Abstract
Single-cell RNA sequencing predominantly employs short-read sequencing to characterize cell types, states and dynamics; however, it is inadequate for comprehensive characterization of RNA isoforms. Long-read sequencing technologies enable single-cell RNA isoform detection but are hampered by lower throughput and unintended sequencing of artifacts. Here we develop Single-cell Targeted Isoform Long-Read Sequencing (scTaILoR-seq), a hybridization capture method which targets over a thousand genes of interest, improving the median number of on-target transcripts per cell by 29-fold. We use scTaILoR-seq to identify and quantify RNA isoforms from ovarian cancer cell lines and primary tumors, yielding 10,796 single-cell transcriptomes. Using long-read variant calling we reveal associations of expressed single nucleotide variants (SNVs) with alternative transcript structures. Phasing of SNVs across transcripts enables the measurement of allelic imbalance within distinct cell populations. Overall, scTaILoR-seq is a long-read targeted RNA sequencing method and analytical framework for exploring transcriptional variation at single-cell resolution.
Similar content being viewed by others
Introduction
Alternative RNA splicing is a key driver of proteome complexity and cellular phenotypic diversity. Approximately 95% of human multi-exon genes are alternatively spliced and 15–25% of human hereditary diseases and cancers are linked to alternative splicing1,2,3. Although short-read RNA sequencing has been widely adopted to measure gene expression, it remains challenging to identify full-length isoforms with only 20–40% of the human transcriptome being assembled using gold standard isoform reconstruction tools4,5,6. In addition, alternative splicing, cleavage and polyadenylation events have been shown to be highly tissue-specific7,8. Thus, to better understand cellular diversity and dynamics in health and disease, isoform-level transcriptomic information is required.
Single-cell RNA sequencing (scRNA-seq) has advanced our understanding of cellular heterogeneity, delivering transformative insights into a wide array of pathologies, including autoimmune diseases9,10, neurological disorders11,12 and cancer13,14. To date, the vast majority of single-cell RNA profiling studies have employed short-read sequencing to measure gene expression which is typically quantified by counting reads derived from the 3’- or 5’-ends of genes. While useful for gene expression analysis, identification of isoforms remains challenging for single-cell short-read sequencing due to limited gene body coverage. To address this, multiple groups have performed long-read sequencing of cDNA from single cells which enables sequencing of full-length molecules15,16,17,18,19,20,21,22. However, to accurately identify cell barcodes (CBs) and unique molecular identifiers (UMIs), most of these strategies have implemented short-read sequencing paired with long-read sequencing to assist CB/UMI demultiplexing or specialized library preparation steps to improve read accuracy at the cost of sequencing throughput. In addition, the majority of these studies have demonstrated sequencing of a relatively small number of cells at low per-cell sequencing depth due to current throughput limitations of long-read platforms15,18,23,24. Recent efforts have been developed to employ hybridization-based capture strategies to enrich selected genes of interest17,25. Gene panel designs utilized in previous studies have typically focused on specific biological questions and encompassed less than 50 target genes, which presents a challenge for cell annotation in complex tissues and requires additional short-read sequencing17,25. A particular issue inherent to single-cell long-read library preparation and sequencing is the presence of unwanted artifacts that consume valuable sequencing throughput. These artifactual reads do not exhibit the expected cDNA structure after reverse transcription and amplification; rather, often contain template-switching byproducts or lack adapter sequences16.
To address the aforementioned shortcomings, we have developed single-cell targeted isoform long-read sequencing (scTaILoR-seq). scTaILoR-seq makes use of commercially available or custom-designed gene panels to enrich for greater than 1000 genes of interest. In addition, scTaILoR-seq mitigates the presence of artifacts common to single-cell RNA-seq cDNA by enriching for molecules with the expected adapter sequence using biotinylated PCR primers. Following both gene panel enrichment and artifact mitigation, nanopore long-read sequencing facilitates the assignment, identification, and quantification of transcript isoforms in thousands of single cells. Using scTaILoR-seq, we characterize transcript composition and their differential expression among three ovarian cancer cell lines and compare it to analogous long-read (LR) untargeted sequencing as well as to both targeted and untargeted short-read (SR) sequencing approaches. Moreover, we evaluate SR-guided and unguided strategies for CB and UMI identification, demonstrating that an unguided assignment approach (wf-single-cell) circumvents the need for supplemental SR sequencing. We then apply scTaILoR-seq with the unguided CB/UMI assignment method to profile dissociated tumor cells (DTCs) from two ovarian cancer patients. This enables the identification of cell-type-specific isoforms, reconstruction of immune repertoires, and detection of expressed single-nucleotide variants (SNVs) at the single-cell level. In addition, long reads enable SNV phasing to assemble haplotypes and estimate allelic imbalance from individual tumor epithelial cells.
Results
Artifact mitigation combined with targeted gene enrichment enables efficient long-read sequencing of single-cell cDNA
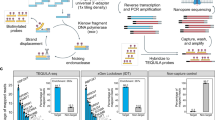
To evaluate gene enrichment, we performed droplet-based single-cell 3’-end RNA sequencing on an equal mixture of three human ovarian cancer lines (SK-OV-3, COV504, and IGROV-1) (Fig. 1). The resulting single-cell cDNA was used to evaluate a variety of strategies aimed at determining optimal conditions for LR sequencing (Fig. 2a, Supplementary Fig. 1, and Supplementary Table 1). Assessment of the targeted approach using a 10x Genomics pan-cancer probe panel demonstrated efficient enrichment of cancer-associated genes with SR sequencing (17.7-fold mean read enrichment). We next sought to maximize the proportion of complete reads (i.e. reads containing both the template switch oligo (TSO) adapter and poly(A) sequences) using LR sequencing. A previously described artifact mitigation (AM) approach was deployed to reduce TSO-TSO byproducts from library preparation16 using biotinylated PCR primers complementary to the Read1 sequence, which enabled streptavidin-coated magnetic bead pull-down and subsequent amplification of complete cDNA constructs. Compared to the targeted approach without AM, the targeted+AM strategy displayed an 11.8% increase in complete read proportion concomitant with a marked decrease in TSO-TSO artifacts (Fig. 2a), which is consistent with improvements observed in a recent single-cell long-read study26. Next, we investigated an orthogonal TSO-TSO depletion approach based on circularization of targeted complete cDNA using rolling circle amplification to concatemeric consensus (targeted+R2C2)19,27 (“Methods”). Compared to targeted+AM, the targeted+R2C2 approach exhibited a slightly higher proportion of complete reads and fewer TSO-TSO artifacts; however, it yielded much lower read throughput: 4.4 M versus 18 M average passed reads per flow cell compared to the targeted+AM approach (Fig. 2b). Therefore, the targeted+AM strategy displayed an optimal balance between increased complete read proportion and higher read throughput. For this reason, the targeted+AM approach became the basis of scTaILoR-seq, which was employed in all subsequent experiments.
