Abstract
DNA polymerase theta (Polθ) is a DNA helicase-polymerase protein that facilitates DNA repair and is synthetic lethal with homology-directed repair (HDR) factors. Thus, Polθ is a promising precision oncology drug-target in HDR-deficient cancers. Here, we characterize the binding and mechanism of action of a Polθ helicase (Polθ-hel) small-molecule inhibitor (AB25583) using cryo-EM. AB25583 exhibits 6 nM IC50 against Polθ-hel, selectively kills BRCA1/2-deficient cells, and acts synergistically with olaparib in cancer cells harboring pathogenic BRCA1/2 mutations. Cryo-EM uncovers predominantly dimeric Polθ-hel:AB25583 complex structures at 3.0-3.2 Å. The structures reveal a binding-pocket deep inside the helicase central-channel, which underscores the high specificity and potency of AB25583. The cryo-EM structures in conjunction with biochemical data indicate that AB25583 inhibits the ATPase activity of Polθ-hel helicase via an allosteric mechanism. These detailed structural data and insights about AB25583 inhibition pave the way for accelerating drug development targeting Polθ-hel in HDR-deficient cancers.
Similar content being viewed by others
Introduction
Mutations in homology-directed repair (HDR) genes, such as BRCA1/2, strongly predispose women to breast and ovarian cancer, and BRCA1/2 (BRCA) mutations are also observed in prostate and pancreatic cancers1,2. Since BRCA-deficient cancer cells are impaired in HDR, they are highly susceptible to DNA damage compared to normal cells2,3,4. Drugs that cause DNA damage and/or inhibit DNA repair, such as Poly (ADP-ribose) polymerase 1 (PARP1) inhibitors, can therefore cause synthetic lethality in BRCA-deficient cells, while sparing normal cells2,3,4. Currently, PARPi have been approved to treat HDR-deficient breast, ovarian, prostate, and pancreatic cancers5,6,7,8. However, a large fraction of patients fail to respond to PARPi and drug resistance is a major problem3,5,9,10,11,12,13. Thus, the development of next-generation precision medicines that selectively kill HDR-deficient cells and suppress PARPi resistance is essential for increasing patient survival rates and ultimately eradicating aggressive and refractory HDR-deficient cancers.
Studies performed in 2015 identified DNA polymerase theta (Polθ) as a potential drug target in HDR-deficient cancers due to its synthetic lethal interaction with BRCA1 and BRCA214,15. Polθ is a large (290 kDa) multi-functional protein containing an N-terminal superfamily 2 (SF2) helicase (Polθ-hel), an unstructured central domain, and a C-terminal A-family polymerase domain (Polθ-pol) which is structurally similar to Klenow fragment and Taq Pol16,17,18,19,20,21,22. Polθ is upregulated in breast tumors and ovarian cancers15,23,24,25,26, and its overexpression correlates with HDR defects and a poor clinical outcome15,23,24,27. Polθ also confers resistance to radiation therapy and genotoxic cancer drugs (e.g., topoisomerase inhibitors, cisplatin), including PARPi28,29,30,31,32.
Polθ promotes double-strand break (DSB) repair via microhomology-mediated end-joining (MMEJ)—also referred to as alternative end-joining and theta-mediated end-joining (TMEJ)14,22,29,33. For example, Polθ-pol facilitates MMEJ of DNA with 3′ single-strand DNA (ssDNA) overhangs containing short tracts (2–6 bp) of microhomology in vitro and suppression of Polθ significantly reduces MMEJ in cells14,21,22,29,34. Inactivation of the ATPase activity of Polθ via site-specific genetic engineering also significantly reduces MMEJ in cells18.
The SF2 DNA helicase domain of Polθ (Polθ-hel) is known to bind many types of DNA substrates, and its ATPase activity is strongly stimulated by ssDNA19. The helicase possesses relatively weak ATP-dependent DNA unwinding in a 3′−5′ direction and promotes ATP-independent ssDNA annealing, similar to some RecQ type helicases17,18. Despite these advances in our understanding of the biochemical activities of Polθ-hel, how this domain contributes to MMEJ remains unclear. Prior biochemical studies found that the ATPase activity of full-length Polθ is dispensable for MMEJ in vitro21. Yet, studies in mammalian cells suggest that Polθ’s ATPase activity promotes MMEJ by dissociating replication protein A (RPA) from 3′ ssDNA overhangs18. The ATPase function of Polθ has also been implicated in removing RAD51:ssDNA nucleoprotein filaments31. However, more recent studies found that Polθ-hel exhibited relatively poor dissociation of RAD51:ssDNA filaments, but instead showed that the helicase displayed efficient and processive ATP-dependent dissociation of RPA from ssDNA, which confirmed prior findings18,35. A recent report supports Polθ-hel displacement of RPA during ssDNA gap repair36.
Although Polθ dependent MMEJ is induced in response to DNA damage caused during the S-phase, DSB repair is largely performed by HDR in S/G2 cell cycle phases, which relies on BRCA1, BRCA2, PALB2, and many other HDR-associated proteins1,37,38,39. Since Polθ-dependent MMEJ and ssDNA gap repair are thought to serve as backup repair pathways for HDR, the synthetic lethal interaction between Polθ and HDR factors is likely due to Polθ’s involvement in MMEJ and ssDNA gap repair36,40. Regardless of the specific functions for Polθ in HDR-deficient cells, suppression or knockout of POLQ in BRCA1 mutant (BRCA1-mut) and BRCA2 mutant (BRCA2-mut) cancer cells causes synthetic lethality14,31,41. In contrast, suppression of POLQ in BRCA wild-type (BRCA-WT) cells has no effect14,31,41. BRCA-deficient cancer cells were also shown to be dependent on Polθ expression for their survival in the presence of genotoxic agents15,31.
Intriguingly, the DNA synthesis and ATPase activities of Polθ were separately shown to promote the survival of Brca1-deficient mouse embryonic stem cells18,31, which suggested that pharmacological inhibition of either Polθ enzymatic domain would selectively kill BRCA-deficient cancer cells. For example, respective inactivation of Polθ-hel and Polθ-pol enzymatic domains via site-specific CRISPR/Cas9 mutagenesis in Braca1-deficient mouse embryonic stem cells significantly reduced colony formation18. CRISPR/Cas9 knockout studies in mouse embryonic fibroblasts (MEFs) also showed that Polθ is synthetic lethal with other DNA repair factors (i.e., Rad54, Ku70/80, and Fancj)34,42.
Recent studies have demonstrated the potential of Polθ inhibitors (Polθi) in BRCA-deficient cells and tumor models. One study repurposed the antibiotic Novobiocin as a Polθ-hel inhibitor43. Although the antibiotic showed the expected activity against HDR-deficient PDX models, Novobiocin was reported to exhibit a relatively high IC50 (~25 μM) against Polθ-hel43. Thus, Novobiocin may exhibit off-target effects at high concentrations44,45,46,47. Another report revealed a potent allosteric inhibitor class of Polθ-pol48. This compound class showed selective killing of BRCA-deficient cells, induction of the DNA damage response (DDR) specifically in BRCA-deficient cells, and the ability to overcome PARPi resistance in BRCA1-mut cells harboring engineered knockouts of the Shieldin complex, which protects DNA ends and enables non-homologous recombination (NHEJ)48. A more recent report revealed a related potent Polθ-pol allosteric inhibitor class that showed selective killing of BRCA2-null HCT 116 cells49. These latter reports revealed successful medicinal chemistry campaigns targeting the polymerase domain with single-digit nanomolar IC50 compounds. Whether Polθ-hel inhibitors can be developed with similar low nanomolar IC50 remains unclear.
