Abstract
Proton-activated chloride (PAC) channels, ubiquitously expressed in tissues, regulate intracellular Cl- levels and cell death following acidosis. However, molecular mechanisms and signaling pathways involved in PAC channel modulation are largely unknown. Herein, we determine that phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] of the plasma membrane inner leaflet is essential for the proton activation of PAC channels. PI(4,5)P2 depletion by activating phosphatidylinositol 5-phosphatases or Gq protein-coupled muscarinic receptors substantially inhibits human PAC currents. In excised inside-out patches, PI(4,5)P2 application to the cytoplasmic side increases the currents. Structural simulation reveals that the putative PI(4,5)P2-binding site is localized within the cytosol in resting state but shifts to the cell membrane’s inner surface in an activated state and interacts with inner leaflet PI(4,5)P2. Alanine neutralization of basic residues near the membrane-cytosol interface of the transmembrane helice 2 significantly attenuates PAC currents. Overall, our study uncovers a modulatory mechanism of PAC channel through inner membrane PI(4,5)P2.
Similar content being viewed by others
Introduction
An acid-sensitive outwardly rectifying (ASOR) Cl− current was initially identified in rat Sertoli cells1, then characterized in HEK293 cells2,3 with permeability to anions in the following order: SCN− > I− > NO3− > Br− > Cl−. Through RNA interference screens, the molecular identity of this channel was recently unveiled as the transmembrane protein 206 (TMEM206 or PACC1) encoding a proton-activated Cl- (PAC) channel2,3. Human PAC (hPAC) channels are variable due to alternative splicing of a single gene, where two main splice variants (hPAC isoform 1 or hPAC1, AK297200.1; hPAC isoform 2 or hPAC2, AK024066.1) exhibited a difference in the N-terminal sequence, with 61 more amino acids in hPAC14. PAC1 splicing variant is less common, while PAC2 is prevalent in most tissues and cell types and contributes to many physiological responses, including endosome acidification, macropinosome shrinkage, and brain damage after ischemia5,6,7.
PAC channels are formed by the homotrimeric association of a subunit comprising two transmembrane (TM) helices (TM1 and TM2) and a large ball-shaped extracellular domain (ECD)8,9. Due to extracellular acidification, clusters of extracellular acidic residues coalesce in the ECD and rearrange TMD helices to open the channel10. Notably, the PAC channel activation threshold is about pH 5.5 at room temperature2. Cryo-EM structural analysis has revealed distinct PAC channel states; conformation obtained at pH 8.0 displayed a closed state, whereas a protonated non-conducting conformation obtained at pH 4.0 indicated a desensitized state8. The open channel conformation of PAC was recently manifested with detergent and lipid nanodisc reconstitution samples at pH 4.510. The channel pore was wider than 3 Å, allowing hydrated Cl- to pass through10.
The acid-activated PAC channel can be regulated by intracellular mediators, such as tyrosine kinase, phosphoinositide 3-kinase (PI3K), and dynamin, but not intracellular Ca2+11. A recent study determined that the membrane phospholipid phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] binds to the extracellular side of the PAC channel’s TMDs and prevents the channel from opening12. Furthermore, applying water-soluble dioctanoyl (diC8)-PI(4,5)P2 to a bath solution similarly inhibited PAC channel activity. Therefore, extracellular PI(4,5)P2 may stabilize desensitized PAC channel states under acidic conditions12. Despite these findings, it is necessary to reconsider PAC channel regulation by intracellular PI(4,5)P2 since PI(4,5)P2 is primarily distributed through the plasma membrane’s inner leaflet13,14. Furthermore, it electrostatically binds to a binding pocket comprising positively charged residues and proximal hydrophobic residues near the membrane–cytoplasm interface15,16,17.
PI(4,5)P2 in the inner leaflet is a prominent target substrate of phospholipase Cβ (PLCβ), an effector activated by Gq-type G protein-coupled receptors (Gq-GPCRs)18. Thus, GPCR-mediated PI(4,5)P2 hydrolysis can modulate the function of many ion channels coupled to PI(4,5)P219,20. In the present study, by manipulating membrane phosphoinositide (PI) levels in intact living cells, we found that inner leaflet PI(4,5)P2, but not other PIs, is required for the proton activation of hPAC channels. Application of diC8-PI(4,5)P2 to the cytosolic side of the plasma membrane led to the increase in hPAC current. Mutagenesis and structural remodeling revealed that PI(4,5)P2 binds to positively charged residues in TM2’s plasma membrane–cytoplasm interface in activated and desensitized channel conformations. We also confirmed that activating muscarinic receptors suppresses PAC channel activity by eliciting PI(4,5)P2 hydrolysis through PLCβ in exogenous and endogenous expression systems. Our results demonstrate that the activation-dependent PI(4,5)P2 coupling is essential for the pathophysiological function and GPCR modulation of PAC channel.
Results
Plasma membrane PI(4,5)P2 regulates human PAC1 and PAC2 channel gating
To investigate the regulatory effects of plasma membrane PI(4,5)P2 on PAC channel activity, the amino acid sequences of two splice variants of human PAC, hPAC1 and hPAC2, and one mouse PAC (mPAC) channels were compared (Supplementary Fig. 1). hPAC1 and hPAC2 exhibited a difference in the N-terminal sequence, with 61 more amino acids in hPAC1. We verified that PAC cDNA extracted from the human embryonic kidney 293 cell line with a plasmid encoding a temperature-sensitive mutant of the SV40 large T antigen (HEK293T) is the shorter hPAC2 isoform. Thus, we produced an hPAC1 channel by inserting 61 amino acids into the N-terminus of hPAC2 cDNA through inverse PCR cloning. We also made five PAC knockout (KO) HEK293T cell lines, KO1 to KO5, via CRISPR/Cas9 (Supplementary Fig. 2a) and confirmed significantly reduced endogenous currents in all KO cell lines at pH 4.6. Among them, KO3 produced almost negligible PAC channel activity (Fig. 1a and Supplementary Fig. 2b).
a Voltage-dependent currents of hPAC channels in extracellular pH 7.4 or 4.6 solutions in wild-type (WT) HEK293T cells expressing PAC channels endogenously and PAC CRISPR knockout KO3 cells. b (left) Representative confocal images of KO3 cells expressing hPAC1-GFP (top) or hPAC2-GFP (bottom) with the plasma membrane marker Lyn-mCherry (Lyn-mCh). The yellow arrow indicates the location and line scanning direction in merged images. Scale bar: 10 μm. (right) Line-scan analyzes of Lyn-mCh and hPAC1-GFP (top) or hPAC2-GFP (bottom). *, peak cytosolic fluorescence intensity. Each experiment was repeated 3 times independently with similar results. c Current density–voltage relationship for hPACs at various pH conditions. n = 5 for pH 7.4, 6.3, 5.3, 5.0; n = 10 for pH 5.5; n = 7 for pH 4.6 in hPAC1; n = 2 for pH 7.4; n = 5 for pH 6.3, 5.5; n = 3 for pH 5.3, 5.0; n = 4 for pH 4.6 in hPAC2. Data are mean ± SEM. d (left) Scheme for PI(4,5)P2 depletion by Dr-VSP. (right) ΔFRETr changes in control (top) or cells transfected with Dr-VSP (bottom) during +100-mV/10-s depolarization pulse. Cells were transfected with the PI(4,5)P2 FRET probes, PHPLCδ1-CFP and PHPLCδ1-YFP. e pH 5.5-induced PAC current traces during test pulse in KO3 cells (top) and transfected with hPAC1 (middle) or hPAC2 (bottom), without (control) or with Dr-VSP. f Current density measured by analyzing the steady-state current at the end of each +100-mV depolarization step in (e). KO3: n = 8; n = 8. hPAC1: n = 6; n = 7. hPAC2: n = 9; n = 6 for −Dr-VSP and +Dr-VSP, respectively. Dots indicate the individual data points for each cell. Data are mean ± SEM. g Average percent inhibition of current densities by PI(4,5)P2 depletion in different pH solutions. h hPAC1 current recovery from Dr-VSP-induced inhibition in cells intracellularly perfused with (top) or without (bottom) ATP. PAC currents were measured at the indicated times after a + 120-mV/10-s depolarization pulse. Dotted lines indicate zero, and dashed lines indicate PAC current amplitude (control) before the depolarizing step. i Average current recovery with (open) or without (filled) of intracellular ATP (n = 6, +ATP; n = 4, -ATP). Data are mean ± SEM. Source data are provided as Source Data file.
