Abstract
Resistance to radiotherapy is a major barrier during cancer treatment. Here using genome-scale CRISPR/Cas9 screening, we identify CD274 gene, which encodes PD-L1, to confer lung cancer cell resistance to ionizing radiation (IR). Depletion of endogenous PD-L1 delays the repair of IR-induced DNA double-strand breaks (DSBs) and PD-L1 loss downregulates non-homologous end joining (NHEJ) while overexpression of PD-L1 upregulates NHEJ. IR induces translocation of PD-L1 from the membrane into nucleus dependent on deglycosylation of PD-L1 at N219 and CMTM6 and leads to PD-L1 recruitment to DSBs foci. PD-L1 interacts with Ku in the nucleus and enhances Ku binding to DSB DNA. The interaction between the IgC domain of PD-L1 and the core domain of Ku is required for PD-L1 to accelerate NHEJ-mediated DSB repair and produce radioresistance. Thus, PD-L1, in addition to its immune inhibitory activity, acts as mechanistic driver for NHEJ-mediated DSB repair in cancer.
Similar content being viewed by others
Introduction
Immune checkpoint blockade has achieved great breakthroughs in the treatment of various cancers1,2,3. Programmed death ligand-1 (PD-L1) (i.e., B7-H1) is encoded by the CD274 gene and has become a critical target in cancers for immunotherapy4. The binding between PD-L1 and PD-1, via the front sheet of PD-1 and three main hot spots on PD-L15, transmits an inhibitory signal that reduces proliferation of T cells and promotes T cell apoptosis leading to suppression of the T cell receptor pathway and T cell-mediated immune activity6,7,8. PD-L1 is extensively overexpressed in various types of cancer, including human lung cancer9,10,11. High expression of PD-L1 not only allows cancers to escape the host immune system7, but also increases tumor aggressiveness with poor prognosis of patients under conventional therapy9,12. Posttranslational modifications, including N-linked glycosylation, phosphorylation and acetylation, have recently been demonstrated to regulate PD-L1 stability, expression level and subcellular localization13,14,15. Glycosylation stabilizes PD-L1 protein and promotes its binding to PD-1. Deglycosylation facilitates GSK-3β-induced phosphorylation of PD-L1 and subsequent β-TrCP E3 ligase-mediated ubiquitination and degradation15.
DNA damage/repair has recently been found to be tightly linked to the immune system, which plays an important role in checkpoint blockade-based immunotherapy16,17. Ionizing radiation (IR)-induced DNA double-strand breaks (DSBs) have been reported to upregulate PD-L1 in cancer cells, which requires ATM/ATR/Chk1 kinases17. Treatment of mice with anti-PD-L1 enhances the efficacy of IR through a cytotoxic T cell-dependent mechanism18. Combination of anti-PD-L1 with radiotherapy overcomes radioresistance in a mouse model of breast cancer19. Non-homologous end joining (NHEJ) and homologous recombination (HR) are two major DSB repair pathways20. The pathway choice between NHEJ and HR is largely dependent on the stage of the cell cycle21. NHEJ can function in all phases of the cell cycle but is dominant in G1 and early S phases while HR operates only in S and G2 phases when a sister chromatid is available during DNA replication20,22,23. The binding between Ku and DSB ends is critical to initiate the NHEJ pathway, which can be promoted by 53BP1, RIF1 and shieldin DNA end resection inhibitors24,25,26. PD-L1 upregulation is associated with activation of the DSB repair pathway27, but the mechanism involved remains unknown. Since IR-induced DSBs are mainly repaired by NHEJ while PD-L1 is upregulated upon IR-induced DSBs17, PD-L1 may play an important role in the NHEJ-mediated repair of IR-induced DSBs and the development of radioresistance in cancer cells.