Ovarian cell lines or dissociated tumor cells are processed using droplet-based single-cell RNA-seq 3’-Gene expression assay to obtain cDNA. Targeted enrichment is performed followed by nanopore sequencing, cell barcode (CB) and unique molecular identifier (UMI) assignment, and read alignment. Downstream analysis enables the measurement of isoforms, SNVs, allelic expression, TCR sequences and gene fusions at single-cell resolution. Figure Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
a Schematic detailing library preparation methods (targeted, targeted+AM, and targeted+R2C2) tested for enrichment using long-read (LR) sequencing. b Complete reads (left), TSO-TSO artifacts (middle), and number of passed reads (QScore ≥7) (right) across library preparation methods. Targeted (n = 3 replicates), Targeted+AM (n = 2 replicates), Targeted+R2C2 (n = 2 replicates). Bars represent mean values across replicates. c Pseudobulk gene-level expression between short-read (SR) untargeted and scTaILoR-seq approach. CPM counts per million. d Metagene max-normalized coverage profiles for untargeted and targeted SR and LR sequencing approaches. e Pseudobulk gene expression correlation for scTaILoR-seq replicates. f Frequency distributions of read counts per gene for untargeted LR and scTaILoR-seq. g Number of genes and transcripts uniquely detected (single dot) or shared (‘joined’ dots) across untargeted LR sequencing and scTaILoR-seq. h Pseudobulk transcript-level expression between untargeted LR and scTaILoR-seq methods. i Number of observed on-target novel transcript models for untargeted LR sequencing and scTaILoR-seq. j Number of on-target fusions identified for untargeted LR sequencing and scTaILoR-seq. Source data are provided as a Source Data file.
scTaILoR-seq increases transcript detection sensitivity while preserving relative expression levels
Targeted gene enrichment with scTaILoR-seq yielded ~95% of usable transcript reads that were mapped to target genes. In contrast, without gene enrichment (i.e., LR untargeted), only 5% of transcript reads mapped to target genes (Supplementary Fig. 2). In general, reads obtained with scTaILoR-seq were slightly longer, consistent with the increased median transcript length of on-target genes (1360 nt) relative to that of off-target transcripts (991 nt) (Supplementary Fig. 3a). scTaILoR-seq also displayed slightly higher relative read depth across exons compared to the untargeted approach (Supplementary Fig. 3b). Next, we investigated the accuracy and reproducibility of scTaILoR-seq by measuring on-target gene expression levels. From read-depth normalized samples, we observe highly correlated mean gene expression (r = 0.92) between scTaILoR-seq and untargeted SR sequencing, indicating that scTaILoR-seq faithfully recapitulates quantitative gene expression (Fig. 2c and Supplementary Fig. 4). LR sequencing provided extended coverage over gene bodies compared to SR sequencing. scTaILoR-seq and untargeted LR sequencing displayed similar coverage (Fig. 2d) and RNA biotype profiles (Supplementary Fig. 5). Gene expression was strongly correlated across replicates (r = 0.95) with 98.8% gene overlap demonstrating the reproducibility of scTaILoR-seq (Fig. 2e). Moreover, scTaILoR-seq resulted in a 16.4-fold increase of on-target reads compared to untargeted LR sequencing, yielding a significant boost in read counts per gene (two-sided Mann–Whitney U test, P = 3.7 × 10−129) (Fig. 2f).
Next, we sought to characterize the improvement in transcript detection provided by target enrichment. scTaILoR-seq identified an additional 279 on-target genes and 2484 annotated transcripts that were not detected in the untargeted LR approach (Fig. 2g and Supplementary Fig. 3c), representing a 4.5-fold increase in the median proportion of detected transcripts per gene (Supplementary Fig. 3d). Transcript expression from scTaILoR-seq was highly correlated with untargeted LR sequencing (r = 0.87) with some saturation observed among highly expressed transcripts (Fig. 2h). Relative to the untargeted LR approach, scTaILoR-seq detected a higher frequency of Novel In Catalog (NIC) and Novel Not In Catalog (NNIC) transcript models associated with targeted genes (Fig. 2i). The NIC transcripts are composed of known annotated splice junctions but occur as novel arrangements and the NNIC transcripts include unannotated splice acceptors and donors.
We also assessed the ability of scTaILoR-seq to detect chimeric fusion transcripts derived from genes on the targeting panel. Using the fusion detection algorithm JAFFAL28, scTaILoR-seq identified 6.7-fold more on-target fusions than untargeted LR sequencing (Fig. 2j). We detected high-confidence and known gene fusions annotated in the Mitelman database29 which displayed enrichments ranging from 3- to 26.5-fold (Supplementary Fig. 6). Interestingly, the majority of fusions identified with scTaILoR-seq comprised an on-target gene fused to an off-target gene partner, suggesting that the probe coverage over one partner is sufficient to identify fusion events.
Detection of alternative splicing at the single-cell level
Higher error rates observed in nanopore sequencing reads can confound CB and UMI assignment, which led to the development of strategies that leverage supplemental high-accuracy SR sequencing data16,24. We compared one such guided method, SiCeLoRe16, with a recently developed unguided approach, wf-single-cell (“Methods”). This unguided method eliminates the requirement for supplemental SR sequencing to assign CBs and UMIs (Supplementary Fig. 7a). We observed a high degree of overlapping CBs between SiCeLoRe and wf-single-cell assignments. These overlapping CBs encompass nearly all of those found in the associated untargeted SR sequencing data (Supplementary Fig. 7b). In addition, UMI counts per CB from SiCeLoRe and wf-single-cell were highly correlated (r = 0.97), with the latter yielding higher counts (Supplementary Fig. 7c). Gene expression for matched cell line populations was also highly correlated (Supplementary Fig. 7d). Taken together, these results indicate that scTaILoR-seq is compatible with current guided and unguided CB/UMI assignment methods.
To assess the ability of target enrichment on distinguishing cell types in the ovarian cell line mixture, we focused on differential expression variation captured by principal component analysis. Because each principal component (PC) explains a fraction of overall transcriptome expression variation (known as the variance ratio), the cumulative variance ratio of ranked PCs provides a measure of usable expression information. For the ovarian cell line mixture, we determined the cumulative variance ratio (PC1-PC10) across SR and LR sequencing as well as for the untargeted and targeted approaches (Fig. 3a). This analysis showed that target enrichment marginally increases the cumulative variance ratio between untargeted and targeted SR methods (0.24 vs. 0.26). However, between the untargeted LR approach and scTaILoR-seq, we observed a marked 3.2-fold increase in cumulative variance ratio (0.06 vs. 0.18). This difference was manifested in cell embeddings using the first three PCs, which showed separation of genetically deconvoluted cell lines when using scTaILoR-seq (Fig. 3b and Supplementary Fig. 8). Ultimately, the expression variation captured by scTaILoR-seq yielded clusters consistent with known cell line genotypes (Fig. 3c). While target enrichment showed marginal value for SR sequencing, we observed a marked improvement in the ability to distinguish cell types using scTaILoR-seq over an untargeted LR sequencing approach.
a Comparative principal component analysis using ovarian cell line mixture (COV504, IGROV-1, and SK-OV-3). SR short-read, LR long-read. b Cell embeddings using the first three principal components for untargeted LR sequencing and scTaILoR-seq. c UMAP visualization of ovarian cell line mixture. d Comparison of on-target genes, UMIs, and transcripts per cell across untargeted and scTaILoR-seq library preparation methods.Violin plots represent kernel density estimates. Box and whisker plots represent the first and thrid quartiles (box), median (dot) and the minimum and maximum values (whiskers). LR untargeted (n = 2100) scTaILoR-seq (n = 2101). e Cluster level PARP2 isoform proportions and single-cell transcript UMAP visualization. Alternative 5’ splice site within exon 2 of PARP2 is indicated by the shaded pink rectangle in the transcript model. UMAP visualization diplays scaled expression. f Cluster level RTKN isoform proportions and single-cell transcript UMAP visualization Alternative 5’-UTR and first exon usage of RTKN is indicated by the shaded pink rectangle in the transcript model. UMAP visualization diplays scaled expression. Source data are provided as a Source Data file.