First, to examine PAC channel cellular distribution, KO3 cells were co-transfected with each hPAC channel labeled with a green fluorescent protein (GFP), hPAC1-GFP, or hPAC2-GFP, and the plasma membrane marker Lyn11-mCherry (Lyn-mCh). We confirmed that the hPAC1 was predominantly colocalized with Lyn-mCh at the plasma membrane (as evidenced by overlapped GFP and mCh line scanning results). In contrast, hPAC2 was primarily distributed in cytosolic organelles and the plasma membrane (Fig. 1b and Supplementary Fig. 3b, c). To further clarify the cytosolic hPAC2 localization, we visualized colocalization with the early endosome marker Rab5 and the lysosomal marker LAMP1. The hPAC2 was mainly colocalized with Rab5 but not LAMP1 (Supplementary Fig. 3d, e). The hPAC2 N-terminus has the endosomal targeting motif YXXφ, composed of 10YQEL13 sequences, where tyrosine (Y) or leucine (L) substitution by alanine (A) causes a relocalization from the vesicle to the cell surface8. Similarly, since the extra 61 hPAC1 amino acids were inserted into the endosomal targeting sequence, hPAC1 was prominently expressed at the cell surface (Supplementary Fig. 3a). hPAC1 and hPAC2 current densities increased under lower extracellular pH. Under pH 5.3, hPAC1 displayed a greater channel activity than hPAC2, coinciding with Y10A or L13A hPAC2 mutant results in a previous study (Fig. 1c)8.
We examined whether the plasma membrane phospholipid PI(4,5)P2 regulated hPAC1 and hPAC2 using the zebrafish voltage-sensing phosphatase (Dr-VSP). This phosphatase dephosphorylates the 5ʹ-phosphate of PI(4,5)P2 at the plasma membrane’s inner leaflet when stimulated by a depolarizing voltage (Fig. 1d, left)21,22. To examine the regulatory effect of the inner leaflet PI(4,5)P2 on PAC activity, a 10-s depolarizing step protocol was applied at +100 mV to elicit PAC Cl- currents and deplete PI(4,5)P2. Using the Förster resonance energy transfer (FRET) approach, we examined the time course of Dr-VSP-mediated PI(4,5)P2 depletion by applying a + 100 mV/10 s depolarizing pulse in cells co-transfected with ECFP- and EYFP-tagged PH domains derived from PLCδ1 (PHPLCδ1-CFP and PHPLCδ1-YFP). Control cells did not exhibit a ΔFRETr signal change during the depolarizing pulse. However, the ΔFRETr fell with a 0.25 ± 0.03 s time constant (τ) in Dr-VSP-expressing cells and remained at lower levels during the depolarizing pulse. This observation suggests that Dr-VSP activation reduced PI(4,5)P2 concentration at the plasma membrane (Fig. 1d, right).
With the 10-s depolarizing step protocol, we tested whether endogenous PAC channel activity was sensitive to membrane PI(4,5)P2 in wild-type (WT) HEK293T cells. Perfusion of the pH 5.5 solution evoked endogenous PAC currents larger than that in the neutral environment of control cells (Supplementary Fig. 4). However, Dr-VSP co-expression almost blocked channel activity at pH 5.5 entirely, suggesting that PI(4,5)P2 depletion by Dr-VSP inhibits the PAC channel. This feature was further examined using PIP5-kinase type-1γ (PIPKIγ), which elevates PI(4,5)P2 levels by phosphorylating phosphatidylinositol 4-phosphate [PI(4)P]. Comparatively, PIPKIγ co-expression slightly increased current density. The current was significantly decreased during Dr-VSP activation, likely due to the inner leaflet’s rapid PI(4,5)P2 resynthesis. We further evaluated the PI(4,5)P2 sensitivity of PAC channels in KO3 cells transfected with hPAC1 or hPAC2. Heterologous hPAC1 and hPAC2 indicated robust currents in control cells without Dr-VSP at pH 5.5 (Fig. 1e). However, Dr-VSP co-expression inhibited the elevation of hPAC1 and hPAC2 currents. The initial weak peak current may occur due to the slow PI(4,5)P2 depletion by Dr-VSP during the depolarizing pulse (Fig. 1d). hPAC1 and hPAC2 current densities at +100 mV were remarkably reduced in cells co-expressing Dr-VSP (Fig. 1f). The Dr-VSP-induced current inhibition of hPAC1 and hPAC2 was also apparent in the pH 5.0 or 4.6 extracellular solution (Fig. 1g).
Next, we tested whether PI(4,5)P2 resynthesis after depletion is needed to recover PAC currents from Dr-VSP-induced suppression. Current recovery was monitored by applying +50 mV/0.5 s test pulses with successively longer delays after Dr-VSP activation starting at 0.5 s, as indicated above the traces23. In the presence of intracellular ATP (3 mM), the hPAC1 current reduced by the depolarizing pulse was restored to initial levels at a recovery time constant τ of 3.7 s (Fig. 1h, i). Since PI(4,5)P2 synthesis from PI(4)P requires intracellular ATP, we attenuated PI 5-kinase activity by dialyzing an ATP-free pipette solution into the cell for 10 min. Without ATP in the pipette solution, the maximum current inhibition from depolarization was not affected, but PAC current recovery was slowed (Fig. 1h, i). These findings together support that PI(4,5)P2 is needed for normal PAC channel activity.
Phosphoinositide regulation of PAC channel responds specifically to PI(4,5)P2
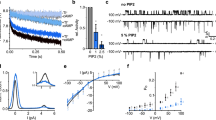
To further assess whether PAC channel activity is regulated by the plasma membrane’s PI(4,5)P2, we applied the rapamycin-inducible translocation of phosphoinositide phosphatases from cytosol to the plasma membrane’s inner surface. As shown in Fig. 2a, we employed the enzymatic fusion proteins of mRFP-FKBP (RF) to manipulate inositol lipids of the plasma membrane as follows: (1) Sac1, PI(4)P 4’-phosphatase; (2) inositol polyphosphate-5-phosphatase E (INPP5E); (3) pseudojanin (PJ), which possesses both INPP5E and Sac1. Using confocal microscopy, we validated whether lipid phosphatases can adequately function at the intracellular side by co-expressing PHOsh1-GFP and PHPLCδ1-GFP as PI(4)P and PI(4,5)P2 biomarkers, respectively (Supplementary Fig. 5).
All experiments were executed at extracellular pH 5.5 conditions. a Schematic of rapamycin (Rapa)-induced poly-phosphoinositide phosphatase translocation on PAC currents. (left) Metabolic pathway of PI(4,5)P2 synthesis by lipid kinases (4-K and 5-K) and lipid phosphatase breakdown (4-P and 5-P). PI, phosphatidylinositol; PI4P, phosphatidylinositol 4-phosphate. (right) Schematic of rapamycin-induced dimerization of FRB and FKBP proteins. This dimerization recruits poly-PI-metabolizing enzymes to the plasma membrane. RF-Dead is a translocatable construct with inactive mutant Sac1 and INPP5E enzymes. b Representative current traces of hPAC1 from cells expressing RF-Dead, RF-Sac1, RF-INPP5E, or RF-PJ before and after rapamycin. Voltages were stepped from –80 to +80 mV in 20-mV intervals. c Summary of whole-cell current vs. voltage relationship. (black) Before rapamycin; (red) after rapamycin. RF-Dead, n = 6; RF-Sac1, n = 6; RF-INPP5E, n = 5; RF-PJ, n = 8. Data are mean ± SEM. d (left) Time-dependent hPAC1 current amplitude changes by rapamycin were measured at +80 mV every 5 s from cells expressing RF-Dead, RF-Sac1, RF-INPP5E, or RF-PJ. Black lines indicate the addition of 100 nM rapamycin for 3 min. (right) hPAC1 current comparisons before (black) and after (red) rapamycin. e Voltage-independence of rapamycin-induced PAC1 channel inhibition (n = 5 for INPP5E; n = 8 for PJ). Student’s two-tailed unpaired t-test: P values are 0.3042 for INPP5E and 0.4410 for PJ. Data are mean ± SEM. f Summary of hPAC1 current inhibition (%) by rapamycin-induced translocation of RF-Dead, RF-Sac1, RF-INPP5E, or RF-PJ to the plasma membrane. Dots indicate the individual data points for each experiment. Bars are mean ± SEM. ns, not significant, compared to RF-Dead. One-way ANOVA followed by Sidak’s multiple comparisons test: P values are 0.5676 for Sac1, <0.0001 for INPP5E, <0.0001 for PJ, and 0.4044 for INPP5E vs. PJ. Source data are provided as Source Data file.