Here, we employed cryo-EM structural determination methods as a tool for understanding the mechanism of inhibition of a previously unreported Polθ-hel small-molecule inhibitor, AB25583. AB25583 exhibits 6 nM IC50 against Polθ-hel and selectively kills BRCA-deficient cells. Utilizing single-particle cryo-EM, we characterized the inhibitor binding site and the mechanism of action of AB25583. The structural studies reveal a binding pocket deep within the helicase central channel, which explains the high specificity and potency of AB25583. The structures, along with biochemical data, indicate that AB25583 acts as an allosteric inhibitor by perturbing ATP-triggered conformational switches of the helicase. Unexpectedly, Polθ-hel dimers were primarily observed, which provides potential insights into how Polθ-hel functions during MMEJ. The high-resolution cryo-EM Polθ-hel: AB25583 complex structures elucidate a previously undescribed small-molecule binding site and are expected to facilitate Polθ-hel inhibitor drug development for BRCA-mutant cancers.
Results
Biochemical and cellular activity of AB25583
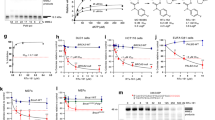
AB25583 was identified and synthesized from a patent application (WO 2020/243459 A1) (Fig. 1a). The IC50 of AB25583 against Polθ-hel was determined using the Promega ADP-Glo assay in triplicate. Here, increasing concentrations of AB25583 were incubated with recombinant human Polθ-hel (residues 1–894) in the presence of ssDNA and 100 μM ATP for 60 min at room temp (Fig. 1b). Next, reactions were subjected to ADP-glo reagents which degrade the remaining ATP, then convert the generated ADP to ATP, and finally quantitation of ATP via an ATP-dependent luminescence reaction was performed. AB25583 exhibited 6 nM IC50, demonstrating the expected potency for a potential drug candidate (Fig. 1c). We next examined the relative selectivity of AB25583 by testing its inhibitory activity against related SF2 helicases, RECQL5, Bloom’s (BLM) helicase, and Werner’s (WRN) helicase, which possess homologous ATPase domains16. The results showed no inhibition of WRN and RECQL5 and very minor inhibition of BLM at the highest concentrations, indicating that AB25583 is relatively selective for Polθ-hel (Fig. 1c). As a comparison, the recently reported repurposed Polθ-hel inhibitor, Novobiocin, exhibited >50 μM IC50 against Polθ-hel (Supplementary Fig. 1a). We examined the IC50 of AB25583 against Polθ-hel in the presence of increasing concentrations of ATP. If AB25583 acted as a competitive inhibitor, it would be expected to lose potency at increasing concentrations of ATP. We observed nearly identical IC50 of AB25583 at multiple concentrations of ATP from 100–600 μM, indicating the small-molecule acts as a non-competitive allosteric inhibitor (Fig. 1d). Additionally, AB25583 did not interfere with Polθ-hel binding to various DNA substrates (Supplementary Fig. 1b–e). Considering that AB25583 exhibits significantly higher potency than Novobiocin, these data characterize AB25583 as a promising scaffold for Polθ-hel drug development.
a Structure of AB25583. b Schematic of Polθ-hel ATPase activity assay. c Scatter plot showing AB25583 inhibition of the indicated SF2 DNA helicases. Data represent the mean of two technical replicates. IC50 of AB25583 against Polθ-hel = 6 nM. d Scatter plot showing AB25583 inhibition of Polθ-hel ATPase activity in the presence of the indicated ATP concentrations. Data represent the mean of two technical replicates. e, f, l, o Statistical significance was measured from a two-sample t-test and P values are indicated. e, f Scatter plot showing % colony survival in the presence of the indicated concentrations of AB25583. Data represent the mean of three biological replicates. n = 3, ±s.e.m; P = 0.002507 for 0.5 μM and P = 0.000044 for 1 μM in HCT pair; P = 0.00278 for 0.5 μM and P = 0.000245 for 1 μM in DLD1 pair. g Structure of AB25595. h Scatter plot showing AB25595 inhibition of Polq-hel. Data represent the mean of three biological replicates. n = 3, ±s.d. i–l Scatter plot showing % colony survival of the indicated cell lines in the presence of the indicated concentrations of AB25595 (i) or AB25583 (j–l). i, k, l Data represent the mean of three biological replicates. n = 3, ±s.e.m; P = 0.00185 for 10 uM for Polq −/− vs sgGFP. j Data represent the mean of two biological replicates. n = 2, ±s.e.m. m Bar plot showing % nuclei with >5 gH2AX foci following treatment with the indicated concentrations of AB25583 (right). Data represent the mean of three biological replicates. n = 3, ±s.e.m. Representative images of gH2AX immunofluorescence following DMSO and AB25583 treatment Magnification 40x; Scale bar, 10 um (left). P = 0.00303 for 5 μM and P < 0.00001 for 10 μM in BRCA2-KO. n Bar plot showing % nuclei with >5 RAD51 foci following DMSO and AB25583 treatment. Data represent the mean of three biological replicates. n = 3, ±s.d. P = 0.019585 for IR treated, DMSO vs AB25583. o Bar plot showing quantitation of MMEJ in U2OS cells indicated by % of GFP/dsRED following DMSO and 20 μM AB25583 treatment. Data represent the mean of three biological replicates. n = 3, ±s.d. P < 0.0001 for DMSO vs 20 μM AB25583. Source data are provided as a Source data file.
Considering that genetic inactivation of Polθ’s ATPase domain was previously shown to significantly reduce MMEJ and the survival of Brca1-deficient mouse embryonic stem cells18, we investigated the effects of AB25583 on the survival of BRCA-deficient cells. We probed AB25583 activity against the DLD1 BRCA2-wild-type (WT) and BRCA2 knockout (KO) isogenic cell pair via colony survival assays. The results showed selective killing of the BRCA2-KO cells by AB25583, with little to no effect on the survival of BRCA2-WT cells (Fig. 1e and Supplementary Fig. 1f). Similar results were observed in the HCT 116 BRCA2-WT and BRCA2-KO isogenic cell pair (Fig. 1f). As a comparison, Novobiocin showed selective killing of BRCA2-KO cells at significantly higher concentrations (Supplementary Fig. 1g).
We next investigated the on-target effect of AB25583 by changing the aromatic polar thiadiazole motif to a slightly larger polar pyridazine, which resulted in a closely related compound (AB25595) with >3000-fold lower inhibition potency against Polθ-hel than AB25583 (Fig. 1g, h). As expected, AB25595 showed no selective killing of BRCA2-KO cells (Fig. 1i). Taken together, these data support the on-target activity of AB25583, which exhibits single-digit nanomolar potency against Polθ-hel in vitro, and as a result, robust selective killing of BRCA2-KO cells.
A recent report revealed differential effects of Brca1 mutations on the cellular sensitivity to the Polθ-pol inhibitor ART55850. We, therefore, examined the activity of AB25583 against multiple BRCA1-deficient cell lines. AB25583 demonstrated selective killing of RPE-1 TP53−/−;BRCA1−/− cells (Fig. 1j). AB25583 also significantly reduced the survival of triple-negative breast cancer (TNBC) MDA-MB-436 cells harboring a pathogenic BRCA1 mutation (5396 + 1 G > A), but showed little to no effect against MDA-MB-231 BRCA1 wild-type (WT) TNBC cells (Fig. 1k). The polymerase domain inhibitor ART558 exhibited similar preferential killing of BRCA1-mutant MDA-MB-436 cells (Supplementary Fig. 1h). Two separate studies showed relatively modest activity of ART558 against mouse embryonic fibroblasts (MEFs) harboring a Brca1 Δ11 mutation which impairs Brca1-mediated DNA resection48,50. AB25583 also exhibited modest activity against Brca1Δ11;Δ11 MEFs (Fig. 1l). As a comparison, previously characterized Brca1Δ11;Δ11;Polq−/− MEFs were mostly resistant to AB25583, which further supports the on-target activity of the inhibitor (Fig. 1l)50. AB25583 also exhibited preferential killing of the previously characterized Brca1cc/cc MEFs, which are defective in BRCA1:PALB2 complex interactions (Supplementary Fig. 1j). Novobiocin showed no preferential killing of these cells up to 10 μM as a comparator (Supplementary Fig. 1i). Novobiocin exhibits >50–100 μM IC50 in BRCA1-deficient cells43,51, thus higher concentrations are likely required to observe the selective killing of Brca1cc/cc MEFs. Taken together, AB25583 exhibits preferential killing of BRCA1- and BRCA2- deficient cells. Future comprehensive genetic studies, however, will be required to fully characterize the possible differential effects of various BRCA1/2 mutations on the activity of AB25583.