CRISPR/Cas9 gene editing technology has provided a powerful tool for genome-scale knockout screening due to the ease of generating gRNAs and the specificity of Cas9-sgRNA complexes in knocking out the expression of target genes28,29,30. Genome-scale CRISPR/Cas9 screening can identify specific genes that confer resistance of cancer cells to radiotherapy31, chemotherapy32, targeted therapy33, and immunotherapy34. Here we identify the CD274 (PD-L1) gene as a major radiation resistance gene in human lung cancer cells by genome-scale loss-of-function screening with the CRISPR/Cas9 system and a gRNA pooled library. IR-induced DSBs facilitate PD-L1 deglycosylation and translocation into the nucleus, leading to resistance of lung cancer to radiotherapy through acceleration of NHEJ-mediated DNA double-strand-break repair.
Results
Genome-scale CRISPR/Cas9 knockout screening of radiation-resistant genes in human lung cancer cells
To systemically investigate the genes that confer resistance to IR in non-small cell lung cancer (NSCLC) cells, we performed CRISPR/Cas9-based loss-of-function genetic screening for negatively selected genes whose loss synergizes with IR to kill lung cancer cells. H460 cells were transduced with the sgRNA GeCKO v2 library for gene knock-out in the human genome as detailed in “Methods”. After puromycin selection, cells were divided into two groups. One group was treated with 4 Gy of IR (X-ray); the second group received no treatment (control). After 7 days in cell culture, genomic DNA was isolated, followed by sequencing the distribution of sgRNAs (Fig. 1a). Hierarchical clustering analysis shows the sgRNAs associated with decreased expression profile (blue gradient) or increased expression profile (red gradient) in cells treated with IR compared to the control (Supplementary Fig. 1). Analysis with volcano plot further screened out the significantly decreased (n = 85) or increased (n = 47) sgRNAs, the corresponding genes of which confer H460 cells resistance to IR (negatively selected) or synergy with IR (positively selected), respectively (Fig. 1b). Several genes that are involved in DSB repair were among the negatively selected genes (Fig. 1b, left), such as NELFB35, DDX1136, SPIDR37, STAG338, and NPM139. Intriguingly, CD274 gene, that encodes PD-L1, was among the negatively selected genes (genes mediating resistance to IR) (Fig. 1b, green spot), indicating that CD274 (PD-L1) loss sensitizes IR to kill lung cancer H460 cells. We also analyzed the negatively selected genes using the RRE algorithm. Among 437 negatively selected genes, CD274 was consistently located at a high rank (# 5) (Fig. 1c and Supplementary Data 1). We further clustered the screened-out genes into different biological process using hypergeometric test (HGT). Results reveal that the genes at high-ranking positions are positive regulators of DNA double-strand breaks, G2/M transition, and transcriptional elongation (Fig. 1d), indicating IR-treated cells are under DNA damaging conditions. Importantly, negative regulators of CD8 positive T cell activation, including the CD274 (PD-L1) gene, were also at high rank positions (Fig. 1d). PD-L1 is a critical immune checkpoint inhibitory protein that binds to PD-1 on T cells and mediates the immune escape of tumor cells40. Our findings from CRISPR loss-of-function genetic screening indicate that PD-L1, in addition to its immune inhibitory activity, may also play a critical role in the development of radioresistance by regulating DSB repair pathway in human lung cancer cells.
a Schematic illustration of the CRISPR/Cas9 loss-of-function screen in human lung cancer H460 cells. b Volcano plot showing both negatively (left) and positively (right) selected genes at day 7 after radiation treatment (4 Gy). CD274 gene that encodes PD-L1 is highlighted in green. c Negative selection of CRISPR screen data ranked based on RRE method. d Differentially selected sgRNAs from (b) were subject to a functional enrichment analysis using over-representation test (ORT) as implemented in MAGeCKFlute.