Rapamycin treatment recruited all the translocatable enzymes from the cytosol to the plasma membrane. This system operates via rapamycin-bound FKBP (FK506 binding protein), forming a complex with FRB (the rapamycin-binding domain of mTOR)24. This complex is anchored to the membrane through myristoylation and palmitoylation sites of Lyn kinase’s 11 N-terminal amino acid residues (Lyn11). No PHOsh1 or PHPLCδ1 translocation change was observed in the overexpression of RF-Dead without Sac1 and INPP5E enzyme activity (Supplementary Fig. 5). Recruiting RF-Sac1 or RF-INPP5E increased PHOsh1 and PHPLCδ1 fluorescence intensity in the cytosol, dephosphorylating 4’-phosphate from PI(4)P and 5’-phosphate from PI(4,5)P2, respectively. We also verified that recreating RF-PJ on the plasma membrane released PHOsh1 or PHPLCδ1 into the cytosol.
We applied these rapamycin-inducible translocation systems to pH 5.5-induced hPAC1 currents to verify phosphoinositide regulation. Supplementing 100 nM rapamycin did not affect hPAC1 activity in cells expressing RF-Dead or RF-Sac1 (Fig. 2b, c). However, recruiting RF-INPP5E or RF-PJ significantly inhibited hPAC1 current density. As a result of observing the real-time changes of hPAC1 currents in a single cell expressing each enzyme, adding rapamycin inhibited pH 5.5-evoked hPAC1 currents in cells expressing RF-INPP5E or RF-PJ by 72 ± 4% and 82 ± 5%, respectively (Fig. 2d, f). This observation proposes that only PI(4,5)P2, but not PI or PI(4)P, is required to activate hPAC1 channels. Furthermore, these results also show that rapamycin-induced current inhibition of hPAC1 was independent of voltages between +40 and +80 mV (Fig. 2e).
To examine the effects of PI(3,4,5)P3 on PAC channels, we applied the chimeric enzyme Ci-VSTPEN15, where the Ci-VSP catalytic domain was replaced with full-length PTEN (a phosphatase and tensin homolog). PTEN dephosphorylates 3’-phosphate of PI(3,4,5)P3. We measured Ci-VSPTEN activity using the FRET approach in cells co-transfected with PH domains derived from Bruton’s tyrosine kinase tagged with ECFP or EYFP (PHBtk-CFP and PHBtk-YFP) (Supplementary Fig. 6a). During the +100 mV/10 s depolarizing pulse, PI(3,4,5)P3 dephosphorylation was proven in cells expressing Ci-VSPTEN, but not in the control group (Supplementary Fig. 6b). With the 10-s depolarizing step protocol, we tested whether the hPAC1 channel is sensitive to membrane PI(3,4,5)P3 depletion. However, no difference in pH 5.5-induced activation of the hPAC1 channel was observed between control and co-expressing Ci-VSTPEN cells (Supplementary Fig. 6c, d). Collectively, we concluded that among the inner leaflet PIs, only PI(4,5)P2 can activate the hPAC1 channel.
Intracellular application of PI(4,5)P2 potentiates PAC channels in excised inside-out membrane patches
To determine whether PI(4,5)P2 alone is sufficient to potentiate PAC channels on the inner side of membranes, we examined the activities of hPAC1 and hPAC2 channels in excised inside-out membrane patches. Both hPAC1 and hPAC2 channels were activated by depolarizing voltage stimulations of more than 50 mV in acidic extracellular pH. Representative current traces evoked by different voltage stimulations from a single excised membrane patch of either hPAC1 or hPAC2 channel are shown in Fig. 3a. However, we were unable to detect single-channel activity from the excised inside-out patches of untransfected PAC channel knockout cells (Supplementary Fig. 7a). The single channel openings of both hPAC1 and hPAC2 channels were highly voltage-dependent, as observed in the macroscopic current traces. Larger depolarization-induced significant increases in the channel open probability (NPO) (Fig. 3b). Like as the whole-cell patch clamp recording of a putative PI(4,5)P2-binding site mutant hPAC1 (R396A), we could not measure voltage-dependent currents of R396A mutant hPAC1 from the excised inside-out patches (three independent patches, Supplementary Fig. 7b). These results suggest that the outwardly rectifying I-V relationships of macroscopic hPAC1 and hPAC2 currents are due to the voltage-dependent gating of channels rather than pore blockade, as cytosolic cellular components were removed in the excised inside-out patch-clamp experiments. Gaussian fitting of channel gating levels (closing and openings) allowed us to assign a unitary channel current at specific membrane voltages. The estimated unitary slope conductance of the hPAC1 and hPAC2 channels were approximately 46.06 pS (n = 5) and 30.23 pS (n = 4), respectively (Fig. 3c). Previously, the chord conductance of a single endogenous PAC channel (PAC2) in HEK293 cells was reported to be 10 pS at +43 mV, 13 pS at +86 mV, and 20 pS at +126 mV25. The extrapolated value of slope conductance from the previous report was approximately 25 pS, which aligned well with our measurements. To confirm that the recorded single-channel activities were from acid-activated Cl− current rather than cationic current, we measured the single-channel activities in either a normal intracellular solution (CsCl and MgCl2) or a cation-replaced solution (NMDG-Cl). The single-channel current amplitudes of both hPAC1 and hPAC2 channels did not significantly differ between the solutions, but the open probabilities decreased under the NMDG-replaced condition, similar to the previous report on the endogenous PAC channel (Supplementary Fig. 8)25.
a Single-channel currents and all-point histograms of hPAC1 (left) and hPAC2 (right) channels at the indicated voltage stimulations and pH (external or pipette solution) recorded in inside-out patch clamp configurations. The dashed red lines indicate current levels of closed or open states, and ‘C’ and ‘O’ represent closed and open states, respectively. The number next to ‘O’ indicates the number of open channels. The scales represent 10 pA and 1 s. All-point histograms of single-channel recordings were obtained from 1-minute recordings at the indicated voltage and pH. The voltage-channel open probability (NPo) relationships (b) and unitary current-voltage plot (c) for hPAC1 (open circle, n = 5) and hPAC2 (filled circle, n = 4) channels at pH 5.5. Data are represented as mean ± SEM. d Time-dependent plot of hPAC1 currents in inside-out configuration (left) at pH 5.5. The internal or bath application of 10 μM diC8-PI(4,5)P2 induced a potentiation of PAC1 currents, but the potentiation was inhibited by a PAC channel inhibitor, 4,4’-diisothiocyano-2,2’-stilbenedisulfonic acid (DIDS). hPAC1 currents were elicited by a + 100 mV step pulse for 10 s from holding potential of 0 mV, and the mean currents were obtained from the average values of the last 10 s. The currents were recorded every 0.5 s. Right, representative current traces (numbers in parentheses, right) show hPAC1 currents at arrow-pointing spots. Source data are provided as Source Data file.
We then examined the effect of intracellular PI(4,5)P2 on the hPAC1 channel by applying diC8-PI(4,5)P2 to the internal side of excised inside-out membrane patches (Fig. 3d, e). After forming an inside-out patch, only a few hPAC1 channel openings were observed at the single-channel level, but the application of 10 μM diC8-PI(4,5)P2 significantly increased ionic currents to a macroscopic current level of ~30 pA. The treatment of a PAC channel inhibitor, 4,4’-diisothiocyano-2,2’-stilbenedisulfonic acid (DIDS)25, abolished PAC current potentiation even in the presence of 10 μM diC8-PI(4,5)P2 (Fig. 3d). The initial low-level activity of the hPAC1 channel might be due to the rapid rundown of the hPAC1 channel caused by dissociation or degradation of PI(4,5)P2 in the isolated membrane patch.
Identification of residues affecting PI(4,5)P2 sensitivity in the intracellular side of PAC channels
Structural analysis has revealed that fundamental residues like lysine and arginine around the membrane-cytosol interface usually interact with PI(4,5)P2 in functionally active ion channels16,17,26. Thus, we conducted alanine-scanning mutagenesis of positively charged residues, including K120 and K124 in the N-terminus and K390, K394, R396, K397, R398, K401, R402, and R403 located in the TM2 cytoplasmic interface of hAPC1 (Fig. 4a). All hPAC1 mutant channels, including the WT hPAC1, were co-localized with Lyn-mCh at the plasma membrane, as indicated by yellow in the merged images (Fig. 4b). Quantifying the co-localization of PAC channels with Lyn-mCh showed non-significant difference in Pearson’s coefficient between WT and mutant channels (Fig. 4d)27. Of the ten single amino acid mutant forms, the pH 5.5-induced current amplitude at +100 mV was dramatically reduced in K394A, R396A, and K397A compared to WT without Dr-VSP expression (control) (Fig. 4c, e). Similarly, we could not measure any significant voltage-dependent currents of R396A mutant PAC1 from the excised inside-out patches (Supplementary Fig. 7b). Triple mutation of three effective basic residues (K394A/R396A/K397A) did not affect the channel’s plasma membrane distribution but predominantly abolished the hPAC1 currents in acidic pH ranges down to 5.0 (Fig. 4f, g). With Dr-VSP, there was minor suppression of currents in the triple mutant form; however, the additional suppression of whole-cell current by Dr-VSP was even found in PAC-KO cells (see Fig. 1f). We also conducted alanine substitution mutations of the positively charged residues K59, K63, K329, K333, R335, K336, R337, K340, R341, and R342 in the hPAC2 channel, corresponding to hPAC1 residues (Supplementary Fig. 9a), and identified cellular distribution and channel activity. Mutant hPAC2 construct distribution was consistent with the WT hPAC2 expression pattern (Supplementary Fig. 9b). Similar to hPAC1, K333, R335, and K336 alanine mutations of hPAC2 substantially impaired channel activity (Supplementary Fig. 8c, d). Moreover, triple mutation (K333A/R335A/K336A) of hPAC2 channel retrained the current generation and Dr-VSP suppression without affecting the cellular distribution. These results suggest that those three basic amino acids in the TM2 domain’s distal end are important for regulating the PI(4,5)P2 regulation in both hPAC1 and hPAC2 channels.