Next, we sought to quantify the enrichment performance of scTaILoR-seq at the single-cell level using the ovarian cell line mixture. Relative to the untargeted LR sequencing approach, scTaILoR-seq exhibited a tenfold median increase in on-target genes per cell and a 29-fold median increase in on-target transcripts per cell (Fig. 3d). Pseudobulk transcript-level expression was highly correlated between untargeted LR sequencing and scTaILoR-seq for each cell line (SK-OV-3: r = 0.87, IGROV-1: r = 0.86, COV504: r = 0.84) (Supplementary Fig. 9). In addition, the top-25 expressed genes from scTaILoR-seq were noticeably depleted of mitochondrial and house-keeping genes that were abundant in the untargeted SR and LR sequencing approaches (Supplementary Fig. 10). Next, we assessed whether scTaILoR-seq can be used to identify alternative splicing events across the ovarian cancer cell lines. Using differential transcript expression (two-sided Welch’s t test), we identified significant cell line-specific isoform usage (Benjamini–Hochberg adjusted P < 0.05; “Methods”) (Supplementary Fig. 11). For example, we identified alternative 5’ splice site usage of exon 2 in PARP2, the frequency of which varied across the three cell lines (Fig. 3e and Supplementary Fig. 12). Exon 2 of PARP2 is localized at the N-terminal region which is known to facilitate activation on DNA single-strand breaks. Alternative splicing within this region may modulate the DNA damage-sensing activity of PARP230. In addition, we identified a predominant alternative 5’-UTR and the first exon usage event in the Rho-binding domain of RTKN specific to SK-OV-3 (Fig. 3f and Supplementary Fig. 13). RTKN is a scaffold protein that interacts with GTP-bound Rho proteins to subsequently regulate cell growth and transformation31. These examples demonstrate the ability of scTaILoR-seq to enrich for genes of interest, which enables the identification of differential isoform usage events and alternative splicing patterns at the single-cell level.
Surveying the transcriptional landscape of an ovarian tumor microenvironment
The tumor microenvironment (TME) is a complex niche characterized by dynamic interactions among diverse cell types, including epithelial, stromal, and immune cells. To quantify differential isoform usage and to annotate cell-type populations within the TME, both pan-cancer and immune enrichment panels were used, targeting a total of 2243 genes. We performed scTaILoR-seq on dissociated tumor cells (DTCs) from two stage-III treatment-naive ovarian cancer patients: P1—high-grade serous ovarian carcinoma (HGSOC), P2—ovarian clear cell carcinoma (OCCC). Sequencing was performed on the PromethION instrument resulting in a total of 371 million reads (Supplementary Table 1) with a median of 4020 and 1867 UMIs per cell for P1 and P2, respectively. We detected 8695 cells derived from the two patient samples and identified several major cell types (B cells, T/NK cells, myeloid, fibroblast and epithelial) (Supplementary Fig. 14). Lineage-specific cell proportions were consistent between scTaILoR-seq and untargeted SR data generated from the same single-cell cDNA (Supplementary Fig. 15). Of particular interest was sample P1 (n = 2482 cells) which was analyzed to a greater extent since it contains a higher number of EPCAM+ tumor epithelial cells (n = 1498) in addition to an even representation of both stromal and immune cells (Fig. 4a and Supplementary Fig. 15). Differential expression analysis identified transcripts from genes that were consistent with annotated cell identity such as expression of EPCAM in epithelial cells, multiple isoforms of COL3A1 and COL1A2 in fibroblasts, CD3E and CD2 in T cells and distinct C1QB isoforms in cells derived from the myeloid lineage (Fig. 4b).
a UMAP visualization of dissociated tumor cells (n = 2482) from a HGSOC patient (P1). Scaled expression of cell-type-specific canonical marker genes are shown as additional UMAPs (right). b Scaled expression of the top-10 differentially expressed transcripts across coarse cell-type groupings. The colored horizontal bar at top corresponds to cell-type annotations in (a). c IL-32 transcript models for theta and beta isotypes and differential IL-32 isoform usage identified across CD8 + T cells and PDGFRa-/b+ Fibroblasts (two-sided Mann–Whitney U test). d Zoomed in UMAP of T cells showing successfully reconstructed TCRs. e Proportion of T cells with TCR chain assignments: no chain identified (N/A), one chain identified (α or β) or both chains identified (α & β). f Higher-order (>2 cells) clonotypes identified within T cells. The inner ring denotes the number of cells while the outer ring denotes individual clonotype frequency. g Projection of cells expressing TP53 chr17:7674241_G > C (HGVS 17:g.7674241 G > C). h Correlation between single-cell MSK HGSOC geneset expression and PROGENy pathway activity scores. i MSK HGSOC Cancer.3 (JAK-STAT/NF-kB/TNF-active) and Cancer.6 (Hypoxia-active) geneset scores mapped to epithelial cell embedding. Source data are provided as a Source Data file.
Provided that alternative splicing events are prevalent in cancer and the associated TME32,33, we analyzed differential isoform usage between all cell types. This analysis identified 43 significant events (Supplementary Table 2) including differential IL-32 isoform usage between CD8 + T cells and PDGFRɑ-/β+ fibroblasts (two-sided Mann–Whitney U test; ENST00000530890 P = 4.6 × 10−9 and ENST00000440815; P = 1.6 × 10−14) (Fig. 4c). Expression of IL-32β isotype (ENST00000440815) was dominant across all cell types; whereas, IL-32θ (ENST00000530890) expression was markedly low in PDGFRɑ-/β+ fibroblasts (Supplementary Fig. 16). IL-32β is associated with hypoxic conditions in solid tumors and IL-32θ inhibits NF-kB which counters the epithelial-mesenchymal transition34.
Next, we turned our attention to the immune component of the TME, where current single-cell TCR/BCR reconstruction with SR sequencing requires supplemental library preparation and is limited to 5’-expression profiling. A recent single-cell study that implemented untargeted LR sequencing was unable to obtain sufficient read depth for low abundance TCR transcripts, indicating a need for increased detection sensitivity35. Thus, we asked whether scTaILoR-seq (3’-expression) would be amenable to TCR repertoire profiling. Reads were processed by TRUST4 which performs single-cell reconstruction of TCR sequences36. Of the barcodes associated with successfully assembled TCRs, 98% corresponded to annotated T cells (Fig. 4d) and 85.9% had at least one chain (α and/or β) identified (Fig. 4e). With scTaILoR-seq, we obtained a TCR α/β chain pairing rate of 49.7%, which is a two-fold improvement over previous targeted and untargeted LR strategies17,27. Within the expanded T-cell population (n = 56 cells), we identified 15 high-order clonotypes with more than two cells sharing identical CDR3 regions. The CDR3 sequence CASSYYSTARAGYTF was detected in seven cells, representing the largest observed clonotype population (Fig. 4f).
High-grade serous ovarian cancer is characterized by near ubiquitous mutation of the tumor suppressor TP53 and approximately two-thirds of mutations occur within exons 5-837,38. Characterization of mutations within this hot-spot region of TP53, specifically in the tumor epithelial cell population, was of particular interest. scTaILoR-seq enables the detection of expressed SNVs that are outside the typical read coverage of SR single-cell sequencing (Fig. 2d and Supplemental Fig 17a). For example, using the LR variant caller Clair339, we detected a SNV in exon 7 of TP53 (chr17:7674241 G > C). This missense variant (S241C; HGVS 17:g.7674241 G > C) alters the DNA-binding domain of TP53 and is annotated as a putative pathogenic ovarian cancer mutation40. Despite TP53 exon 7 detection across several other cell types (Supplemental Fig. 17b), this SNV was exclusive to a subpopulation of tumor epithelial cells (Fig. 4g). Within this subpopulation, 97% of reads harbored the variant whereas only 3% of reads derived from non-epithelial cells exhibit the same variant (Supplemental Fig. 17c). Closer inspection revealed that detection of the SNV in the remaining tumor epithelial cells was hindered due to lack of coverage of TP53 exon 7. In general, when compared to untargeted LR sequencing, scTaILoR-seq improved SNV detection in targeted genes by ~2.5-fold (673 vs. 1669) (Supplementary Fig. 18).