We next examined the effects of AB25583 on the DNA damage response (DDR). Consistent with the ability of AB25583 to induce synthetic lethality in BRCA2-KO cells by suppressing DNA repair, AB25583 promoted a significant increase in phosphorylation of γH2AX exclusively in BRCA2-KO cells (Fig. 1m). Similar results were observed for the polymerase domain inhibitor ART558 in prior studies48. Suppression of Polθ was previously shown to cause a significant increase in the recruitment of RAD51 to DNA damage induced by ionizing radiation (IR)31. Thus, we envisaged that AB25583 treatment would lead to an increase in RAD51 foci following IR. Indeed, we observed a significant increase in RAD51 foci in IR-exposed cells following AB25583 treatment (Fig. 1n). Considering that Polθ-hel was reported to counter HR and RAD51 foci formation31, these data further support the on-target activity of AB25583. Prior studies showed that site-specific genetic inactivation of Polθ ATPase function significantly reduced MMEJ, which confirmed the involvement of Polθ-hel in end-joining18. Consistent with this, we observed that AB25583 treatment significantly reduced MMEJ using a previously characterized GFP MMEJ reporter (Fig. 1o)39. Taken together, these data demonstrate that the small-molecule inhibitor suppresses MMEJ, induces DNA damage in BRCA-deficient cells, and increases RAD51 foci in cells exposed to IR.
Importantly, knockdown of Polθ or specific inhibition of Polθ-pol has been shown to potentiate the effects of PARPi in HDR-deficient cells31,48. We therefore examined possible synergistic activity between AB25583 and olaparib in BRCA-mutant cancer cell lines. We first tested the combination of AB25583 with olaparib in the ovarian cancer cell line PE01 which has a homozygous BRCA2 mutation (BRCA2.5193 C > G), and a second mutation (BRCA2.5192 A > T) which is thought to cause BRCA2 reactivation52. Although AB25583 exhibited limited activity as a single agent in this BRCA2-mutant cell line, synergistic activity was observed with olaparib (Fig. 2a). We additionally observed synergistic activity between AB25583 and olaparib in BRCA1-mutant MDA-MB-436 TNBC cells (Fig. 2b). These data support further preclinical evaluation of Polθ-hel inhibitors with PARPi for treating HDR-deficient cancers.
a, b Scatter plot showing % colony survival of PE01 cells (a) and MDA-MB-436 cells (b) in the presence of the indicated concentrations of Olaparib and AB25583 or DMSO (left). Data represent the mean of two biological replicates performed in triplicate ±s.e.m. Plots generated by Combenefit software showing synergy between AB25583 and Olaparib (right). Source data are provided as a Source data file.
Dimeric and tetrameric Polθ-hel structures in complex with AB25583
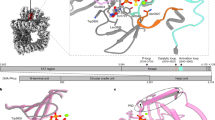
Structure biology is an important method for accelerating drug development and determining the mechanism of action of small-molecule inhibitors. In order to elucidate the binding site and mechanism of action of the Polθ-hel inhibitor, we utilized cryo-EM technology to determine the atomic resolution structure of Polθ-hel bound to AB25583. We resolved the cryo-EM structures of Polθ-hel in complex with AB25583 in two unique oligomeric states: a dimeric form at 3.0 Å and a tetrameric form at 3.2 Å resolution (Fig. 3, Table 1, and Supplementary Figs. 2–5). Predominantly, the dimeric form was observed in Polθ-hel particles, comprising ~95% of the total population (Fig. 3a, b). This was unexpected considering that prior X-ray structures of Polθ-hel were solved as tetramers19. The tetrameric form was observed and classified as a minor species, accounting for about 5% of the total. This cryo-EM data suggests that Polθ-hel can exist as both a dimer and a tetramer in solution, with the dimeric form being significantly more stable and prevalent. The formation of the dimeric interface is mediated through the interactions between two neighboring subdomains D4 (D4–D4 contacts), one of the five subdomains of Polθ-hel structure (Fig. 3c, d), and the tetrameric structure is assembled by two dimers through the same D4 domains, but with different D4–D4 interfaces (Fig. 3b).
a Representative 2D class averages of dimer and tetramer forms of Polθ-hel in complex with AB25583. The dimer population dominates in the Polθ-hel particles occupying about 95% of the particles. b 3D cryo-EM reconstructions of dimer and tetramer forms of Polθ-hel in complex with AB25583 at 3.0 and 3.2 Å resolution, respectively. c Cartoon representation of the five subdomains (D1–D5) Polθ-hel (residues 1–894), each colored in a discrete color that matches the 3D structure in panel-d. D1 (yellow) and D2 (green) are the two tandem RecA-like helicase domains. D3 (orange): winged helix (WH); D4 (magenta): contains the “Rachet helix” for ssDNA translocation. D5 (light blue): contains the helix-loop-helix (HLH). The N-terminal disordered region (residues 1–67) is indicated in a gray line. d Two orthogonal views of the 3D cryo-EM reconstruction map (top) and the atomic structure (bottom) of the Polθ-hel dimer bound to AB25583. AB25583 is drawn in spheres with carbon atoms in cyan. The D4 mediates the dimer formation, and two dimers contact each other via additional but less D4–D4 interactions to form the tetramer.
The protomer structure within both the dimeric and tetrameric forms exhibit high similarity, as evidenced by an average root mean square deviation (r.m.s.d.) of 0.001 when superimposed with each other. When a Polθ-hel monomer is aligned with the crystal structures of apo Polq-hel PDB 5A9J vs. the ADP-bound PDB 5A9F and AMP-PNP-bound PDB 5AGA Polθ-hel forms, the average r.m.s.d. values are 0.96, 1.38, and 1.72, respectively, indicating that the Polθ-hel monomer overlaps better with the apo Polθ-hel structure than to the NTP-bound Polθ-hel structures. The monomeric Polθ-hel structure is comprised of five subdomains D1–D5 (Fig. 3c), which are arranged in a twisted ring shaped-structure, forming a narrow central-channel between D1–D4 subdomains for ssDNA binding and translocation (Fig. 4a). The characteristic RecA-fold motor domain is exhibited by subdomains D1 (yellow) and D2 (green), which together form a functional protein motor with a pocket for ATP-binding and hydrolysis at the interface between the two subdomains (Supplementary Fig. 5). This ATP-binding and hydrolysis mediated by D1 and D2 motor domains stimulates conformational changes of the helicase that are coupled to DNA unwinding and the translocation of ssDNA within the central-channel integrately formed by D1–D4. Subdomain D3 (orange) folds back to interact with D1, generating a helical bundle and extending to the helical domain D4 (magenta), which houses the “ratchet” helix along the central-channel responsible for ssDNA interaction and translocation (Fig. 4a–c). D5 (light blue) forms a smaller helical domain and interacts with D2 and D4 on the outer periphery of the twisted ring structure.
a Structure of a Polθ-hel monomer with bound-AB25583. The five domains (D1–D5, domain colors matching those in Fig. 1c, d) of a Polθ-hel monomer form a twisted ring architecture. The inhibitor AB25583 binds deep inside the Polθ-hel central-channel surrounded by four subdomains (D1, D3, D4, and D5), but interact directly with D1 and D4. b–d Close-up view of the AB25583 (sticks with carbon atoms in cyan) binding pocket. AB25583 binding residues are shown in the sticks. Notably, AB25583 interacts with several residues in the rachet helix of D4 (magenta, panels b, c), and at the same time bridges with multiple residues from D1 (yellow) and D3 (orange). Well-featured electron density around the AB25583 is shown in (c, d). e Detailed interaction between AB25583 and all protein residues from D1 and D3 of Polθ-hel. f The surface electrostatic potential of the AB25583 binding pocket (PDB: 9BP9), showing a generally positively charged groove to accommodate the AB25583. The surface area is colored according to the calculated electrostatic potential from −10.0 kT/e (red) to +10.0 kT/e (blue).