a Sequence of positively charged residues clustering in hPAC1. Purple lines indicate N- and C-terminal sections. Green lines indicate transmembrane domain (TM) 1 and 2 sections. Positively charged residues indicated in blue are selectively mutated to alanine (A). b Representative confocal images of KO3 cells expressing wild-type (WT) or alanine-substituted hPAC1 mutants with the plasma membrane marker Lyn-mCh. Scale bar: 10 μm. Scatter plots in right panels display a 2D intensity histogram between hPAC1-GFP and Lyn-mCh pixels in the confocal image. Each experiment was repeated 3 times independently. c Representative current traces of WT or mutant channels elicited at +100 mV in the control (left) and cells expressing Dr-VSP (right). d Pearson’s correlation coefficients (R) for the colocalization between hPAC1-GFP and Lyn-mCh confocal images. n = 6 for WT; n = 10 for K120A, K390A, K397A, R398A, K401A; n = 9 for K124A, K394A, R396A, K394/R396A/K397A, K402A, K403A. Dots indicate the individual data points for each cell. Data are mean ± SEM. e hPAC1 current densities at +100 mV for control and cells expressing Dr-VSP in pH 5.5. The dashed gray line indicates the standard WT level. n = 6 for WT, K394A, K402A; n = 7 for K120A; n = 4 for K124A; n = 5 for K390A, R396A, K397A, K394/R396A/K397A, R398A, K403A; n = 3 for K401A. Data are mean ± SEM. ns, not significant compared to WT. One-way ANOVA followed by Dunnett’s multiple comparisons test. P values: 0.9993, K120A; 0.1938, K124A; 0.9994, K390A; 0.0009, K394A (***P < 0.001); <0.0001, K396A (****P < 0.0001); 0.0103, K397A (*P < 0.05); <0.0001, K394/R396/K397A (****P < 0.0001); 0.8065, K398A; 0.9938, K401A; 0.9999, R402A; 0.9249, R403A. f Families of hPAC1 current of WT (left) and K394A/R396A/K397A mutant (right) elicited by the voltage steps shown at the top under different extracellular pH condition. g pH-dependent hPAC1 current measured at +100 mV in cells expressing hPAC1 WT (n = 6 for pH 7.4, 5.8, 5.5; n = 5 for pH 6.3; n = 7 for pH 5.3, 5.0) or K394A/R396A/K397A (n = 5 for pH 7.4, 6.3, 5.8, 5.5, 5.3, 5.0). Data are mean ± SEM. Source data are provided as Source Data file.
Verification of PI(4,5)P2-PAC interaction from structural modeling
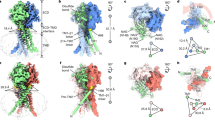
The PAC channel’s cryo-EM structure can be divided into resting, activated, and desensitized states10. Since this structure has been identified in the PAC2 channel, we investigated K333, R335, and K336 residue positions in the hPAC2 channel. Through the simulation using I-TASSER and the cryo-EM structure, we obtained the resting state conformation of the hPAC2 channel. The Ramachandran plot of the simulation was strikingly similar to that of hPAC2’s cryo-EM structure (Supplementary Fig. 10a). The model revealed that the three fundamental residues (K333, R335, and K336) and their side chains were only located in the cytosolic region (Supplementary Fig. 10b). However, we determined that the three basic residues were translocated and resided at the membrane–cytosolic interface in both activated and desensitized conformations (Fig. 5a).
a Cryo-EM structure models of resting, activated, and desensitized PAC2 channel conformations (Protein Data Bank ID: resting, 7SQG; activated, 7SQF; desensitized, 7SQH). Two black lines indicate the outer and inner plasma membrane surfaces. Blue, red, and green boxes indicate the location of putative PI(4,5)P2-binding residues K333, R335, and K336, respectively. b Molecular docking between K333, R335, and K336 of the activated chain A conformation and PI(4,5)P2 (top) or PI(4)P (bottom). (left) Autodock Vina and UCSF Chimera were used for GIRK PI(4,5)P2 or PI(4)P to the cryo-EM structure of activated PAC2. (right) Nine conformation superpositions of PI(4,5)P2 or PI(4)P. c Docking between K333, R335, and K335 of the desensitized chain A conformation and PI(4,5)P2. (top) Autodock Vina and UCSF Chimera were used for GIRK PI(4,5)P2 to the cryo-EM structure. (bottom) Nine conformation superpositions of PI(4,5)P2. d Hypothetical model of a cytosolic PI(4,5)P2 binding site depending on resting, activated, or desensitized PAC channel conformation.
During the structural rearrangement from the resting to the activated state, the TM2 domain is raised in the extracellular direction by 10Å10. This conformational change in the activated form translocates K333, R335, and K336A to the plasma membrane’s inner surface. With this structural feature, we assessed the PI(4,5)P2 coupling of K333, R335, and K336 residues in activated and desensitized hPAC2 channels via molecular docking simulation. Using the PI(4,5)P2 molecule extracted from the G protein-gated inwardly rectifying potassium (GIRK) K+ channel28, we identified three salt bridges between the docked PI(4,5)P2 molecule and the putative PI(4,5)P2 binding sites in activated form: 1) R335 side chains interact with 5’-phosphate of PI(4,5)P2 through two salt bridges, and 2) 4’-phosphate of PI(4,5)P2 coordinates the K336 residue through another salt bridge (Fig. 5b top). For PI(4)P, the charge–charge interaction between 5’-phosphate and R335 disappeared, whereas the interaction between 4’-phosphate and K336 remained (Fig. 5b bottom).
Although PI(4,5)P2 sensitivity was not confirmed in the desensitized PAC channel, we simulated molecular docking to examine PI(4,5)P2 binding to the three positively charged residues in a desensitized state. The docked PI(4,5)P2 has two salt bridges with the R335 side-chain in a desensitized conformation (Fig. 5c). In the hPAC2 resting conformation at natural pH, even the basic residues were located in the cytosol away from the plasma membrane. Our docking simulation indicated that PI(4,5)P2 does not interact with the K333, R335, or K336 side-chain (Supplementary Fig. 10c). Together, these results suggest that the hPAC2 channel in the resting state does not interact with PI(4,5)P2, but can couple with the cytoplasmic leaflet PI(4,5)P2 when the TM2 domain is outwardly shifted after activation (Fig. 5d). The PI(4,5)P2 coupling of the TM2 domain remains stable during the desensitized state.