To assess cancer-associated expression patterns among the epithelial cells, we performed pathway activity analysis using PROGENy41, which identified two signatures: JAK-STAT/NF-κB/TNFɑ and hypoxia (Fig. 4h). These two pathways were correlated with single-cell RNA-seq expression patterns characteristic of tumor cells from treatment-naive HGSOC patients: “Cancer.3” and “Cancer.6”13. PROGENy signatures and associated gene expression were localized to distinct cell subsets within the epithelial cell embedding (Fig. 4i and Supplementary Fig. 19). Collectively, these data suggest that a large fraction of the epithelial cells exhibit distinctive cancer signaling pathways consistent with ovarian cancer.
Identifying structural transcript variation associated with expressed SNVs
After determining the ability of scTaILoR-seq to detect SNVs, we asked whether these expressed variants were associated with differential transcript structures in HGSOC, as reported in several other tissues and cell lines42. We utilized a deep learning-based model called SpliceAI43 to predict and score cryptic splicing events associated with detected SNVs from targeted transcripts within the tumor epithelial cell population (Fig. 5a). For the 82 hits from 1669 SNVs (SpliceAI score >0.1) (Fig. 5b), we identified transcript structure variation by assessing the coverage divergence (1 − r2) between reads matching the reference base (REF) or the alternative base (ALT) for a given SNV site (Fig. 5a). Among the 82 queried hits, 44 displayed coverage divergence, indicating a difference of transcript structure between REF and ALT alleles. scTaILoR-seq produced a 1.5-fold increase in detected SNVs associated with alternative transcript structures compared to the untargeted LR sequencing approach (Supplementary Fig. 20a). In addition, the divergence scores for alternative transcript structures found in both scTaILoR-seq and the untargeted LR approach were strongly correlated (r = 0.94) (Supplementary Fig. 20b).
a Workflow for detecting SNV-associated differential transcript structures: (i) Obtain reads from ovarian tumor epithelial cells (scTaILoR-seq), (ii) Determine SNVs using Clair3, (iii) Predict cryptic splice events using SpliceAI, (iv) Compute coverage divergence between reference (REF) or alternative (ALT) variant reads, (v) Identify differential transcript structures among SpliceAI hits using coverage divergence. b SNVs of genes exhibiting SpliceAI score above the threshold value of 0.1. Each SNV is colored by SpliceAI score and whether a hit also displays coverage divergence between REF and ALT. c Hierarchical clustering based on extent of divergence at transcript structural elements (CDS and UTR/Intron) of 44 “Hit and divergent” SNVs. d, e Plots for ELF3 and STAT1, respectively: normalized coverage tracks for REF and ALT and corresponding transcript model. Source data are provided as a Source Data file.
Using scTaILoR-seq reads, the local coverage divergence along transcripts was used to classify transcript structural events into two categories: “CDS” for protein-coding and “UTR/Intron” for untranslated regions and introns (Fig. 5c). We detected differential ELF3 transcript structures associated with chr1:202011127 A > C in exon 2 for which flanking introns were retained among REF reads (1130 UMIs in 574 cells); whereas, ALT reads (2962 UMIs in 1013 cells) exhibited normal splicing (Fig. 5d). ELF3 is a transcription factor highly expressed in epithelial tissue and has been shown to inhibit the epithelial-to-mesenchymal transition13,44 while supporting angiogenesis45. Another example of differential transcript structures linked to an SNV (chr2:190975811 C > A) was observed with the transcription factor STAT1, which exhibited distinct allele-specific events (REF = 2501 UMIs in 525 cells and ALT = 1254 UMIs in 353 cells) that spanned both CDS and UTR/Intron (Fig. 5e). In summary, scTaILoR-seq can provide insight into variation of transcript structures associated with SNVs, leading towards an improved understanding of transcriptional complexity associated with genetic alteration in cancerous cells.
Phasing of expressed SNVs reveals allelic imbalance within tumor epithelial cells
Given the LR output of scTaILoR-seq, we reasoned that transcripts containing multiple SNVs could be used for haplotype reconstruction and subsequent allele-specific expression analysis42,46,47. We observed that the median number of SNVs per gene was two (Supplemental Fig. 21a) and the median distance between SNVs of the same gene was 511 nucleotides (Supplementary Fig. 21b). Using multi-SNV reads, haplotypes were revealed by iteratively phasing SNVs along a given gene (“Methods”). Two haplotypes were reconstructed for 370 genes with scTaILoR-seq, for which 94.6% of transcript reads had the majority of SNVs match a haplotype sequence. In comparison, the LR untargeted approach yielded only 124 genes with reconstructed haplotypes, 117 of which were also detected with scTaILoR-seq (Supplementary Fig. 22a). Thus, these haplotypes are generally representative of observed allele-specific transcripts.
Among the haplotypes, human leukocyte antigen (HLA) alleles were noteworthy given their diversity and function in adaptive immunity48. Consistent with their well-known polymorphism, a large number of SNVs were detected in the HLA genes, ranging from 46 in HLA-A to 8 in HLA-DRA (Supplementary Fig. 21c). We observed uneven mapping of transcript reads between the two alleles; HLA-DRA exhibited a striking 3.6-fold bias for transcripts mapping to haplotype 1 (H1) versus haplotype 2 (H2) (Fig. 6a). Allele-specific expression is recognized as a pervasive feature of cancer, potentially stemming from alterations such as genomic structural rearrangement and dysfunctional epigenetic regulation49. Here, in the context of HGSOC, we sought to systematically characterize the allele-specific expression differences between tumor epithelial cells and the residual TME cell populations. We identified 33 genes displaying imbalanced allelic expression within the epithelial cell population but not in the remaining cells (two-sided Mann–Whitney U test; Benjamini–Hochberg adjusted P < 10−6 and P > 0.05, respectively) (Fig. 6b). Differential allele expression across epithelial and non-epithelial cell subsets correlated well between scTaILoR-seq and the LR untargeted approach (Pearson r = 0.76) (Supplementary Fig. 22b). Among genes exhibiting epithelial-specific imbalanced allelic expression, VEGFA and CD276 are therapeutic targets for treatment of ovarian malignancies like HGSOC50 (Fig. 6c, d). With scTaILoR-seq, phasing of SNVs permitted high-quality haplotype reconstruction and enabled quantitation of allele-specific expression among cellular populations within ovarian tumor samples.
a Phased (H1—blue, H2—green) single-molecule read tracks for HLA-DRA. b Allelic expression (H1/H2) and proportions of cells detected for epithelial and non-epithelial groups ranked by magnitude of imbalance. c, d Plots for HLA-DRA and VEGFA, respectively: UMAP visualization of haplotype expression with epithelial cells highlighted in red and non-epithelial cells highlighted in blue. Violin plots show significant cell-type-specific allelic imbalance (two-sided Mann–Whitney U test). Source data are provided as a Source Data file.
Discussion
Recent improvements in nanopore sequencing chemistries, basecalling accuracy and bioinformatic tools have enabled single-cell LR sequencing, which can deliver unprecedented insights into cell-type-specific transcriptional diversity. However, there remain key challenges to overcome such as considerably lower throughput and template switch artifacts. Here, we developed scTaILoR-seq to address these challenges using targeted enrichment and artifact mitigation. Fortunately, several expert-curated commercial gene panels—like the ones used in this study—are available for a range of biological applications. The use of custom gene panels further expands the versatility of scTaILoR-seq to facilitate diverse biological inquiries. This approach was developed to optimally allocate sequencing reads to hundreds or a few thousand genes of interest; whereas, current methods shallowly survey the whole transcriptome18,51 or deeply examine a narrow set of target genes17,25. With our approach, we demonstrate improved transcript detection sensitivity for applications ranging from differential isoform expression analysis to the discovery of sequence variants.