In both the dimeric and tetrameric forms, each Polθ-hel protomer binds to a single AB25583 inhibitor with identical binding interactions. The AB25583 inhibitor is situated deeply within the Polθ-hel central-channel, which is surrounded by subdomains D1, D2, D3, and D4, but the inhibitor interacts bonds tightly with multiple amino acid residues from subdomains D1 and D4, effectively cementing these two subdomains together to freeze the protein conformation in the inhibitor-bound state (Fig. 4a–e).
Detailed interactions of Polθ-hel with AB25583
AB25583 establishes direct interactions with a total of 17 amino acid residues of Polθ-hel, with 10 of these residues coming from D1 and the remaining 7 from D4 (Fig. 4b–e). In addition to its interactions with the side chains, the inhibitor also forms Van der Waals interactions with the main-chain atoms of Polθ-hel. This comprehensive inhibitor–protein interaction allows the inhibitor to nestle snugly deep within the central-channel, effectively obstructing it like a cork to prevent the conformational switch within the central-channel. The interactions between the inhibitor and the protein are a combination of hydrophobic packing with side chains and main-chain atoms, polar interactions, and hydrogen bonds, which include some weaker hydrogen bonds between the -CH of the inhibitor and the oxygen atoms of the protein. Specifically, the chlorobenzene portion interacts typically with E204, K206, and H180 via the chloride, and with Y171, F181, and R200, and L201 side chain via hydrophobic packing (Fig. 4b–e). The thiadiazole ring in the middle portion of AB25583, connected to the chlorobenzene through an ether linkage, fits into a tight space of its binding pocket to pack with the main-chain atoms of G196 and Y197 and interacts with the side chains of two arginines R193 and R200 of D1 (Fig. 4d, e). This tight-space fitting and specific interactions with the aromatic polar thiadiazole motif observed in this structure offer a mechanistic explanation for the >3000-fold lower inhibition potency by changing it in AB25583 to a slightly larger pyridazine in the compound AB25595 (Fig. 1a, g, h). The pyridine and methoxybenzene ring, linked to the thiadiazole via an amide bond, interacts with multiple residues, including G173, S174, S620, S621, S622, V757, G760, M761, and V764 (Fig. 4b, e). This extensive interaction network unveiled by the complex structure aligns with the well-featured electron density of AB25583 and the observed highly potent 6 nM IC50 (Fig. 4c, d). The Polθ-hel binding pocket for AB25583 is characterized by a predominantly positively charged surface mixed with minor neural and negatively charged areas (Fig. 4f).
Molecular mechanism of inhibition of Polθ-hel by AB25583
The structure of Polθ-hel in complex with the inhibitor AB25583 elucidates two potential molecular mechanisms of inhibition. The first mechanism postulates that the binding of AB25583 to Polθ-hel blocks the conformational changes of Polθ-hel that is essential to couple ATP-binding/hydrolysis to the translocation of ssDNA within its channel. As AB25583 is lodged between subdomains D1 and D4 (Fig. 4b, e), the elaborate bonding interactions between AB25583 and both D1 and D4 subdomains are likely to bond the two subdomains together. Such strong bonding of the inhibitor with D1 and D4 subdomains deep inside the central-channel is expected to prevent the conformational switches of the central-channel formed by D1–D4 subdomains. As the conformational changes of D1–D4 is likely required for the cyclic ATP hydrolysis to occur, AB25583 binding to Polθ-hel is expected to inhibit ATP hydrolysis in a non-competitive manner, which is consistent with our experimental observation (Fig. 1d) The second possible inhibition mechanism posits that the binding of AB25583 deep inside the central-channel blocks the ssDNA translocation through the channel (Fig. 5a, b and Supplementary Fig. 6a–d). The pivotal “ratchet” helix of D4 subdomain (Fig. 4b, c and Supplementary Fig. 6d), located along the central-channel, is proposed to bind and translocate ssDNA along the centra-channel. In the superposition between the structures of Polθ-hel/AB25583 structure and its homolog HEL308/DNA (PDB: 2P6R), the ssDNA bound inside the central-channel of HEL308 also runs through the central-channel of Polθ-hel along the rachet helix without significant clash (Supplementary Fig. 6a, d). AB25583 binds deep inside the central-channel to the left end of the rachet helix, which is in contrast to Novobiocin, that binds near the right end of the rachet helix which is the entry point for the ssDNA (Supplementary Fig. 6d, e). This difference in the binding modes of AB25583 and Novobiocin provides a plausible explanation why Novobiocin inhibits Polθ-hel ssDNA binding whereas AB25583 does not (Supplementary Fig. 1b)53. Specifically, Novobiocin binds at the entry point of the central-channel, blocks the binding of the ssDNA to the channel53. In contrast, AB25583 binds deeper inside the central-channel, leaving most part of the channel accessible for DNA substrates to bind (Supplementary Fig. 1b–e).
a DNA-bound model of Polθ-hel dimer. The bound-DNA with 3′-overhang ssDNA (light and dark gray tubes) was modeled from a homolog structure of HEL308 (PDB ID: 2P6R) by superimposition HEL308 to the Polθ-hel:AB25583 structure (see Supplementary Fig. 6a). AB25583 is shown in spheres with carbon atoms in cyan. Duplex DNA is outside the helicase ring channel, and the 3′-overhang ssDNA passes through the central-channel. b Location of AB25583 in a DNA-bound model. AB25583 lies in the path of the 3′-overhang ssDNA, which is predicted to block DNA translocation. c Comparison of AB25583-bound cryo-EM structure (pink, this study) and the inhibitor-free crystal structure of Polθ-hel (blue, PDB ID: 5AGA). The two structures were superimposed based on the D4 subdomain that mediates the dimerization of Polθ-hel. The largest displacement was observed for the D1 subdomain with up to 9 Å shift in the main-chain atoms between the two structures. d The same comparison of the two structures around the AB25583-binding site as in panel-c, but with a zoom-in view around the rachet helix. In the AB25583-bound structure (pink), the R-helix containing two arginines (R193 and R200) in the D1 subdomain shifted toward the ratchet helix in the D4 subdomain, resulting in tighter packing of D1 and D4 subdomains than that in the inhibitor-free structure (blue).
In support of the first inhibitory mechanism in which AB25583 blocks ATPase activity of the helicase via an allosteric mechanism, we demonstrate that AB25583 inhibits Polθ-hel ATPase activity even regardless of the presence or absence of the ssDNA substrate (Supplementary Fig. 1j). Hence, binding of the AB25583 deep within the central-channel prevents the ability of the helicase to effectively hydrolyze ATP, supporting an allosteric inhibitory mechanism. The allosteric mechanism of inhibition is also consistent with the ability of AB25583 to display nearly identical IC50 values in the presence of increasing ATP concentrations (Fig. 1d).
Discussion
Despite our limited understanding of how Polθ-hel functions in MMEJ and other potential DNA repair mechanisms, the results presented herein unequivocally show that Polθ-hel inhibition by a potent and selective small-molecule inhibitor AB25583 exhibits selective killing of BRCA-deficient cells and shows synergistic activity with PARPi olaparib. Hence, these data validate Polθ-hel as an important precision oncology drug target in HDR-deficient cancers. Biochemical assays demonstrate that AB25583 exhibits 6 nM IC50 against Polθ-hel while exhibiting little to no inhibition of other related SF2 helicases, WRN, BLM and RECQL5. We further find that AB25583 selectively kills BRCA-deficient cells which is consistent with the synthetic lethal interaction between Polθ and BRCA1/2 previously reported14,18,31,41,42,48.