The PAC channel is modulated by Gq-coupled muscarinic receptors
The plasma membrane PI(4,5)P2 level is dynamically regulated by signaling pathways, and Gq-GPCR activation reduces PI(4,5)P2 by hydrolyzing it into DAG and IP3 via PLCβ. First, we assessed whether Gq-GPCR can function in a pH 5.5 acidic environment by monitoring PHPLCδ1-GFP translocation, a PI(4,5)P2 probe in KO3 cells co-transfected with the M1 muscarinic receptor (M1R). As shown in Fig. 6a, M1R stimulation with oxotremorine-M (Oxo-M) translocated PHPLCδ1-GFP from the plasma membrane to the cytosol at pH 7.4 and 5.5. However, PHPLCδ1-GFP’s elevated cytosolic intensity at pH 5.5 was slightly lower than at pH 7.4 (Fig. 6b), likely due to the low agonist binding affinity to muscarinic receptors in acidic pH conditions29,30. To test whether receptor activation can modulate PAC channel activity, KO3 cells expressing hPAC1 or hPAC2 with M1R were stimulated with Oxo-M, which considerably and reversibly reduced pH 5.5-induced hPAC1 and hPAC2 currents (Fig. 6c). The maximum inhibitions by M1R activation were 84 ± 4 % and 87 ± 3 % with the time constants τ = 6 ± 2 s and τ = 5 ± 1 s for hPAC1 and hPAC2, respectively (Fig. 6d, e).
a Live confocal images of PHPLCδ1-GFP in HEK293T cells transfected with M1R before (control) and during ( + Oxo-M) application of 50 μM Oxo-M at pH 7.4 (black) or 5.0 (red). Scale bars are 10 μm. b Time-dependent changes in cytosolic fluorescence intensity by PHPLCδ1–GFP translocation upon M1R activation (n = 10, pH 7.4; n = 12, pH 5.5). Data are mean ± SEM. c (left) PAC current modulation by M1R activation with a 15-s Oxo-M application (50 μM) in cells expressing hPAC1 (top) or hPAC2 (bottom). Current amplitudes were measured at +100 mV every 5 s. (right) Representative current traces at different time points in time course recording. d Percent inhibition of pH 5.5-induced PAC currents by 50 μM Oxo-M (n = 6 for hPAC1; n = 5 for hPAC2). Data are mean ± SEM. e Time constants (τ) of current inhibition by muscarinic simulation in hPAC1 (n = 6) and hPAC2 (n = 5). Dots indicate the individual data points for each cell. Data are mean ± SEM. Student’s two-tailed unpaired t-test: P value, 0.5674. f Identifying PAC gene expression via RT-PCR analysis with RNA extracted from wild-type HEK293T or SH-SY5Y cell lines. The experiment was repeated 4 times independently with similar results. g Representative voltage-dependent whole-cell currents of endogenous PAC channels in SH-SY5Y cells under pH 7.4, pH 5.5, and pH 5.0 solutions. h Current density-voltage relationship of endogenous PAC in SH-SY5Y cells at the indicated pH values (n = 5, pH 7.4; n = 5, pH 5.5; n = 4, pH 5.0). Data are mean ± SEM. i, Endogenous PAC current modulation by muscarinic stimulation with a 15-s Oxo-M application (30 μM) in SH-SY5Y cells at pH 5.5 (blue) or 5.0 (red). PAC current amplitude was measured at +100 mV every 5 s. j Percent (%) inhibition of pH 5.5- or 5.0-induced endogenous PAC currents by 30 μM Oxo-M (n = 8 for pH 5.5; n = 6 for pH 5.0). Dots indicate the individual data points for each cell. Data are mean ± SEM. Student’s two-tailed unpaired t-test: P value, 0.0013 (**P < 0.01). Source data are provided as Source Data file.
PAC channels are ubiquitously expressed in all tissues and cell types11,31,32,33. According to the Human Protein Atlas (https://www.proteinatlas.org/), RNA expression of the PAC channel is the highest in the SH-SY5Y cell line, a human neuroblastoma (Supplementary Fig. 11). We compared hPAC1 and hPAC2 gene expressions in HEK293T and SH-SY5Y through RT-PCR analysis using sequence-specific designed primers (Supplementary Table 1). Both cell types expressed hPAC2 (but not hPAC1) with a relatively higher density in SH-SY5Y cells (Fig. 6f). We then examined innate PAC2 channel regulation by the endogenous M3 muscarinic receptor (M3R) in SH-SY5Y cells34. There were potent Cl- currents in the acidic extracellular solution with outward rectification at positive membrane potentials (Fig. 6g, h). Oxo-M treatment at pH 5.5 and pH 5.0 diminished endogenous PAC currents by 79 ± 1% and 68 ± 2%, respectively (Fig. 6i, j). These results demonstrate that PAC channels can be modulated by activating endogenous Gq-GPCRs in native cells.
Discussion
Despite the pathophysiological significance of PAC channels, their modulatory mechanisms through signaling pathways remain largely unknown. In this study, using the voltage-sensing phosphatase Dr-VSP and rapamycin-induced translocation of phosphatases, we verified that PI(4,5)P2 in the inner leaflet of the plasma membrane is essential for the acid-induced activation of PAC channels. Mutagenesis and structural modeling suggest that the putative interacting site for PI(4,5)P2 is located near the cytosolic TM2 interface in the PAC channel. However, the PI(4,5)P2 interacting site initially resides within the cytosolic area in a resting state and is translocated to the plasma membrane’s inner surface in an activated state. This observation indicates that PAC channel PI(4,5)P2 interaction only occurs when the channels are activated by proton binding. Our data also show that PI(4,5)P2 coupling of the PAC channel is required for proton-induced channel opening, as channel activity diminished by nearly 90% without intracellular PI(4,5)P2. Finally, we confirmed that endogenous GPCRs modulate PAC channels; thus, muscarinic receptor activation substantially suppresses PAC channel activity through PI(4,5)P2 hydrolysis.
Our study has revealed activation-dependent PI(4,5)P2 coupling as a modulatory mechanism of PAC channels (Fig. 4d): (1) Resting state, PAC channel is PI(4,5)P2-free due to the cytosolic positioning of fundamental amino acids in the TM2 domain; (2) When extracellular H+ binds to the PAC channel, TM2 shifts towards the extracellular side and facilitates hPAC2 channel to PI(4,5)P2 binding at the membrane-cytosol interface; (3) Desensitized conformation, TM1 undergoes a 45° shift to the left, while TM2 tilts at an angle of 22.8°8,10. Despite these structural alterations, PI(4,5)P2 continuously interacts with the hPAC2 channel’s binding site. In inside-out patch clamp recordings, PAC currents were dramatically increased to and sustained at a macroscopic current level during the application of diC8-PI(4,5)P2. These results demonstrate that the charge–charge coupling of TM2 C-terminal basic residues with the inner leaflet PI(4,5)P2 phosphate groups is integral for converting inactive PAC channel to an activated structure through proton binding.
It was recently reported that PI(4,5)P2 in the plasma membrane’s outer leaflet inhibits PAC channel gating12. They observed that the application of soluble PI(4,5)P2 from extracellular side inhibits PAC channel activity and the degree of PI(4,5)P2-mediated PAC inhibition correlates with channel desensitization. Thus, the prevalence of desensitized PAC state facilitates the inhibitory effect of PI(4,5)P2 on PAC. Structural analysis further revealed that the PIP2 binding site in desensitized PAC is located on the extracellular side of the TMD12. In contrast, our results unveil that PI(4,5)P2 in the inner leaf is more important in the activation of PAC from the resting state through the interaction with the cytosolic side of TM2. Therefore, depending on its location, PI(4,5)P2 may have dual effects on PAC channel: Inner leaflet PI(4,5)P2 favors increasing the open probability by stabilizing the open state of PAC, while outer leaflet PI(4,5)P2 achieves inhibitory control by altering the free energy landscape to favor the desensitized state from open state of the PAC channel.
Our simulation study indicated that alanine mutations of K394, R396, or K397 residues along the membrane–cytoplasm interface of the hPAC1 channel TM2 domain selectively decreased current amplitude and PI(4,5)P2 regulation. This observation suggests that basic PAC channel residues interact with PI(4,5)P2 in the inner leaflet and that GPCR-mediated PI(4,5)P2 dynamics can modulate PAC channel gating. In addition, the putative PI(4,5)P2-interacting residues were conserved in hPAC2. The transient decrease in the outer leaflet PI(4,5)P2 level induced by flipping to the inner bilayer after depletion in the inner membrane PI(4,5)P2 may be not the cause of PAC channel suppression because the decrease in the outer leaflet PI(4,5)P2 level should result in the elevation of PAC current amplitudes12. Therefore, consistent with other channels, hPAC1 and hPAC2 are regulated by inner leaflet PI(4,5)P2 levels via basic residue coupling at the plasma membrane–cytoplasm interface17,26,35,36,37.
According to the NCBI reference sequences (RefSeq) database38, there are four isoforms in the PAC channel via alternative splicing. hPAC1 is the longest isoform with an internal exon segment 1 in the N-terminus (Supplementary Fig. 1). In contrast, hPAC2 lacks exon segment 1 but has an intact endosome-targeting signal sequence in the N-terminus (Supplementary Fig. 3). Herein, we verified that hPAC1 channels are highly distributed through the plasma membrane because exon segment 1 is inserted into the endosome-targeting signal sequence, while many hPAC2 channels are recruited to endomembranes. However, our results suggest that many hPAC2 channel proteins are expressed at the plasma membrane and generate PAC2 currents. Thus far, PAC channel studies were primarily accomplished with PAC210,12,39. Furthermore, we found that HEK293T and SH-SY5Y cells only express PAC2 channels. Nevertheless, our results display potent current generation in cells expressing hPAC1 channels. These results demonstrate that, though PAC1 channel is a less common splice variant in the tissues, it may detect acid stimuli and trigger downstream cell death signaling like PAC2.