Fundamental to scRNA-seq is the ability to resolve reads by individual CBs and UMIs, which enables cell-specific quantification of gene expression. Given the higher basecalling error rates of nanopore sequencing compared to SR sequencing, prior single-cell LR approaches have relied on supplemental SR data to improve CB/UMI assignment accuracy16,24. Despite the inherent errors of nanopore reads, we demonstrate that scTaILoR-seq paired with wf-single-cell CB/UMI assignment and IsoQuant transcript quantification is capable of accurately producing single-cell transcriptomes from a complex tumor tissue without supplemental SR data.
Using scTaILoR-seq, we demonstrate that targeted single-cell LR sequencing is an effective approach to generate TCR repertoires from a complex tumor tissue. Despite T cells comprising only 12% of the total cell population, scTaILoR-seq provided sufficient coverage of the TCR locus for sequence reconstruction. While TCR analysis typically requires specialized library preparation and/or 5’ RNA-seq with SR approaches, we demonstrate TCR reconstruction with scTaILoR-seq using conventional 3’ RNA-seq. In addition, we were able to identify unique and expanded clonotypes which could provide insight into TME-specific T-cell interactions and tumor antigens. Both the immune repertoires and the extent of clonal expansion are key determinants of the anti-tumor response and outcomes52,53,54.
Greater read depth and broader transcript coverage enabled more comprehensive detection of expressed SNVs, which was fundamental for the characterization of transcript structure alterations. Because many of these SNVs are proximal to annotated splice junctions, we suspect sequence-specific impacts on spliceosome function may contribute to the altered transcript structures. In some cases, a SNV and its associated site of structural divergence are within an A/T-rich region, which may be susceptible to internal priming during reverse transcription and/or second-strand synthesis55. While such artifacts would be considered false positives, we also observed opposing examples in which structural divergence was detected in non-A/T-rich regions. Taken together, scTaILoR-seq facilitates the characterization of SNV-associated transcript structures which may be particularly impactful in evaluating the functional consequences of cancer mutations.
By reconstructing haplotypes from multi-SNV reads, we identified imbalanced allelic expression within tumor epithelial cells. Of potential therapeutic relevance is the observed allele-specific expression of VEGFA, which is the target of bevacizumab (Avastin) for the treatment of platinum-resistant recurrent epithelial ovarian cancer56,57. In addition, CD276 showed imbalanced allelic expression and is under clinical development as a cancer immunotherapy50. Thus, beside the fundamental biological insights afforded by scTaILoR-seq, the ability to simultaneously characterize cell- and allele-specific transcriptional variation has the potential to impact diagnostic and therapeutic approaches.
Recently, Oxford Nanopore Technologies introduced “adaptive sampling” as a method for enrichment or depletion of specific sequences in real time. This method has been applied for the detection of expressed SNVs in single cells58 and established the use of adaptive sampling in a transcriptomics application. However, comprehensive analysis of its efficiency and utility for quantitation of expression is lacking. Recent long-read metagenomic studies have suggested that enrichment with adaptive sampling for fragment lengths less than about 2 kb is not efficient and may negatively impact sequencing throughput59,60.
While we have highlighted key capabilities of scTaILoR-seq, there are several ways to further improve this method. First, abortive reverse transcription hinders analysis due to truncated long reads and may be addressed with alternative reverse transcription strategies. Second, the analyses presented here focuses primarily on annotated transcripts. We showed that scTaILoR-seq improved sensitivity for discovering novel isoforms and fusions; however, to enable broad detection of novel transcripts, probe designs may need to be expanded beyond the sequence space of annotated transcripts. Finally, we expect that this approach can be adapted to many of the emerging and existing commercial scRNA-seq platforms (e.g. droplet, nanowell, and combinatorial indexing) in addition to synergistic technologies like spatial transcriptomics. With these adaptations in mind, scTaILoR-seq presents an attractive option for efficient exploration of full-length transcriptomes, especially for large-scale single-cell atlasing initiatives. Overall, scTaILoR-seq efficiently allocates sequencing throughput to improve detection and quantitation of transcripts of interest at multiple resolutions: from exon structure down to single-nucleotide variants.
Methods
The described research complies with all relevant Genentech ethical regulations. Ovarian cancer dissociated tumor cells from de-identified consented patients were purchased through Discovery Life Sciences (Huntsville, AL) for research purposes only. Sex or gender analysis was not relevant for this proof-of-concept study.
Single-cell isolation and 10X Genomics 3’ cDNA generation
Cell lines
To evaluate the sensitivity and the robustness of our method, we obtained three ovarian cancer cell lines SK-OV-3 (ATCC [HTB-77]), IGROV-1 (NCI-FC DCTD), and COV504 (ECACC [07I007]). SK-OV-3 and IGROV-1 were maintained in RPMI-1640 medium, supplemented with 10% fetal bovine serum (FBS) and 2 mM l-Glutamine. COV504 cells were maintained in DMEM supplemented with 10% FBS and 2 mM l-Glutamine. Cells from each cell line were prepared following the 10x Genomics Cell Preparation Guide (CG000053_CellPrepGuide_RevC) and combined at equal cell concentrations prior to loading onto the 10x Genomics Chromium Controller at a concentration of 1000 cells/μL. cDNA generated through the single-cell platform was then split for single-cell targeted LR enrichment (see below) or for generating scRNA-seq SR libraries using the 10x v3.1 protocol (CG000204_ChromiumNextGEMSingleCell3’v3.1_RevD).
Dissociated tumor cells
Ovarian cancer dissociated tumor cells were purchased through Discovery Life Sciences (Huntsville, AL). Both samples were from untreated female ovarian cancer patients with stage-III cancer subtypes of Clear Cell Carcinoma and High-Grade Serous Carcinoma. Cells were thawed and prepared following the recommended 10x Genomics Cell Preparation Guide shown above with minor adjustments. Cells were thawed for 2 min and placed into 15 mL of warm RPMI media containing 10% FBS media. Cells were spun at 300× g for 5 min. DNase I was added after the first spin to prevent clumping. Three additional spins were performed with 1X PBS with 0.04% BSA to ensure proper removal of DNase I prior to 10x loading. Cells were counted and checked for viability using Vi-Cell XR (Beckman Coulter). The viability was 88.3% and 82.5% and the target capture was for 6000 cells prior to injection. Both the cell lines and primary tumor cells were run on Chip G using the 10x v3.1 kit for generating the cDNA (CG000204_ChromiumNextGEMSingleCell3’v3.1_RevD). The cDNA amplification step was modified by extending the elongation time to 2 min rather than the recommended 1 min. cDNA generated through the droplet single-cell platform was then split for either LR enrichment or for preparing scRNA-seq SR libraries using the 10x v3.1 protocol (CG000204_ChromiumNextGEMSingleCell3’v3.1_RevD).
Illumina library generation and sequencing
Whole transcriptome SR libraries were dual-indexed and sequenced paired-end on the Illumina NovaSeq 6000 p with the recommended 10x run parameters (Read 1—28 cycles, i7—10 cycles, i5—10 cycles, and Read 2—90 cycles). Targeted SR libraries were dual-indexed and sequenced paired-end on the Illumina NextSeq 2000 following the same run parameters as shown above.
Single-cell-targeted gene enrichment for long-read sequencing
Pre-amplification
In total, 10 ng of the 10x cDNA derived from the cell lines were split into two reactions and amplified an additional five cycles of PCR using two customized primers: (1) TruSeq Read1 forward primer 5’ (Fwd_partial_read1) and (2) partial TSO reverse primer (Rev_partial_TSO) (Supplementary Table 3). The PCR reaction was carried out using 2X LongAmp Taq (NEB) with the following PCR parameters 94 °C for 3 min, with five cycles of 94 °C 30 s, 60 °C 15 s, and 65°C for 3 min, with a final extension of 65 °C for 5 min. The cDNA was then purified using 0.8× SPRI beads to remove unwanted primers and eluted in 30 μL H2O to yield ~300 ng total.