Notably, AB25583 only showed modest preferential killing of Brca1 Δ11 MEFs, which are defective in Brca1-mediated DNA end resection. The previously reported inhibitor of the C-terminal Polθ-pol domain, ART558, showed similar moderate activity in Brca1 Δ11 MEFs in two separate studies48,50. Hence, these observations support the idea that particular Brca1 mutations confer differential vulnerabilities to Polθ inhibitors regardless of whether they target the helicase or polymerase domain. Despite the limited activity of AB25583 in Brca1 Δ11 MEFs, the inhibitor showed more robust preferential killing of MDA-MB-436 cells, which are also defective in DNA end resection. Hence, the molecular basis underlying the synthetic lethal relationship between BRCA1 and Polθ may be more nuanced than previously appreciated and warrants further investigation. AB25583 also induced relatively strong synthetic lethality in two different BRCA2-KO cancer cell lines, and the inhibitor showed synergistic activity with olaparib in two different cancer cell lines harboring pathogenic BRCA mutations. Hence, these data characterize AB25583 as a promising scaffold for drug development.
We applied cryo-EM methods to study the binding interactions of AB25583 with Polθ-hel and its mechanism of inhibition. Surprisingly, our structural studies reveal atomic resolution structures of Polθ-hel binding to AB25583 in both dimeric and tetrameric forms, with dimers being the predominant form in solution. Interestingly, Polθ-hel was previously reported to be a tetrameric form via X-ray crystallography19. Our results suggest both forms exist, even though the dimer form is more stable under our experimental conditions.
Our structures of Polθ-hel:AB25583 complexes reveal that the AB25583 binding pocket is located deep inside the central-channel of the helicase. The AB25583 binding pocket is surrounded by four out of the five subdomains of Polθ-hel, and AB25583 directly bonds with multiple side-chains and main-chain atoms of the motor domain D1 and rachet domain D4 (Fig. 4b–e). The extensive molecular interactions of Polθ-hel with AB25583 is consistent with the well-defined electron density observed for the bound inhibitor (Fig. 4c, d), and also explains the single-digit nanomolar IC50 and specificy of AB25583 for Polθ-hel inhibition. The motor domain D1 and rachet domain D4 are critical for ATP-binding/hydrolysis to trigger conformational switches of the helicase and, hence, for translocating ssDNA through the helicase channel. Therefore, binding of AB25583 at such a strategic location within the central-channel is expected to fully inhibit helicase conformational switches that are coupled to ATP-binding and hydrolysis, and as a result, inhibit the motions necessary for active ssDNA translocation. The location of the inhibitor binding site and biochemical data showing identical potency of AB25583 in the presence of increasing concentrations of ATP strongly support an allosteric mechanism of inhibition. This mechanism is further supported by the inhibitor’s ability to suppress Polθ-hel ATP hydrolysis even in the absence of DNA, which is in contrast to Novobiocin, that acts by blocking Polθ-hel ssDNA binding53.
Elucidation of the inhibitor’s binding mode by cryo-EM also offers a possible perspective on how Polθ-hel might function during the MMEJ process. An intriguing observation made related to this process involves the more stable Polθ-hel dimer form, which was also observed in a recent BioRxiv report54. This dimeric form might indeed be the active state for Polθ-hel during MMEJ, given its role in the repair of double-stranded breaks that necessitate the joining of two ends. Interestingly, the close conformational similarity between Polθ-hel and another homologous helicase HEL308 was revealed by the superposition of the structures of Polθ-hel and HEL308/DNA complex55. This closely overlapped structure showed that the ssDNA segment passes through Polθ-hel’s central-channel (Supplementary Figs. 6a, 7a, b). In its dimeric form, the two 3’-ssDNA ends might pass through the central-channel of each monomer and exit at locations that are in close proximity near the dimer interface (Supplementary Fig. 7a, c1), which could conceivably help the sampling and pairing of microhomologous sequences along 3’-ssDNA ends. The transiently annealed microhomologous dsDNA could then serve as the template for one of the Polθ polymerase domains to synthesize dsDNA in one direction, which could be followed by subsequent polymerization in the other direction by a second polymerase (Supplementary Fig. 7c2–5). These actions would complete the majority of the MMEJ process by enabling 3’-ssDNA overhang synapsis, microhomologous ssDNA annealing, and subsequent extension of the minimally paired 3’-ssDNA overhangs. Because the Polθ-hel is connected to the C-terminal Polθ-pol via a long flexible central domain, the Polθ-pol from one Polθ protomer could, in principle, interact with the Polθ-hel in cis or in trans to coordinate the microhomology search and subsequent polymerization. This process may require the assistance of other cellular factors, as no specific interactions between the helicase and polymerase domain have been observed. However, to fully evaluate this model of Polθ-hel dimer activity during MMEJ, additional structural, biochemical, and cellular studies will be required.
In summary, we have solved high-resolution cryo-EM structures of Polθ-hel bound to the small-molecule inhibitor AB25583 as a dimer and tetramer, which reveals detailed interactions underlying Polθ-hel:inhibitor binding, and provides insight into the mechanism of action by which the inhibitor suppresses Polθ-hel helicase activity. We also characterized the biochemical and cellular activities of AB25583, which revealed its ability to selectively kill BRCA-deficient cells and act synergistically with olaparib in BRCA-deficient cancer cells. Hence, these studies reveal AB25583 as a promising scaffold for preclinical drug development, and show strong potential for Polθ-hel inhibitors as anti-cancer agents. The cryo-EM structural methods and results described herein will be important for accelerating the development of additional Polθ-hel small-molecule inhibitor classes toward preclinical drug candidates.
Methods
Synthesis pathway of AB25583
The General procedure for the synthesis and preparation of AB25583 are described in Supplementary Notes.
Protein expression and purification for cryo-EM
The His6-SUMO-PreScissionProteaseSite-Polθ-hel (1–894) was cloned into the pSUMO vector. The recombinant vector was transformed into the Escherichia coli strain Rosetta 2 (DE3) pLysS. The E. coli cells harboring the expression vectors were grown in an LB medium at 37 °C until the OD600 (optical density at 600 nm) reached 0.3. The protein expression was induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside(IPTG) at 18 °C for 18–20 h. The cell pellets were resuspended with Buffer L (25 mM Tris-HCl (pH 8.5), 500 mM NaCl, 10% glycerol, 0.5 mM Tris (2-carboxyethyl) phosphene (TCEP), 1 tablet of Roche complete inhibitor set per 100 mL. The resuspended cell was mixed with 2 mM phenylmethylsulfonyl fluoride (PMSF) and lysed by sonication, and cellular debris was removed by centrifugation. The supernatant containing His6-SUMO-PP-Polθ-hel was loaded onto the Ni-NTA agarose column (QIAGEN). The nickel column was extensively washed with Buffer W (25 mM tris-HCl, pH 8.5, 0.5 M NaCl, 10% glycerol, 40 mM imidazole, 0.5 mM TCEP). The His6-SUMO-tag was cleaved by incubating with ~50 units of PreScission Protease in a one-bed volume of Buffer L overnight. The Polθ-hel was eluted in three-bed volumes of Buffer L and subjected to HiTrap Heparin HP affinity column (Cytiva). The proteins were eluted with a NaCl gradient of 0.2 to 2.0 M. The eluted proteins were further purified using Superdex 200 Increase 10/300 GL column equilibrated with Buffer C (25 mM tris-HCl, pH 8.5, 0.8 M NaCl, 0.5 mM TCEP). The peak fraction was isolated, concentrated, and stored at −80 °C for cryo-EM work.
Negative-stain EM
About 5 µl of 0.02 mg/ml Polθ-hel sample was applied onto glow-discharged ultrathin formvar/carbon supported copper 400-mesh grids (Electron Microscopy Sciences), blotted and stained with 2.0% uranyl acetate. Negative-stained grids were imaged on a Talos F200C transmission electron microscope (Thermo Fisher Scientific) operated at 200 kV.