The recent identification of the PAC channel’s cryo-EM structure has provided valuable insights into conformational changes under different pH conditions. These structures include a resting conformation at pH 7.4 and activated and desensitized conformations at pH 4.510. The PAC2 structure at pH 4.0 indicates a desensitized state8. In our study, experiments at pH 5.5, where the channel exhibited less desensitization, enabled us to compare the PI(4,5)P2 docking simulation on the PAC channel between resting and activated conformations (Fig. 4). The activated PAC channel conformation is characterized by the striking TM1 and TM2 structural rearrangement creating the channel pore. TM2 shifts approximately 10 Å toward the extracellular side relative to the TM1 of the same subunit. This TM2 rearrangement relocated the three basic amino acids into the membrane–cytoplasm interface, resulting in a new interaction with PI(4,5)P2 in the plasma membrane’s inner leaflet. The activated PAC channels slowly shift into the inactive desensitized form39. Based on our simulation, two basic residues, R335 and K336, in the hPAC2 TM2 were coupled with the phosphate groups of PI(4,5)P2 in the activated state. During the mutagenesis experiments, the mutation of K333 to alanine also attenuated the current amplitude (Supplementary Fig. 7). This may be caused by the structural change of the adjacent putative PI(4,5)P2 binding sites R335 and K336 in the K333A mutant form.
Among the five muscarinic receptor subtypes (M1R to M5R), M1R, M3R, and M5R are predominantly coupled with Gq/11 proteins, activating PLCβ to hydrolyze PI(4,5)P2 into DAG and IP340. IP3 increases intracellular Ca2+ by releasing Ca2+ from the endoplasmic reticulum (ER) and elevated DAG and Ca2+ activate protein kinase C (PKC) enzymes. However, the Capurro group reported that neither intracellular Ca2+ nor phosphorylation by protein kinases significantly modulate PAC channels11. This observation implies that PAC current suppression by muscarinic receptors is independent of intracellular Ca2+ and PKC phosphorylation. We confirmed that M1R and M3R muscarinic PAC channel modulation is due to PI(4,5)P2 hydrolysis by activating PLCβ in the signaling pathway. Several diseases, including brain nervous injury, ischemic stroke, seizure, and hyperglycemia, can cause acidosis as low as an extracellular pH 6.041,42,43. Cell death was significantly reduced when PAC channel activity was blocked by the Cl- channel blocker 4,4’-diisothiocyano-2,2’-stilbenedisulfonic acid (DIDS) or PAC channel KO2,44. In conclusion, our study highlights PI(4,5)P2 as a crucial regulator of PAC channel activity and provides a mechanistic understanding of the pathophysiological implications in phosphoinositide PAC channel regulation.
Methods
Cell culture and transfection
HEK293T (large T antigen transformed tsA201 cells) and SH-SY5Y cells were obtained from Bertil Hille (University of Washington School of Medicine, Seattle, Washington) and the Korean Cell Line Bank (Seoul National University, Seoul), respectively. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone, ThermoFisher Scientific) with 10% fetal bovine serum (HyClone, ThermoFisher Scientific) and 0.2% penicillin/streptomycin (HyClone, ThermoFisher Scientific) at 37°C with 5% CO2. PAC knockout (KO) and SH-SY5Y cells were transiently transfected using Lipofectamine 2000 (Invitrogen) with various cDNAs. For expression, cells were transiently transfected with plasmids (e.g., 200 ng human PAC1-GFP and human PAC2-GFP, 50 ng Lyn-mCh, 1000 ng Dr-VSP, 800 ng PIPKIγ, 300 ng of RF-Dead, Sac1, INPP5E or PJ, and 300 ng LDR, 1000 ng M1R) per 35-mm plate when reaching 50–60% confluency. Transfected cells were plated onto poly-L-lysine (0.1 mg/ml, Sigma)-coated chips and used for voltage-clamp recordings and imaging experiments 24 to 36 h after transfection.
Knockout cell line generation
To disrupt PAC gene expression in HEK293T cells, we adhered to the CRISPR/Cas9-mediated gene deletion methods of Yang’s study2. sgRNAs were cloned into the pSpCas9(BB)-2A-GFP vector (PX458, obtained from Addgene plasmid #48138). The Cas9 vector-sgRNA all-in-one vector was transfected into HEK293T cells. Cells expressing green fluorescence were sorted by FACSAria III and seeded onto 96-well plates. After culturing cells until sphere formation for 2–3 weeks, cells were scaled sequentially in 6-well plates and 100-mm cell culture dishes. Genomic DNA was isolated from cells, and frameshift mutations were confirmed through sequencing (Macrogen).
Plasmids and chemicals
Dr-VSP was obtained from Yasushi Okamura (Osaka University, Osaka, Japan). The rapamycin-inducible dimerization system RF-Dead, RF-Sac1, RF-INPP5E, RF-PJ, and Lyn11-FRB (LDR) were provided by Takanari Inoue (Johns Hopkins University, Baltimore, Maryland) and Gerald R. Hammond (University of Pittsburgh School of Medicine, Pittsburgh, PA). PHPLCδ1-GFP was supplied by Ken Mackie (University of Washington, Seattle, WA). PIPKIγ was provided by Yoshikatsu Aikawa and Thomas F. Martin (University of Wisconsin, Madison, WI). Full-length PTEN conjugated with the VSD of Ci-VSP (Ci-VSPTEN) was provided by Christian Halaszovich (University of Marburg, Marburg, Germany). PH-Btk originated from Tamas Balla (NIH, Bethesda) was subcloned into pECFP-C1 and pEYFP-C1 vectors by Dongil Keum (DGIST, Daegu, Korea). The following compounds were similarly obtained: Oxotremorine M, Na2ATP, CaCl2, and other chemicals (Sigma-Aldrich).
Constructs and molecular cloning
The cDNA of human PAC2 was acquired from HEK293T cells. Forward primer and reverse primer targeted the sequences encoding human PAC: forward primer: 5’-CTC GAG ATG ATC CGG CAG GAG CGC TC-3’ and reverse primer: 5’-GAA TTC CGC TTA TGT GGC TCG TTG CCT GAC-3’. Human PAC2 was subcloned into the pEGFP-N1 vector using XhoI and EcoRI restriction sites, and the sequence was identical to NCBI NP_060722.2. Human PAC1 was generated by inserting 61 amino acids in the N-terminal domain (NP_001185791.1). All single amino acid mutations were constructed from WT hPAC1 or hPAC2 through the inverse PAC method using Pfu Turbo DNA polymerase (Agilent Technologies). Plasmid DNA was digested by Dpn I (Agilent Technologies), the PCR product was 5’-phosphorylated by T4 polynucleotide kinase (Enzynomics), and the PCR product was ligated by the T4 DNA ligase (NEB). All mutations were verified through DNA sequencing (Macrogen).
Solutions
The extracellular solution for PAC channel current recording and confocal imaging contained 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 10 mM HEPES, adjusted to pH 7.4 with NaOH. pH 4.6, 5.0, 5.3, 5.5, and 6.3 solutions were adjusted with 10 mM MES. The pipette (intracellular) solution contained 130 mM CsCl, 1 mM MgCl2, 10 mM EGTA, and 3 mM Na2ATP or no added ATP, adjusted to pH 7.35 with CsOH.
Current recording
Patch-clamp recording using the whole-cell configuration was performed at room temperature (22–25°C) using the HEKA EPC-10 amplifier with Pulse software (HEKA Elektronik)45. Electrodes pulled from borosilicate glass micropipette capillaries (Sutter Instrument) by a Flaming/Brown micropipette puller (P-97, Sutter Instrument Co.) had 2–5 MΩ resistances, and series-resistance errors were compensated by >60%. Fast and slow capacitances were compensated for before the test pulse. During recording, the external solutions were delivered to a Quick Change Chamber Narrow Slotted Bath (RC-46SNLP, Warner Instruments) using a Six-channel Pinch Valve Controller System (VC-6, Warner Instruments). Complete solution exchange was achieved within 1 s. Dr-VSP experiments used step depolarizations to 120 mV for 1 s to activate Dr-VSP and deplete PI(4,5)P2 in cells. We used Pulse/Pulse Fit software with an EPC-10 patch clamp amplifier (HEKA Elektronik) and Igor Pro (WaveMetrics, Inc.) to acquire and analyze data.