Targeted gene panels
Enrichment was performed with the Human Pan-Cancer Panel (PN-1000247) and Human Immunology Panel (PN-1000259) from 10x Genomics. Biotinylated probes from these panels are 120 bases in length and tile across annotated exons at 1× density.
R2C2
Post-enriched cDNA was used for input into an R2C2 reaction following the protocol previously described19. Briefly, 100 ng of the targeted cDNA was circularized using Gibson assembly (NEBBuilder HiFi DNA assembly mix) with a custom splint that is compatible with 10x cDNA containing both the Read1 (10X_UMI_Splint_Forward) and TSO sequences (10X_UMI_Splint_Reverse) (Supplementary Table 3). Any non-circularized byproducts were then digested using an exonuclease mixture of Lambda, Exo I and Exo III (NEB) and incubated at 37 °C overnight. Post overnight digestion the reaction was cleaned up with 0.8X SPRI and eluted in 30 μL. The circularized product was separated into three different reactions and amplified using rolling circular amplification using Phi29 (NEB) and incubated at 30 °C overnight. To debranch the Phi29 product, a T7 endonuclease (NEB) digestion was performed and incubated at 37 °C on a thermal shaker at 1000 RPM for 2 h. A final 0.5× SPRI purification is performed to enrich longer molecules >500 bp, about 1 μg should be recovered.
Cell lines
The pan-cancer gene panel (n = 1253 genes) was designed by 10x Genomics containing 120 bp probes tiled across known annotated exons covering both sense and antisense strands. The cDNA hybridization using the pre-designed panels was performed following the 10x protocol (CG000293_TargetedGeneExpression_SingleCell_UG_RevF) with minor changes. We incorporated TSO blockers (1 μM) during Step 1.1 in the pre-hybridization pooling and drying step (Supplementary Table 3). The pre-hybridization was carried out using 300 ng of cDNA, 20 μL of COT DNA, 0.8 μL of TSO blockers, and 2 μL of Universal Blockers. The samples were dried using the SpeedVac Savant DNA120 concentrator (Thermo Fisher Scientific) on “Medium” setting. Following the hybridization, five cycles of PCR were performed using the same cDNA primers described in the Pre-Amplification step (1) Fwd_partial_read1 and (2) Rev_partial_TSO to amplify molecules off the bead. The following PCR conditions were the same as described in the Pre-Amplification step.
Dissociated tumor cells
For the primary tumor samples, the gene enrichment was performed as discussed above with the exception that the same cDNA was separated into two enrichments one using the pan-cancer gene panel (n = 1253 genes) and the other with the Immune gene panel (n = 1056 genes). The samples were targeted following the 10x protocol (CG000293_TargetedGeneExpression_SingleCell_UG_RevF) with minor changes as indicated above incorporating TSO blockers. Following the hybridization, 5 cycles of PCR were performed using the non-biotinylated primers (1) Fwd_3580_partial_read1_defined and (2) Rev_PR2_partial_TSO_defined from the single-cell ONT protocol (Supplementary Table 3, single-cell-transcriptomics-10x-SST_v9148_v111_revB). The PCR reaction was carried out using 2× LongAmp Taq (NEB) with the following PCR parameters 94 °C for 3 min, with five cycles of 94 °C 30 s, 60 °C 15 s, and 65 °C for 3 min, with a final extension of 65 °C for 5 min. The post cDNA hybridized product was then purified with 0.8× SPRI beads to remove unwanted primers and eluted in 40 μL of H2O. cDNA concentration was measured using Qubit dsDNA HS kit and the size distribution analyzed using Tapestation D5000 Screen Tape (Agilent Technologies). A portion of the 10× cDNA generated from the dissociated tumor cells was not subjected to any gene enrichment (i.e. untargeted) but was cleaned up to remove unwanted TSO artifacts using the TSO artifact mitigation method discussed below using biotinylated primers.
TSO artifact mitigation
Post-hybridization artifact mitigation was performed using the biotinylated version of the forward primer from the ONT protocol, [Btn]Fwd_3580_partial_read1_defined (Supplementary Table 3). The PCR reaction was carried out using 2× LongAmp Taq (NEB) with the following PCR parameters 94 °C for 3 min, with three cycles of 94 °C 30 s, 60 °C 15 s, and 65 °C for 3 min, with a final extension of 65 °C for 5 min. Full-length cDNA was captured using 15 μL M270 streptavidin beads (Thermo Fisher Scientific) that were washed three times with SSPE buffer (150 mM NaCl, 10 mM NaH2PO4, and 1 mM EDTA) and resuspended in 10 μL of 5× SSPE buffer (750 mM NaCl, 50 mM NaH2PO4, and 5 mM EDTA). The cDNA obtained from the gene enrichment step was combined with 10 μL M270 beads and incubated at room temperature for 15 min. After incubation, the cDNA-bead conjugate was washed twice with 1 mL of 1× SSPE. A final wash was performed with 200 μL of 10 mM Tris-HCl (pH 8.0) and the beads bound to the sample were resuspended in 10 μL H2O. A final PCR was performed on-bead using the cDNA primers (cPRM) from the SQK-PCS111 kit following the PCR conditions from the single-cell ONT protocol (single-cell-transcriptomics-10x-SST_v9148_v111_revB). The cDNA was cleaned up with 0.8X SPRI and eluted in 15 μL. The concentration and quality of the sample was evaluated with Qubit dsDNA HS kit and Tapestation D5000 Screen Tape (Agilent Technologies). The expected recovery was above 50 ng.
ONT library preparation and nanopore sequencing
Cell lines
For the mixed ovarian cell lines, library preparation for nanopore sequencing was performed according to the LSK-109 protocol (ONT). For the targeted mixed ovarian cell line samples, the final libraries (targeted, targeted+AM, and targeted+R2C2) were loaded onto a total of seven MinION flowcells (FLO-MIN106D). Approximately 25–30 fmol of the library was loaded for each run. The samples were sequenced for 72 h and basecalled using Guppy v6.0.1. For the untargeted sample, library preparation was performed according to the LSK-110 protocol. A total of 125 fmol was loaded onto a single PromethION flowcell (FLO-PR002), sequenced for 72 h and basecalled using Guppy v6.0.1.
Dissociated tumor cells
After post enrichment and artifact mitigation the rapid adapter addition was performed following SQK-PCS111 protocol. Final libraries (125 fmol per library) across both patient samples were loaded onto a total of 4 PromethION flowcells (FLO-PRO002). The samples were sequenced for 72 h and basecalled using Guppy v6.0.1. Libraries from the untargeted cDNA were prepared using the SQK-PCS111 protocol and loaded onto two PromethION flowcells (FLO-PR002). The samples were sequenced for 90 h and were basecalled using Guppy v6.01.
Long-read CB and UMI assignment
“SiCeLoRe” (https://github.com/ucagenomix/sicelore/commit/b057aa0f7948d2e8f64140b8ec99c2f3bb4b6d53) was used with default settings to process reads when companion SR data were available. When considering complete reads, 79.5% (average of two replicates) could be assigned to a known cell barcode. Of those, ~68% were matched to UMIs identified from SR data. These values are consistent with recent single-cell nanopore LR sequencing efforts. Next, for CB/UMI assignment without companion SR data, we used wf-single-cell (https://github.com/epi2me-labs/wf-single-cell; v0.1.5) with default settings. UMI-deduplication of the resultant tagged bam file was performed using UMI-tools (v1.1.0) with the following settings for the group: --per-cell --per-gene --extract-umi-method=tag --umi-tag=UB --cell-tag=CB --gene-tag=GN. The longest read was retrained from each UMI group. In general, we used the GRCh38 human reference genome and GENCODE v32/Ensembl 98 annotations provided by 10x Genomics (2020-A; July 7, 2020; https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest).