Cryo-EM data acquisition
The purified 1.0 mg/ml (10 μM) Polθ-hel and AB25583 were mixed by 1:1 molar ratio in Buffer C containing 2% DMSO and incubated on ice for 10 min. About 4 ul aliquots of Polq-hel/AB25583 mixture was applied to UltrAu foil R1.2/1.3 gold 300-mesh grids (Electron Microscopy Sciences). Grids were then blotted and vitrified in liquid ethane using Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM data of Polθ-hel:AB25583 complex was collected in Glacios (Thermo Fisher Scientific) equipped with Falcon-4 direct electron detector operated at 200 kV in electron counting mode. Movies were collected at a nominal magnification of 150,000× and a pixel size of 0.92 Å in EER format. A total dose of 58 e-/Å2 per movie was used with a dose rate of 5–6 e-/Å2/sec. About 4500 movies were recorded by automated data acquisition with EPU.
Cryo-EM data processing
A total of 4500 movies were imported into cryoSPARC software package56 and subjected to patch motion correction and CTF estimation in cryoSPARC. Reference-free manual particle picking in a small subset of data was performed to generate 2D templates for auto-picking. A total of 2,932,534 particles were picked initially, extracted, and down-sampled by a factor of 4, on which 2D classification was performed. 1,670,743 particles from 2D class averages with clear features, including dimer- and tetramer-like shapes were selected. We noticed that the tetramer-like classes were present in a subset of 2D classes with low abundance. The particles were re-extracted with full resolution. 3D ab initio reconstruction was then performed to generate eight initial volumes. To further classify the 3D volumes, heterogeneous refinement was performed with two copies of each initial volume, yielding 16 classes. The top two classes, containing 40% of the particles, showed a dimer form with clear secondary structure features. Non-uniform refinement57 was then performed with C2 symmetry to yield the final 3.0 Å resolution dimer map. Among 16 classes from the heterogeneous refinement, a single class containing 5% of the particles showing a tetramer shape, which resembles the previously reported Polθ-hel tetramer, was identified. Non-uniform refinement was then performed with D2 symmetry to yield the final 3.2 Å resolution tetramer map. All resolution evaluation was performed based on the gold-standard criterion of the FSC coefficient at 0.14358.
Model building and refinement
An atomic model derived from crystal structures of Polθ-hel/AMP-PNP complex (PDB ID: 5AGA) was docked into the cryo-EM map using UCSF Chimera59. The model was refined with the phenix.real_space_refine module in Phenix, with secondary structure restraints and geometry restraints60,61. We then manually adjusted the protein side-chain conformation and, when necessary, moved the main chains to match the density map using COOT62. The atomic models went through iterative cycles of real-space refinement in Phenix63. The ligand model and restrains were generated by Phenix eLBOW64 and docked into the cryo-EM map in COOT and real-space refined with the restrains. The final atomic models were validated using the comprehensive cryo-EM validation tool implemented in Phenix (Table 1)65. All structural figures were generated with UCSF ChimeraX66.
Protein purification for biochemical assays
Recombinant Polθ-hel (residues 1–894) was purified as described18. RECQL5 helicase was a gift from Dr. Erik Debler. A DNA fragments encoding catalytic ATPase domains of human Werner syndrome DNA helicase (WRN, residues 500 to 946) and Bloom’s syndrome DNA helicase (BLM, residues 636–1298) were amplified from plasmids pLX209-neo-active WRN (a gift from Francisca Vazquez, Addgene plasmid # 125788; http://n2t.net/addgene:125788; RRID:Addgene_125788) and pEGFP-BLM (a gift from Chris Kok-Lung Chan, Addgene plasmid # 110299; http://n2t.net/addgene:110299; RRID:Addgene_110299), respectively. The fragments were recloned into a bacterial vector pE-SUMOstar expressing N-terminal 6HIS-SUMO tagged protein versions. Recombinant WRN was purified as described below. Briefly, the expression construct was transformed into BL21(DE3) cells, freshly grown colonies were resuspended, added to 4 L of LB with 50 μg/mL kanamycin, and grown at 37 oC until OD600 ~0.5, then the shaker temperature was turned to 18 oC, and the cells were growing for the next 1 h followed by addition of IPTG to a final concentration of 0.2 mM. The cells were further shaken overnight, pelleted in a centrifuge at 4 oC (30 min at 3000×g), and resuspended in lysis buffer containing 50 mM HEPES pH 8.0, 0.5 M NaCl, 10 mM imidazole pH 8.0, 5 mM βME, 0.1 % IGEPAL CA-630 supplemented with 2 mM PMSF and SIGMAFAST EDTA-free protease inhibitor cocktail (Sigma). The cells were sonicated on ice and centrifuged for 60 min at 25,000×g. The cleared lysate was loaded onto a 5 mL HisTrap FF crude column (Cytiva) and washed with lysis buffer with 30 mM imidazole. The bound protein was eluted with lysis buffer with 200 mM imidazole. The fractions containing 6HIS-SUMO-WRN were pooled and dialyzed against 1 L of dialysis buffer (50 mM Tris-HCl pH 7.5, 0.4 M NaCl, 5% glycerol, 5 mM imidazole, 5 mM βME, 0.005% IGEPAL CA-630) with the addition of 50 U of SUMOstar protease (LifeSensors) overnight at 4 oC. The protein was then loaded onto a 5 mL HisTrap HP column (Cytiva), and the flow-through fractions containing SUMOstar protease-cleaved untagged WRN were collected, pooled, concentrated on a spin concentrator Amicon Ultra with 30,000 MWCO (Sigma), aliquoted and frozen at −80 oC. Purification of BLM protein was performed essentially as described for WRN.
DNA helicase IC50 determination assays
ADP-Glo kinase luminescence assay (Promega Corp) was applied to determine IC50 of AB25583 against four human DNA helicases, Polθ-hel, WRN, BLM, and RECQL5. All ATP hydrolysis reactions were performed in 1x helicase reaction buffer (20 mM tris-HCl, pH 7.5, 5 mM MgCl2, 30 mM NaCl, 5% glycerol, 0.1 mg/mL BSA, and freshly added 1 mM DTT). Typically, reactions contained 5 nM enzyme, 50 nM ssDNA (RP316, 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTT), 100 uM ATP, and twofold serial dilutions of AB25583 in DMSO. Control low-signal reactions contained DMSO only, and control high-signal reactions contained an enzyme and DMSO (instead of AB25583). Reactions were done according to the manufacturer’s procedure. First, serial dilutions of the compound or DMSO were added to a mixture containing a 1.25x concentration of an enzyme in a 1.25x helicase reaction buffer. After 5 min incubation at room temperature, to initiate the reaction, a 5x mixture of ssDNA and ATP (0.25 uM ssDNA and 0.5 mM ATP) in water were added to the tubes followed by 40–80 min incubation at room temperature, depending on the particular enzyme’s ATPase activity. Next, a first kit reagent, ADP-Glo, was added to stop the ATPase activity of a helicase and to remove the remaining ATP from the reaction. After 60 min incubation at room temperature, the second kit reagent, Kinase Detection Reagent, was added to convert the generated ADP to ATP and to provide an ATP-dependent luminescence reaction with the luminescence signal directly proportional to the initial ADP concentration. After 60 min incubation at room temperature, the mixtures were transferred to a white solid 384-well plate (Greiner), and endpoint luminescence measurements were performed using microplate reader CLARIOstar Plus (BMG LABTECH). The experiments were done in triplicates and plotted as mean with ±s.d using GraphPad Prism9 software. About 100, 400, or 600 μM ATP were used to determine AB25583 IC50 against Polθ-hel. Essentially the same protocol was used to measure Novobiocin IC50 against Polθ-hel and WRN. However, concentrations of Novobiocin used were significantly higher than for testing IC50 for AB25583.