Inside-out patch-clamp recordings
Inside-out patch-clamp recordings were conducted on PAC knockout HEK293T cells that were transiently transfected with 1.0 μg of hPAC1-GFP/pEGFP-N1 or hPAC2-GFP/pEGFP-N1 plasmid DNA using polyethylenimine (PEI) transfection reagent following the manufacturer’s instructions (Polysciences, Warrington, PA). After 8 h post-transfection, the cells were transferred to the poly-L-lysine (PLL)-coated glass chips. Electrophysiological recordings were conducted 24 to 30 h post-transfection. Patch pipettes were made from borosilicate glass (World Precision Instruments) using a P-1000 micropipette puller (Sutter Instruments) and then fire-polished with an MF-900 microforge (Narishige) to achieve pipette resistances ranging from 3 to 5 MΩ. Inside-out patch clamp experiments were carried out on cells showing green fluorescence utilizing a MultiClamp 700B amplifier and an AxonTM Digidata® 1550B digitizer (Molecular Devices, San Jose, CA). The currents were sampled at 10 kHz and low-pass-filtered at 4 kHz with a four-pole Bessel filter.
The bath (internal) solution consisted of 130 mM CsCl, 1 mM MgCl2, and 10 mM HEPES, adjusted to pH 7.4 with CsOH. The pipette (external) solution contained 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 10 mM HEPES, adjusted to pH 7.4 with NaOH, or 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 10 mM MES, pH 5.5 with NaOH. For cation replacement experiment, the NMDG-internal solution contained 130 mM NMDG-Cl, 10 mM EGTA, and 10 mM HEPES, adjusted to pH 7.4 with NMDG. To evaluate voltage-dependent single-channel activities of PAC1 and PAC2 channels, 1-minute voltage steps ranging from −100 mV to +100 mV in 25 mV increments were applied from the holding potential at 0 mV for 0.5 seconds. To study the effect of PI(4,5)P2 to the excised inside-out patches of PAC-expressed cells, the bath solution with or without 10 μM diC8-PI(4,5)P2 (Echelon Biosciences, Salt Lake City, UT) was perfused to the internal side of membrane patches through a multi-barrel manifold tip (World Precision Instruments, Sarasota, FL). To record the hPAC1 activities in response to the diC8-PI(4,5)P2 application, a holding potential of 0 mV was applied for 0.5 s, followed by stimulation at +100 mV for 10 s. Single-channel events were analyzed by using Clampfit 11.2 software (Molecular Devices, San Jose, CA) and Origin 9.1 software (OriginLab Corporation, Northampton, MA). For visualizing purposes, the single-channel current traces were digitally filtered at 0.5 (for PAC1 channel) or 0.1 kHz (for PAC2 channel) in Clampfit 11.2 software.
Forster resonance energy transfer (FRET)
FRET data were simultaneously performed with current recordings on the PAC channel’s Dr-VSP or Ci-VSPTEN effects in a single cell. FRET signals were acquired and calculated in real-time by a homemade program as previously described46. The FRET calculated ratio (cFactor = CFP/YFP = 0.45) was used to adjust the raw YFP emission signal. FRETr was calculated as follows:
Confocal imaging
HEK293T and SH-SY5Y cells were imaged 1–2 days following transfection on poly-L-lysine coated chips with a Carl Zeiss LSM 700 or LSM 800 confocal microscope (Carl Zeiss) at room temperature. Using digital zoom, cell images were scanned using a 40× (water) objective lens at 1024 × 1024 pixels. 512 × 512 pixels were used for time-course experiments, and image processing was carried out using Zeiss ZEN 2.3 SP1 software. Cytosolic fluorescence intensity during the time-course experiments was assessed by forming relative values from regions of interest drawn in the cytoplasmic region. PHPLCδ1 was normalized to minimum and maximum intensities. To analyze colocalization, we performed quantitative colocalization analysis using Fiji software47 with the Just Another Colocalization plugin (JACoP) to determine Pearson’s correlation coefficient48. Coefficient R values “1” and “0” correspond to perfect colocalization and random (no) colocalization, respectively. Pixel intensities were presented as 2D intensity histograms with a linear regression line and bar graphs with mean R values. All images were transferred from LSM4 to a JPEG format. Raw data were processed with Excel 2019 (Microsoft) and Igor Pro (WaveMetrics, Inc.).
PAC structures predicted with I-TASSER
Molecular modeling was executed using PAC2’s cryo-EM structure as template10 from Protein Data Bank (PDB; http://www.rcsb.org/pdb) to predict PAC2’s three-dimensional (3D) structure in a resting conformation. We submitted the entire sequence of human PAC2 (NP_001185791.1) and the predicted PAC2 as models to the I-TASSER online server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/)49,50. Cryo-EM and simulated PAC2 were superimposed by MatchMaker of the University of California, San Francisco (UCSF) Chimera. Structure visualization and modifications were made using the UCSF Chimera.
Molecular docking in UCSF Chimera
Autodock Vina51 was used for the docking simulation of PI(4,5)P2 to PAC2’s activated and desensitized conformations based on the PDB ID 7SQF and 7SQH crystal structure of PAC2. We used a simulated PAC2 structural model using I-TASSER for the resting conformation. We extracted the PI(4,5)P2 molecule from the crystalized GIRK channel structure (PDB ID 3SYA)52. For docking simulation, water molecules were removed, hydrogens were added to the PAC2 structure, and PI(4,5)P2 was extracted during the “Dock prep” step in Chimera. Then, files were generated in a mol2 format (i.e., PAC2 activated.mol2) to initiate Autodock Vina. Docking simulation could begin after setting up the grid box’s size and position on putative PI(4,5)P2-binding residues. When Autodock Vina was completed, docking results were derived with a root-mean-square deviation (RMSD). Based on docking outcomes, we verified and visualized the hydrogen bonding between the PAC2 channel and PI(4,5)P2 using the FindHBond program in UCSF Chimera.
RNA extraction and RT-PCR
Total RNA was extracted from HEK293T or SH-SY5Y cell lines. Cells collected in Dulbecco’s phosphate-buffered saline (DPBS) were lysed by RNAgents denaturing solution (Promega) after centrifuging and removing the supernatant. Then, the aqueous phase separated by centrifuging was obtained after adding a 2 M sodium acetate and phenol:chloroform:isoamyl alcohol mixture (Sigma-Aldrich). RNA was precipitated through isopropanol of the same volume as the aqueous phase. Total RNA pellets were dried and suspended in RNAse-free water. The total RNA concentration purity was quantified with a NanoDrop 2000 spectrophotometer (Thermo Scientific) using the A260/280 absorbance ratio. RNA was reverse-transcribed to cDNA using the TOPscript cDNA synthesis kit (Enzynomics) for 1 h at 50°C and 5 min at 92°C. β-actin as the control, PAC1, and PAC2 were amplified using forward and reverse primers (Supplementary Table 1). The PCR products were separated through 1% agarose gel electrophoresis. Gel photographs were taken via ChemiDoc XRS+ (Bio-Rad) and documented using Image lab software 6.1 (Bio-Rad).
Statistical analysis
All data were analyzed using Microsoft Office Excel 2019 (Microsoft), IGOR Pro 6.37 (WaveMetrics, Inc.), or GraphPad Prism 7.02 (GraphPad Software). Statistics in text or figures represent mean ± SEM. Statistical comparisons between groups were analyzed using Student’s t-test. The significance of observations among more than two groups was assessed through One-way ANOVA followed by Sidak’s and Dunnett’s post-hoc test. Differences were considered significant at the *p < 0.05, **p < 0.01, and ***p < 0.001 levels.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The source data underlying Fig. 1c, d, f, i; 2c, e, f; 3b, c; 4d, e, g; 6b, d-e, h, j; Supplementary Figs. 3c, e; 4b; 5b, d; 6b, d; 7c; 9d are provided as a Source Data file. Structure factors for cryo-EM TMEM16A (PDB ID: 5oyb), GIRK channel (PDB ID: 3SYA), and Kir2.2 channel (PDB ID: 3SPI) were deposited in the Protein Data Bank. All data supporting the findings of this study are also available from the corresponding author upon request. Source data are provided with this paper.
References
Auzanneau, C., Thoreau, V., Kitzis, A. & Becq, F. A novel voltage-dependent chloride current activated by extracellular acidic pH in cultured rat Sertoli cells. J. Biol. Chem. 278, 19230–19236 (2003).
Yang, J. et al. PAC, an evolutionarily conserved membrane protein, is a proton-activated chloride channel. Science 364, 395–399 (2019).
Ullrich, F. et al. Identification of TMEM206 proteins as pore of PAORAC/ASOR acid-sensitive chloride channels. eLife 8, e49187 (2019).
Ota, T. et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36, 40–45 (2004).
Osei-Owusu, J., Yang, J., Del Carmen Vitery, M., Tian, M. & Qiu, Z. PAC proton-activated chloride channel contributes to acid-induced cell death in primary rat cortical neurons. Channels 14, 53–58 (2020).