Single-cell data analysis
Cell-by-transcript count matrices were generated directly from SiCeLoRe or from IsoQuant (v3.1.0) with default settings using CB-tagged bam files generated by wf-single-cell. The count matrices were processed using scanpy (v1.9.1) as follows: (1) normalize counts per cell (target_sum=106), (2) log1p transform, and (3) scale to unit variance and zero mean. Unsupervised clustering of cell subgroups was performed using the Leiden algorithm applied to the neighborhood graph of principal components. Cluster annotation was based on marker geneset expression. Differential expression of both genes and transcripts computed using two-sided Welch’s t test (method = “t-test_overestim_var”). Geneset expression scores were calculated using the score_genes function from scanpy. Pathway activity scores were calculated using the progeny function (z_scores=TRUE, organism = “Human”, top=300, perm=100) from PROGENy (v1.18.0). For genetic-deconvolution of cell line identity, we used souporcell (v2.0) with known genotypes provided as a BCFtools-merged Clair3-derived.vcf file. Cell multiplets were identified using Scrublet (v0.2.3), implemented within scanpy with default settings. To integrate the expression matrices from the pan-cancer and immune panels, we applied a scalar offset. From the transcripts of 258 genes shared between the two panels, the scalar offset was computed as the mean slope of tenfold cross-validated (CB-shuffled) linear regression slopes (sklearn v1.0.1) using mean transcript expression (cell count-normalized and log1p-transformed). Subsequently, single-cell transcript expression values corresponding to the response variables were multiplied by the scalar offset. To construct the integrated expression matrix, scaled transcript expression values private to the response variables were joined (CB-matched) to the expression matrix corresponding to the predictor variables. Pseudobulk expression was calculated as mean log(CPM + 1).
T-cell receptor reconstruction
The immune enrichment panel design comprises probes targeting the constant TCR genes: TRAC, TRDC, TRBC2, and TRGC1. The TRBC1 and TRGC2 genes were not included in the panel as they have high homology to selected probes. scTaILoR-seq reads were processed by TRUST4 using the parameters --ref human_IMGT + C.fa --barcode CB --UMI UB.
Fusion analysis
Fusion analysis was performed with JAFFA using the default parameters of JAFFAL.groovy. Only fusions with the classification “HighConfidence” were considered for enumeration across untargeted LR sequencing and scTaILoR-seq. Fold-change enrichments were determined from fusions classified as “HighConfidence”, contained within the Mitelman database (known = “True” designation) and had ≥2 spanning reads.
Variant analysis
Cell subpopulation reads were aggregated from CB-tagged bam records using pysam (v0.16.0.1), and then variants were called using Clair3 (v0.1-r11) with pretrained model r941_prom_hac_g360 + g422 (--platform=ont --enable_phase --fast_mode). For analysis of variants associated with transcript structural divergence, Clair3-derived variant calls were filtered (within cancer panel exons, DP > = 100 and QUAL > = 15). Variant calls were scored for cryptic splicing using SpliceAI (v1.3.1, -D 500). Then, for each variant, aligned reads were partitioned by observed base-matching REF or ALT values (via pysam). Read coverage of resultant REF- and ALT-specific bam files were computed using bamCoverage (v3.5.0, --binSize 1). The Pearson correlation coefficient (r) between REF- and ALT-specific read coverage was calculated (minimum depth > =50). The degree of transcript structural divergence was defined as the variance unexplained (1 − r2). For variants exhibiting non-zero coverage divergence, linear regression residuals between REF- and ALT-specific coverage at single-base resolution were mapped to annotated transcript structural features: CDS and UTR/Intron. Then, the proportion of bases with residual z-score >0.5 within each structural feature was max-normalized per variant before agglomerative hierarchical clustering (method = “ward”, metric = “euclidean”) using SciPy (v1.7.3).
Haplotype analysis
Only reads with at least two detected SNVs were considered for haplotype reconstruction. For each gene, the observed variant status of each read was encoded as a vector of position-sorted SNV sites (n=number of detected SNVs within the gene) with the following values: undetermined=0, REF = 1, ALT = 2. The SNV vector with the highest read count was used as the seed haplotype. For each element in this seed vector equal to 0 (i.e., undetermined), the variant status was determined as follows:
-
1.
Identify all reads that contain at least one determined SNV site (REF = 1 or ALT = 2) from the current SNV vector in addition to the undetermined site.
-
2.
Update variant status at undetermined site based on highest frequency nucleotide identity (REF or ALT) at that position.
Haplotype reconstruction was complete when all SNV sites were determined. Then, allele-specific reads with a majority of SNVs (>50%) matching the haplotype were masked before a second haplotype was determined as outlined above. The final allele-specific read annotations were similar to above (i.e., majority of haplotype-matching SNVs per read) but omits SNV sites with shared identity between H1 and H2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw Illumina (short-read) and Oxford Nanopore Technologies (long-read) sequencing data were deposited to NCBI Sequence Read Archive (SRA) under the BioProject accession PRJNA993664. Supplementary Fig. source data will be made available on request. Source data are provided with this paper.
Code availability
The code used to analyze data from this study has been deposited in the GitHub repository: https://github.com/danledinh/sctailor-tools61.
References
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008).
Jiang, W. & Chen, L. Alternative splicing: human disease and quantitative analysis from high-throughput sequencing. Comput. Struct. Biotechnol. J. 19, 183–195 (2021).
Sterne-Weiler, T. & Sanford, J. R. Exon identity crisis: disease-causing mutations that disrupt the splicing code. Genome Biol. 15, 201 (2014).
Steijger, T. et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10, 1177–1184 (2013).
Tilgner, H. et al. Accurate identification and analysis of human mRNA isoforms using deep long read sequencing. G3 3, 387–397 (2013).
Sharon, D., Tilgner, H., Grubert, F. & Snyder, M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 31, 1009–1014 (2013).
Xu, Q., Modrek, B. & Lee, C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 30, 3754–3766 (2002).
Olivieri, J. E. et al. RNA splicing programs define tissue compartments and cell types at single-cell resolution. eLife 10, e70692 (2021).
Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019).
Argyriou, A. et al. Single cell sequencing identifies clonally expanded synovial CD4 T cells expressing GPR56 in rheumatoid arthritis. Nat. Commun. 13, 4046 (2022).
Olah, M. et al. Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat. Commun. 11, 6129 (2020).
Smajić, S. et al. Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain 145, 964–978 (2022).
Vázquez-García, I. et al. Ovarian cancer mutational processes drive site-specific immune evasion. Nature 612, 778–786 (2022).
Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021).
Byrne, A. et al. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 8, 16027 (2017).
Lebrigand, K., Magnone, V., Barbry, P. & Waldmann, R. High throughput error corrected Nanopore single cell transcriptome sequencing. Nat. Commun. 11, 4025 (2020).
Singh, M. et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun. 10, 3120 (2019).
Gupta, I. et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. https://doi.org/10.1038/nbt.4259 (2018).
Volden, R. et al. Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. Proc. Natl. Acad. Sci. USA 115, 9726–9731 (2018).
Rebboah, E. et al. Mapping and modeling the genomic basis of differential RNA isoform expression at single-cell resolution with LR-Split-seq. Genome Biol. 22, 286 (2021).