EMSA
About 160 nM of Polθ-hel and indicated concentrations of AB25583 were mixed in reaction buffer (25 mM Tris-HCl pH 7.5, 5 mM MgCl2, 30 mM NaCl, 5% glycerol, 0.1 mg/mL BSA, 1 mM DTT) and incubated 10 min at room temperature followed by the addition of 10 nM of fluorescently labeled DNA substrates. After 5–10 min incubation at room temperature, the samples were resolved in non-denaturing 8% PAAG with 0.5X TBE buffer, and DNA was visualized using Typhoon PhosphorImager. Aurintricarboxylic acid (ATA) was used as a protein-DNA binding inhibition control. The following DNA oligonucleotides were used to obtain single-stranded (ssDNA), double-stranded (dsDNA), or partially double-stranded (pssDNA) fluorescently labeled templates (5′-to 3′ sequences): ssDNA (RP316, FAM-TTTTTTTTTTTTTTTTTTTTTTTTTTTTT); dsDNA (RP348, Cy3-CACTGTGAGCTTAGGGTTAGAGCCGG/RP348c, CCGGCTCTAACCCTAAGCTCACAGTG); pssDNA (RP348, Cy3-CACTGTGAGCTTAGGGTTAGAGCCGG/RP343, CTAAGCTCACAGTG; RP469D, CTGTCCTGCATGATG/RP486, Cy5-CACTGTGAGCTTAGTCACATTTCATCATGCAGGACAG).
Cell lines
U2OS cells with MMEJ reporter (EJ2-GFP) was a kind gift from Dr. Jeremy Stark (City of Hope) and were generated and described in prior studies67. They were cultured in Dulbecco’s Modified Eagle Medium (DMEM, GIBCO) supplemented with 15% fetal bovine serum (Cytivia), 2 mM l-glutamine (Sigma), and penicillin/streptomycin (Sigma). DLD1 BRCA2 −/− and DLD1 Parental were obtained from Horizon Discovery, Waterbeach, UK. HCT 116 BRCA2 −/− and HCT 116 Parental were obtained from Cancertools, London, UK. MEF BRCA1 −/− (CC) and MEF Parental (Wildtype) was a kind gift from Dr. Neil Johnson (Fox Chase Cancer Center). MDA 436 BRCA1 mut and MDA 231 (used as wild-type control for MDA 436) cells were obtained from ATCC, Manassas, VA. DLD1 BRCA2 −/−, DLD1 Parental, MDA 436 BRCA1 mut and MDA 231 were cultured in RPMI supplemented with 10% fetal bovine serum, 2 mM l-glutamine, non-essential amino acids, and penicillin/streptomycin. HCT 116 BRCA2 −/−, HCT 116 Parental, MEF BRCA1 −/− and MEF Parental, were cultured in DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, non-essential amino acids, and penicillin/streptomycin.
Colony survival assays
About 800 cells/well of BRCA null, and 200 cells/well of wildtype were plated for DLD1, HCT, and RPE-1 pair on 24-well plates. About 500 cells/well of MDA 436 and 100 cells of MDA 231 were plated on 24-well plates. For MEFs, 300 cells per well in a six-well plate were seeded. The medium was replaced every 2 or 3 days until the colonies were ready for staining. Colonies are typically ready for staining in 10–12 days. For staining: Medium was removed from plates, and cells were rinsed with PBS. Fixation was carried out with—Water: Ethanol: Acetic acid (5:4:1) for 30 min followed by staining of colonies with 0.5% crystal violet in Water: Ethanol (3:2) for 2 h at room temperature. The plates were rinsed with water and left for drying overnight at room temperature. Colonies were then counted manually, and response curves are shown as mean colony formation ± S.E.M.
Immunofluorescence
Immunofluorescence of γH2AX
Cells were plated on six-well plates with glass coverslips and treated with AB25583 a day after plating. Four days after treatment, cells were fixed with 4% (v/v) paraformaldehyde for 20 min at 4 oC, washed with PBS, permeabilized with 0.5% (v/v) Triton X for 10 min and blocked with PBS containing 3% BSA. Cells were incubated with primary antibody (rabbit anti-gamma H2AX [p Ser139] antibody, Bethyl Lab #A700-053, 1:500 dilution in 1% BSA in PBS) overnight at 4 oC followed by 3x washes with PBS and then 1 h incubation with secondary antibody (Goat anti-Rabbit IgG (H + L) Secondary Antibody, DyLight 488 (Thermo #35552) 1:2000 dilution in 1% BSA in PBS). After 3x washing in PBS for 3 min, slides were mounted in 20 ul Prolong antifade with DAPI (LifeTechnologies) to counterstain the nuclei. Cells were visualized and imaged using a Nikon A1R Confocal microscope at a 63X objective magnification, and images were analyzed using ImageJ software. For quantification, >50 cells were counted for all conditions from three independent experiments.
Immunofluorescence of RAD51
Mouse embryonic fibroblasts (MEFs) were incubated with DMSO or 10 µM AB25583 for 24 h. Cells were then subject to 2 Gy γ-irradiation (IR) and fixed at 0 and 6 h post-IR. Immunofluorescence microscopy was performed as follows. Cells were fixed at room temp for 10 min with 4% paraformaldehyde and treated for 10 min with 1% Triton X-100 in PBS. Primary Rad51 antibody (Abcam, ab133534) was incubated overnight at 4 °C in 5% goat serum in PBS. Alexa Fluor 488 conjugated secondary antibody (Thermo Fisher Scientific, A-11034) was incubated for 1 h at room temp and slides were mounted using Vectashield antifade mounting media with DAPI (Vector Laboratories). Z-stack images were captured using a Stellaris 5 confocal microscope, and projection images were generated. An ImageJ macro was used for the quantification of foci-positive cells, which were defined as nuclei containing more than 5 Rad51 foci. Percentage of foci-positive cells are presented as mean and SEM from three independent experiments with the average of each biological replicate shown by open circle data points. A minimum of five images and 200 nuclei were collected and analyzed per sample in each replicate.
MMEJ GFP reporter assay
The GFP MMEJ reporter assay was performed as described68. Briefly, U2OS cells carrying one copy of the previously described E2J-GFP MMEJ reporter cassette39 were sorted for GFP-positive cells, followed by treatment with 20 μM AB25583 for 24 h before transfection. Pretreated cells were co-transfected with I-SceI cDNA, and dsRED-Mito cDNA (control for transfection efficiency) using lipofectamine 2000. Ninety-six hours post-transfection, GFP+ and dsRed+ frequencies were analyzed by flow cytometer (Facscanto, BD). Transfection efficiency was corrected using dsRed+ frequency and % MMEJ was calculated as the ratio of GFP + /dsRed+ cells. Data represent the mean of three biological replicates ± SD.
Statistical analysis and reproducibility
Data were expressed as mean ± SEM from at least three independent experiments with triplicates for each condition unless stated otherwise. A two-tailed unpaired t-test was used for conducting a comparison between the two groups. Significance was assumed at p < 0.05. Asterisks in the figures indicate significance, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Statistically significant p values and number of replicates are indicated in the Figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic models have been deposited in the PDB with accession codes: 9BP9 (Polθ-hel:AB25583 dimer) and 9BPA (Polθ-hel:AB25583 tetramer). The cryo-EM maps have been deposited in the EMDB with accession codes: EMD-44765 (Polθ-hel:AB25583 dimer) and EMD-44766 (Polθ-hel:AB25583 tetramer). Raw electron microscopy data files have been deposited in the Electron Microscopy Public Image Archive (EMPIAR) with accession code EMPIAR-11711. Source data are provided with this paper.
References
Moynahan, M. E. & Jasin, M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11, 196–207 (2010).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Sonnenblick, A., de Azambuja, E., Azim, H. A. Jr. & Piccart, M. An update on PARP inhibitors-moving to the adjuvant setting. Nat. Rev. Clin. Oncol. 12, 27–41 (2015).
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005).
Lord, C. J. & Ashworth, A. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017).
Mateo, J. et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015).
Smith, T. J. Olaparib in metastatic castration-resistant prostate cancer. N. Engl. J. Med. 384, 1175 (2021).
Golan, T. et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 381, 317–327 (2019).
Kim, Y. et al. Reverse the Resistance to PARP Inhibitors. Int. J. Biol. Sci. 13, 198–208 (2017).
Guillemette, S. et al. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 29, 489–494 (2015).
Sakai, W. et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120 (2008).
Edwards, S. L. et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451, 1111–1115 (2008).
Barber, L. J. et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J. Pathol. 229, 422–429 (2013).