Osei-Owusu, J. et al. Proton-activated chloride channel PAC regulates endosomal acidification and transferrin receptor-mediated endocytosis. Cell Rep. 34, (2021).
Zeziulia, M., Blin, S., Schmitt, F. W., Lehmann, M. & Jentsch, T. J. Proton-gated anion transport governs macropinosome shrinkage. Nat. Cell Biol. 24, 885–895 (2022).
Ruan, Z., Osei-Owusu, J., Du, J., Qiu, Z. & Lü, W. Structures and pH-sensing mechanism of the proton-activated chloride channel. Nature 588, 350–354 (2020).
Deng, Z. et al. Cryo-EM structure of a proton-activated chloride channel TMEM206. Sci. Adv. 7, eabe5983 (2021).
Wang, C., Polovitskaya, M. M., Delgado, B. D., Jentsch, T. J. & Long, S. B. Gating choreography and mechanism of the human proton-activated chloride channel ASOR. Sci. Adv. 8, eabm3942 (2022).
Capurro, V. et al. Functional analysis of acid-activated Cl− channels: properties and mechanisms of regulation. BBA-Biomembranes 1848, 105–114 (2015).
Mihaljević, L., Ruan, Z., Osei-Owusu, J., Lü, W. & Qiu, Z. Inhibition of the proton-activated chloride channel PAC by PIP2. eLife 12, e83935 (2023).
Balasubramanian, K. & Schroit, A. J. Aminophospholipid asymmetry: a matter of life and death. Annu. Rev. Physiol. 65, 701–734 (2003).
van Meer, G., Voelker, D. R. & Feigenson, G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008).
Lacroix, J. et al. Controlling the activity of a phosphatase and tensin homolog (PTEN) by membrane potential. J. Biol. Chem. 286, 17945–17953 (2011).
Hansen, S. B. Lipid agonism: The PIP2 paradigm of ligand-gated ion channels. BBA-Mol. Cell Biol. Lipids 1851, 620–628 (2015).
Le, S. C., Jia, Z., Chen, J. & Yang, H. Molecular basis of PIP2-dependent regulation of the Ca2+-activated chloride channel TMEM16A. Nat. Commun. 10, 1–12 (2019).
Oude Weernink, P. A., Han, L., Jakobs, K. H. & Schmidt, M. Dynamic phospholipid signaling by G protein-coupled receptors. BBA-Biomembranes 1768, 888–900 (2007).
Hille, B., Dickson, E. J., Kruse, M., Vivas, O. & Suh, B. C. Phosphoinositides regulate ion channels. BBA-Mol. Cell Biol. Lipids 1851, 844–856 (2015).
Duncan, A. L., Song, W. & Sansom, M. S. P. Lipid-dependent regulation of ion channels and G protein–coupled receptors: insights from structures and simulations. Annu. Rev. Pharmacol. Toxicol. 60, 31–50 (2020).
Murata, Y., Iwasaki, H., Sasaki, M., Inaba, K. & Okamura, Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 435, 1239–1243 (2005).
Okamura, Y., Murata, Y. & Iwasaki, H. Voltage-sensing phosphatase: actions and potentials. J. Physiol. 587, 513–520 (2009).
Ko, W. & Suh, B. C. Differential regulation of Ca2+-activated Cl− channel TMEM16A splice variants by membrane PI(4,5)P2. Int. J. Mol. Sci. 22, 4088 (2021).
Banaszynski, L. A., Liu, C. W. & Wandless, T. J. Characterization of the FKBP·Rapamycin·FRB ternary complex. J. Am. Chem. Soc. 127, 4715–4721 (2005).
Lambert, S. & Oberwinkler, J. Characterization of a proton-activated, outwardly rectifying anion channel. J. Physiol. 567, 191–213 (2005).
Gao, S., Yao, X. & Yan, N. Structure of human Cav2.2 channel blocked by the painkiller ziconotide. Nature 596, 143–147 (2021).
Zinchuk, V., Wu, Y. & Grossenbacher-Zinchuk, O. Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci. Rep. 3, 1365 (2013).
Ko, W. et al. Allosteric modulation of alternatively spliced Ca2+-activated Cl− channels TMEM16A by PI(4,5)P2 and CaMKII. Proc. Natl. Acad. Sci. USA 117, 30787–30798 (2020).
Anthony, B. L. & Aronstam, R. S. Effect of pH on muscarinic acetylcholine receptors from rat brainstem. J. Neurochem. 46, 556–561 (1986).
Anthony, B. L. & Aronstam, R. S. High-affinity agonist binding to rat brainstem muscarinic receptors is eliminated by low pH. Neurosci. Lett. 69, 84–88 (1986).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Thul, P. J. et al. A subcellular map of the human proteome. Science 356, eaal3321 (2017).
Karlsson, M. et al. A single–cell type transcriptomics map of human tissues. Sci. Adv. 7, eabh2169 (2021).
Willars, G. B. & Nahorski, S. R. Quantitative comparisons of muscarinic and bradykinin receptor-mediated Ins(1,4,5)P3 accumulation and Ca2+ signalling in human neuroblastoma cells. Br. J. Pharmacol. 114, 1133–1142 (1995).
Yu, K., Jiang, T., Cui, Y., Tajkhorshid, E. & Hartzell, H. C. A network of phosphatidylinositol 4,5-bisphosphate binding sites regulates gating of the Ca2+-activated Cl− channel ANO1 (TMEM16A). Proc. Natl. Acad. Sci. USA 116, 19952–19962 (2019).
Pant, S. et al. PIP2-dependent coupling of voltage sensor and pore domains in Kv7.2 channel. Commun. Biol. 4, 1–14 (2021).
Damian, M. et al. Allosteric modulation of ghrelin receptor signaling by lipids. Nat. Commun. 12, 3938 (2021).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 (2007).
Osei-Owusu, J. et al. Molecular mechanism underlying desensitization of the proton-activated chloride channel PAC. eLife 11, e82955 (2022).
Maeda, S., Qu, Q., Robertson, M. J., Skiniotis, G. & Kobilka, B. K. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019).
Smith, M.-L., von Hanwehr, R. & Siesjö, B. K. Changes in extra- and intracellular pH in the brain during and following ischemia in hyperglycemic and in moderately hypoglycemic rats. J. Cereb. Blood Flow Metab. 6, 574–583 (1986).
Nemoto, E. M. & Frinak, S. Brain tissue pH after global brain ischemia and barbiturate loading in rats. Stroke 12, 77–82 (1981).
Zha, X., Xiong, Z. G. & Simon, R. P. pH and proton-sensitive receptors in brain ischemia. J. Cereb. Blood Flow Metab. 42, 1349–1363 (2022).
Cai, R., Tang, J. & Chen, X. Z. Ion permeation controlled by hydrophobic residues and proton binding in the proton-activated chloride channel. iScience 24, 103395 (2021).
Keum, D., Baek, C., Kim, D. I., Kweon, H. J. & Suh, B. C. Voltage-dependent regulation of CaV2.2 channels by Gq-coupled receptor is facilitated by membrane-localized β subunit. J. Gen. Physiol. 144, 297–309 (2014).
Keum, D., Kruse, M., Kim, D. I., Hille, B. & Suh, B. C. Phosphoinositide 5- and 3-phosphatase activities of a voltage-sensing phosphatase in living cells show identical voltage dependence. Proc. Natl. Acad. Sci. USA 113, E3686–E3695 (2016).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Dunn, K. W., Kamocka, M. M. & McDonald, J. H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol.-Cell Physiol. 300, C723–C742 (2011).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Whorton, M. R. & MacKinnon, R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell 147, 199–208 (2011).
Acknowledgements
We thank the laboratories for kindly providing plasmids. This research was supported by grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1A6A3A01086672 to W.K) and the Ministry of Science, Information and Communications Technology, and Future Planning (2022R1A2C1006560 and RS-2024-00351000 to B.C.S, 2021R1A2C1004884 to H.H.L.), and the KBRI basic research program through Korea Brain Research Institute (24-BR-01-02 to H.H.L.).
Author information
Authors and Affiliations
Contributions
B.C.S. and W.K. conceptualized the project and designed the experiments; W.R. and J.E.K. performed the whole-cell patch and confocal experiments and analyzed the results; E.L. and H.H.L. performed the inside-out patch experiments. W.K. and H.H.L. wrote the original draft. B.C.S. supervised the project and wrote the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Peer review
Peer review information
Nature Communications thanks Zhaozhu Qiu, Huanghe Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ko, W., Lee, E., Kim, JE. et al. The plasma membrane inner leaflet PI(4,5)P2 is essential for the activation of proton-activated chloride channels. Nat Commun 15, 7008 (2024). https://doi.org/10.1038/s41467-024-51400-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51400-y
- Springer Nature Limited