Karlsson, K. & Linnarsson, S. Single-cell mRNA isoform diversity in the mouse brain. BMC Genomics 18, 126 (2017).
Shiau, C.-K. et al. High throughput single cell long-read sequencing analyses of same-cell genotypes and phenotypes in human tumors. Nat. Commun. 14, 4124 (2023).
Hardwick, S. A. et al. Single-nuclei isoform RNA sequencing unlocks barcoded exon connectivity in frozen brain tissue. Nat. Biotechnol. 40, 1082–1092 (2022).
Tian, L. et al. Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol. 22, 310 (2021).
Thijssen, R. et al. Single-cell multiomics reveal the scale of multilayered adaptations enabling CLL relapse during venetoclax therapy. Blood 140, 2127–2141 (2022).
Cortés-López, M. et al. Single-cell multi-omics defines the cell-type-specific impact of splicing aberrations in human hematopoietic clonal outgrowths. Cell Stem Cell 30, 1262–1281.e8 (2023).
Volden, R. & Vollmers, C. Single-cell isoform analysis in human immune cells. Genome Biol. 23, 47 (2022).
Davidson, N. M. et al. JAFFAL: detecting fusion genes with long-read transcriptome sequencing. Genome Biol. 23, 10 (2022).
Mitelman, F., Johansson, B. & Mertens, F. Mitelman database of chromosome aberrations and gene fusions in cancer. https://mitelmandatabase.isb-cgc.org (2023).
Riccio, A. A., Cingolani, G. & Pascal, J. M. PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucleic Acids Res. 44, 1691–1702 (2016).
Liu, C.-A., Wang, M.-J., Chi, C.-W., Wu, C.-W. & Chen, J.-Y. Rho/Rhotekin-mediated NF-kappaB activation confers resistance to apoptosis. Oncogene 23, 8731–8742 (2004).
Bonnal, S. C., López-Oreja, I. & Valcárcel, J. Roles and mechanisms of alternative splicing in cancer-implications for care. Nat. Rev. Clin. Oncol. 17, 457–474 (2020).
Bradley, R. K. & Anczuków, O. RNA splicing dysregulation and the hallmarks of cancer. Nat. Rev. Cancer 23, 135–155 (2023).
Shim, S. et al. A paradoxical effect of interleukin-32 isoforms on cancer. Front. Immunol. 13, 837590 (2022).
Dondi, A. et al. Detection of isoforms and genomic alterations by high-throughput full-length single-cell RNA sequencing in ovarian cancer. Nat. Commun. 14, 7780 (2023).
Song, L. et al. TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods 18, 627–630 (2021).
Ahmed, A. A. et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol. 221, 49–56 (2010).
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Zheng, Z. et al. Symphonizing pileup and full-alignment for deep learning-based long-read variant calling. Nat. Comput. Sci. 2, 797–803 (2022).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Schubert, M. et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 (2018).
Glinos, D. A. et al. Transcriptome variation in human tissues revealed by long-read sequencing. Nature 608, 353–359 (2022).
Jaganathan, K. et al. Predicting splicing from primary sequence with deep learning. Cell 176, 535–548.e24 (2019).
Subbalakshmi, A. R. et al. The ELF3 transcription factor is associated with an epithelial phenotype and represses epithelial-mesenchymal transition. J. Biol. Eng. 17, 17 (2023).
Seo, S. H. et al. Hypoxia-induced ELF3 promotes tumor angiogenesis through IGF1/IGF1R. EMBO Rep. 23, e52977 (2022).
Deonovic, B., Wang, Y., Weirather, J., Wang, X.-J. & Au, K. F. IDP-ASE: haplotyping and quantifying allele-specific expression at the gene and gene isoform level by hybrid sequencing. Nucleic Acids Res. 45, e32 (2017).
Hagemann-Jensen, M. et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 38, 708–714 (2020).
Altmann, D. M. & Trowsdale, J. Major histocompatibility complex structure and function. Curr. Opin. Immunol. 2, 93–98 (1989).
Robles-Espinoza, C. D., Mohammadi, P., Bonilla, X. & Gutierrez-Arcelus, M. Allele-specific expression: applications in cancer and technical considerations. Curr. Opin. Genet. Dev. 66, 10–19 (2021).
MacGregor, H. L. et al. High expression of B7-H3 on stromal cells defines tumor and stromal compartments in epithelial ovarian cancer and is associated with limited immune activation. J. Immunother. Cancer 7, 357 (2019).
Shi, Z.-X. et al. High-throughput and high-accuracy single-cell RNA isoform analysis using PacBio circular consensus sequencing. Nat. Commun. 14, 2631 (2023).
Biasco, L. et al. Clonal expansion of T memory stem cells determines early anti-leukemic responses and long-term CAR T cell persistence in patients. Nat. Cancer 2, 629–642 (2021).
Valpione, S. et al. The T cell receptor repertoire of tumor infiltrating T cells is predictive and prognostic for cancer survival. Nat. Commun. 12, 4098 (2021).
Tsuji, T. et al. Clonality and antigen-specific responses shape the prognostic effects of tumor-infiltrating T cells in ovarian cancer. Oncotarget 11, 2669–2683 (2020).
Sessegolo, C. et al. Transcriptome profiling of mouse samples using nanopore sequencing of cDNA and RNA molecules. Sci. Rep. 9, 14908 (2019).
Byrne, A. T. et al. Vascular endothelial growth factor-trap decreases tumor burden, inhibits ascites, and causes dramatic vascular remodeling in an ovarian cancer model. Clin. Cancer Res. 9, 5721–5728 (2003).
Hu, L. et al. Vascular endothelial growth factor trap combined with paclitaxel strikingly inhibits tumor and ascites, prolonging survival in a human ovarian cancer model. Clin. Cancer Res. 11, 6966–6971 (2005).
Grimes, S. M. et al. Single-cell multi-gene identification of somatic mutations and gene rearrangements in cancer. NAR Cancer 5, zcad034 (2023).
Weilguny, L. et al. Dynamic, adaptive sampling during nanopore sequencing using Bayesian experimental design. Nat. Biotechnol. 41, 1018–1025 (2023).
Martin, S. et al. Nanopore adaptive sampling: a tool for enrichment of low abundance species in metagenomic samples. Genome Biol. 23, 1–27 (2022).
Byrne, A. et al. Single-cell long-read targeted sequencing reveals transcriptional variation in ovarian cancer. Danledinh/sctailor-Tools: Initial Release. Zenodo https://doi.org/10.5281/ZENODO.12587533 (2024).
Acknowledgements
The authors would like to thank Chris Rose, Carolina Galan, Andrew McKay, and Spyridon Darmanis for critical review of the manuscript. The authors would also like to thank Oleg Mayba and Bing Wu for helpful discussions regarding analysis.
Author information
Authors and Affiliations
Contributions
A.B. and W.S. conceived the study. K.S. provided ovarian cell lines for single-cell RNA-seq. H.M. assisted with single-cell RNA-seq. A.B., W.S., and J.L. performed long-read sequencing. N.P., J.L., A.X.-M., and M.S. performed short-read sequencing. D.L., A.B., W.S., S.V., and T.S.-W. performed analysis. W.S., Z.M., and Y.L. supervised the project. W.S., D.L., Z.M., and A.B. prepared the manuscript. All authors discussed the results and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing interests: All the authors are current or previous employees and shareholders of Roche/Genentech.
Peer review
Peer review information
Nature Communications thanks James Brenton who co-reviewed with Thomas Bradley and Ana Conesa for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Byrne, A., Le, D., Sereti, K. et al. Single-cell long-read targeted sequencing reveals transcriptional variation in ovarian cancer. Nat Commun 15, 6916 (2024). https://doi.org/10.1038/s41467-024-51252-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51252-6
- Springer Nature Limited