pneumonia: a randomized controlled-trial. Arch. Intern. Med. 172, 922–928 (2012).
Falguera, M. et al. Prospective, randomised study to compare empirical treatment versus targeted treatment on the basis of the urine antigen results in hospitalised patients with community-acquired pneumonia. Thorax 65, 101–106 (2010).
Darie, A. M. et al. Fast multiplex bacterial PCR of bronchoalveolar lavage for antibiotic stewardship in hospitalised patients with pneumonia at risk of Gram-negative bacterial infection (Flagship II): a multicentre, randomised controlled trial. Lancet Respir. Med. 10, 877–887 (2022).
Gadsby, N. J. et al. Discordance between semi-quantitative nucleic acid detection of bacteria and quantitative bacteriology in sputum from patients with pneumonia. J. Infect. 86, 607–609 (2023).
Schweitzer, V. A. et al. Narrow-spectrum antibiotics for community-acquired pneumonia in Dutch adults (CAP-PACT): a cross-sectional, stepped-wedge, cluster-randomised, non-inferiority, antimicrobial stewardship intervention trial. Lancet Infect. Dis. 22, 274–283 (2022).
Demars, Y. et al. Utility of polymerase chain reaction in nasopharyngeal swabs for identifying respiratory bacteria causing community-acquired pneumonia. Microbiol. Spectr. 10, e0037922 (2022).
Serigstad, S. et al. Diagnostic utility of oropharyngeal swabs as an alternative to lower respiratory tract samples for PCR-based syndromic testing in patients with community-acquired pneumonia. J. Clin. Microbiol. 61, e0050523 (2023).
Chan, K. F., Ma, T. F., Ip, M. S. & Ho, P. L. Invasive pneumococcal disease, pneumococcal pneumonia and all-cause pneumonia in Hong Kong during the COVID-19 pandemic compared with the preceding 5 years: a retrospective observational study. BMJ Open 11, e055575 (2021).
Mandell, L. A. et al. Community-acquired pneumonia in Canada during coronavirus disease 2019. Open Forum Infect. Dis. 9, ofac043 (2022).
Amin-Chowdhury, Z. et al. Impact of the coronavirus disease 2019 (COVID-19) pandemic on invasive pneumococcal disease and risk of pneumococcal coinfection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): prospective national cohort study, England. Clin. Infect. Dis. 72, e65–e75 (2021).
Abelenda-Alonso, G. et al. Impact of comprehensive molecular testing to reduce antibiotic use in community-acquired pneumonia (RADICAP): a randomised, controlled, phase IV clinical trial protocol. BMJ Open 10, e038957 (2020).
R Core Team. R: a language and environment for statistical computing. https://www.R-project.org/ (R Foundation for Statistical Computing, 2023).
Wickham, H., François, R., Henry, L., Müller, K. & Vaughan, D. dplyr: a grammar of data manipulation. R package version 1.1 (2023).
Carmezim, J. et al. REDCapDM: REDCap data management. R package version 0.8 (2023).
Wassmer, G. & Pahlke, F. rpact: confirmatory adaptive clinical trial design and analysis. R package version 3.4.0 (2023).
Subirana, I., Sanz, H. & Vila, J. Building bivariate tables: the compare Groups package for R. J. Stat. Softw. 57, 1–16 (2014).
Acknowledgements
This study was supported by a grant (201808-10) by the Fundació La Marató de TV3, Barcelona, Spain. The funder of the study had no role in the study design, data collection, data analysis, data interpretation, or the writing of the report. G.A.A. is supported by a Rio Hortega predoctoral grant (CM21/00047) by the Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation, Madrid, Spain. A.R. is supported by a predoctoral grant (PFIS contract FI18/00183) by the Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation, Madrid, Spain.
Author information
Authors and Affiliations
Contributions
J.C. was the lead investigator. G.A.A., A.P., C.T., C.A., and J.C. contributed to the study design and development concept. G.A.A. and J.C. drafted the manuscript. P.S. and C.T. performed statistical analysis. J.C. obtained funding. G.A.A., A.R., Y.M., I.O., N.S., A.D., J.A., and C.G. recruited patients for the study and participated in coordination. L.C., J.N., J.L., and C.A. performed the microbiological studies. G.A.A., P.S., C.T., C.A., and J.C. had full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to the acquisition, analysis or interpretation of data, and performed critical revision of the manuscript for intellectual content.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Abelenda-Alonso, G., Calatayud, L., Rombauts, A. et al. Multiplex real-time PCR in non-invasive respiratory samples to reduce antibiotic use in community-acquired pneumonia: a randomised trial. Nat Commun 15, 7098 (2024). https://doi.org/10.1038/s41467-024-51547-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51547-8
- Springer Nature Limited