Mateos-Gomez, P. A. et al. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015).
Ceccaldi, R. L. et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature https://doi.org/10.1038/nature14184 (2015).
Black, S. J., Kashkina, E., Kent, T. & Pomerantz, R. T. DNA polymerase theta: a unique multifunctional end-joining machine. Genes 7, 67 (2016).
Ozdemir, A. Y., Rusanov, T., Kent, T., Siddique, L. A. & Pomerantz, R. T. Polymerase theta-helicase efficiently unwinds DNA and RNA-DNA hybrids. J. Biol. Chem. 293, 5259–5269 (2018).
Mateos-Gomez, P. A. et al. The helicase domain of Poltheta counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol. 24, 1116–1123 (2017).
Newman, J. A., Cooper, C. D. O., Aitkenhead, H. & Gileadi, O. Structure of the helicase domain of DNA polymerase theta reveals a possible role in the microhomology-mediated end-joining pathway. Structure 23, 2319–2330 (2015).
Zahn, K. E., Averill, A. M., Aller, P., Wood, R. D. & Doublie, S. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 22, 304–311 (2015).
Black, S. J. et al. Molecular basis of microhomology-mediated end-joining by purified full-length Poltheta. Nat. Commun. 10, 4423 (2019).
Kent, T., Chandramouly, G., McDevitt, S. M., Ozdemir, A. Y. & Pomerantz, R. T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat. Struct. Mol. Biol. 22, 230–237 (2015).
Lemee, F. et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl Acad. Sci. USA 107, 13390–13395 (2010).
Higgins, G. S. et al. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget 1, 175–184 (2010).
Arana, M. E., Seki, M., Wood, R. D., Rogozin, I. B. & Kunkel, T. A. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 36, 3847–3856 (2008).
Seki, M. et al. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 23, 4484–4494 (2004).
Begg, A. POLQ in breast cancer. Oncotarget 1, 161–162 (2010).
Chandramouly, G. et al. Poltheta promotes the repair of 5’-DNA-protein crosslinks by microhomology-mediated end-joining. Cell Rep. 34, 108820 (2021).
Yousefzadeh, M. J. et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10, e1004654 (2014).
Higgins, G. S. et al. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 70, 2984–2993 (2010).
Ceccaldi, R. et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 518, 258–262 (2015).
Dai, C. H. et al. Co-inhibition of pol theta and HR genes efficiently synergize with cisplatin to suppress cisplatin-resistant lung cancer cells survival. Oncotarget 7, 65157–65170 (2016).
Koole, W. et al. A polymerase theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 5, 3216 (2014).
Wyatt, D. W. et al. Essential roles for polymerase theta-mediated end joining in the repair of chromosome breaks. Mol. Cell 63, 662–673 (2016).
Schaub, J. M., Soniat, M. M. & Finkelstein, I. J. Polymerase theta-helicase promotes end joining by stripping single-stranded DNA-binding proteins and bridging DNA ends. Nucleic Acids Res. 50, 3911–3921 (2022).
Belan, O. et al. POLQ seals post-replicative ssDNA gaps to maintain genome stability in BRCA-deficient cancer cells. Mol. Cell 82, 4664–4680 e4669 (2022).
San Filippo, J., Sung, P. & Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008).
Li, X. & Heyer, W. D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18, 99–113 (2008).
Truong, L. N. et al. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl Acad. Sci. USA 110, 7720–7725 (2013).
Ramsden, D. A., Carvajal-Garcia, J. & Gupta, G. P. Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining. Nat. Rev. Mol. Cell Biol. 23, 125–140 (2022).
Mengwasser, K. E. et al. Genetic screens reveal FEN1 and APEX2 as BRCA2 synthetic lethal targets. Mol. Cell 73, 885–899 e886 (2019).
Feng, W. et al. Genetic determinants of cellular addiction to DNA polymerase theta. Nat. Commun. 10, 4286 (2019).
Zhou, J. et al. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat. Cancer 2, 598–610 (2021).
Constantinou, A., Henning-Chubb, C. & Huberman, E. Novobiocin- and phorbol-12-myristate-13-acetate-induced differentiation of human leukemia cells associated with a reduction in topoisomerase II activity. Cancer Res. 49, 1110–1117 (1989).
Edenberg, H. J. Novobiocin inhibition of simian virus 40 DNA replication. Nature 286, 529–531 (1980).
Cotten, M., Bresnahan, D., Thompson, S., Sealy, L. & Chalkley, R. Novobiocin precipitates histones at concentrations normally used to inhibit eukaryotic type II topoisomerase. Nucleic Acids Res. 14, 3671–3686 (1986).
Dlugosz, A. & Janecka, A. Novobiocin analogs as potential anticancer agents. Mini Rev. Med. Chem. 17, 728–733 (2017).
Zatreanu, D. et al. Poltheta inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 12, 3636 (2021).
Bubenik, M. et al. Identification of RP-6685, an orally bioavailable compound that inhibits the DNA polymerase activity of Poltheta. J. Med. Chem. https://doi.org/10.1021/acs.jmedchem.2c00998 (2022).
Krais, J. J. et al. Genetic separation of Brca1 functions reveal mutation-dependent Poltheta vulnerabilities. Nat. Commun. 14, 7714 (2023).
Oh, G. et al. POLQ inhibition elicits an immune response in homologous recombination-deficient pancreatic adenocarcinoma via cGAS/STING signaling. J. Clin. Invest. https://doi.org/10.1172/JCI165934 (2023).
Stordal, B. et al. BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Mol. Oncol. 7, 567–579 (2013).
Syed, A. et al. Novobiocin blocks nucleic acid binding to Poltheta and inhibits stimulation of its ATPase activity. Nucleic Acids Res. 51, 9920–9937 (2023).
Guo, H. et al. Cryo-EM structure of DNA polymerase θ helicase domain in complex with inhibitor novobiocin. Preprint at bioRxiv https://doi.org/10.1101/2023.01.20.524915 (2023).
Buttner, K., Nehring, S. & Hopfner, K. P. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 14, 647–652 (2007).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Moriarty, N. W., Grosse-Kunstleve, R. W. & Adams, P. D. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Bennardo, N., Cheng, A., Huang, N. & Stark, J. M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008).
Vekariya, U. et al. DNA polymerase theta protects leukemia cells from metabolically induced DNA damage. Blood 141, 2372–2389 (2023).
Acknowledgements
The research was funded by National Institutes of Health grants R01GM130889 and R35GM152198 to R.T.P., National Institutes of Health grants R01CA214799 and R01CA262466 to N.J., and Z.L. was a recipient of University of Southern California (USC) dean’s fellowship, and F.I. was a former fellowship awardee from Nakajima Foundation. We thank Dr. Debler for providing recombinant RECQL5 helicase. Electron microscopy data were collected at the Core Center of Excellence in Nano Imaging (CNI) at USC. Cryo-EM data were computed at the Center for Advanced Research Computing (CARC) at USC. We thank Htet Khant, Carolyn Marks, and John Curulli at USC for assisting with the operation and maintenance of transmission electron microscopes at CNI, and Tomek Osinski for assisting computing work with CARC at USC.
Author information
Authors and Affiliations
Contributions
R.T.P. and X.S.C. conceived and supervised the project. F.I. and Z.L. purified Polθ-hel and performed cryo-EM structural studies. L.M. purified DNA helicases and performed biochemical assays. G.C., M.T., B.T., R.B., J.J.K., U.V. and M.C. performed cellular assays. N.J. and T.S. provided support for cellular assays. X.S.C. and R.T.P. wrote the initial manuscript draft, and all authors contributed to the revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
R.T.P. is a cofounder and CSO of Recombination Therapeutics, LLC. X.S.C. is a cofounder of Recombination Therapeutics, LLC. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ito, F., Li, Z., Minakhin, L. et al. Structural basis for a Polθ helicase small-molecule inhibitor revealed by cryo-EM. Nat Commun 15, 7003 (2024). https://doi.org/10.1038/s41467-024-51351-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51351-4
- Springer Nature Limited