PD-L1 contributes to radioresistance, and depletion of PD-L1 restores radiosensitivity and retards DSB repair
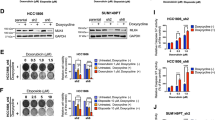
PD-L1 is widely expressed in various types of cancer, including lung cancer, breast cancer, melanoma, colon cancer, bone osteosarcoma, etc15,17. (Supplementary Fig. 2a. See complete unedited blots in Source Data file). According to our CRISPR/Cas 9 screening data (Fig. 1), PD-L1 confers lung cancer cells resistance to IR. To further test the role of PD-L1 in IR resistance, we have established the ionizing radiation-resistant A549 (A549-IRR) cell line as described previously41. Compared to A549 parental cells, increased levels of PD-L1 were observed in A549-IRR (Supplementary Fig. 2b, c). Intriguingly, knockout of PD-L1 from A549-IRR cells restored radiosensitivity (Supplementary Fig. 2d–f). These findings further indicate that PD-L1 confers lung cancer cells resistance to IR. To further test the effect of endogenous PD-L1 on radiosensitivity in various human lung cancer cells, endogenous PD-L1 in H358 and H460 cells was stably depleted by knockout using CRISPR/Cas9 system, followed by IR exposure and colony formation assay. Depletion of endogenous PD-L1 not only reduced cell growth without IR, but also sensitized H460 and H358 cells to IR at various doses (1–4 Gy) (Fig. 2a–f). Importantly, the PD-L1 knockout-induced radiation sensitization could be alleviated by expression of exogenous PD-L1 (Fig. 2a–f). Similar findings of PD-L1 reduction of radiosensitivity in human breast cancer, colorectal cancer and NSCLC cells were also reported by other investigators42,43, indicating this is a general response and not a cell type-specific phenomenon.
a Endogenous PD-L1 was knocked out from H460 cells using the CRISPR/Cas9 system. PD-L1 was rescued by transfection with exogenous WT PD-L1. PD-L1 expression levels were analyzed by western blot. b, c H460 parental, H460 PD-L1 knockout cells and H460 PD-L1 knockout cells expressing exogenous WT PD-L1 were treated with increasing doses of IR, followed by colony formation analysis. Error bars represent ± s.d., n = 3 per group. ***P < 0.001, ****P < 0.0001, by two-tailed t test. d Endogenous PD-L1 was knocked out from H358 cells. PD-L1 was rescued by transfection with exogenous WT PD-L1. PD-L1 expression levels were analyzed by western blot. e, f H358 parental, H358 PD-L1 knockout cells and H358 PD-L1 knockout cells expressing exogenous WT PD-L1 were treated with increasing doses of IR, followed by colony formation analysis. Error bars represent ± s.d., n = 3 per group. ***P < 0.001, by two-tailed t test. g, h H460 parental and PD-L1 knockout H460 cells were treated with 4 Gy of IR at 2 Gy/min dose rate, followed by immunofluorescence analysis of γH2AX foci and DAPI at various time points. Foci ≥5/cell were counted as foci-positive cells. Error bars represent ± s.d., n = 3 per group. ***P < 0.001, by two-tailed t test. Source data are provided as a Source Data file.
To assess whether the effect of PD-L1 on radiation resistance results from its role in regulating DSB repair, γH2AX foci in both H460 parental and H460 PD-L1 knockout cells were compared at various time points following IR (4 Gy) exposure. In parental H460 cells, γH2AX signals rapidly vanished within 2 h after IR treatment (Fig. 2g, h). In contrast, about 50% of γH2AX signals could still be observed in H460 PD-L1 knockout cells at 4 h after IR (Fig. 2g, h), suggesting that depletion of endogenous PD-L1 suppresses DSB repair. Similarly, knockout of endogenous PD-L1 from H358 cells resulted in retardation of DSB repair (Supplementary Fig. 2g, h). Comet assay confirmed that loss of PD-L1 negatively regulated the repair of IR-induced DNA damage in H460 cells (Supplementary Fig. 3), which supports the findings from γH2AX staining. These results suggest that PD-L1 can accelerate DSB repair after IR exposure, which may be the mechanism of IR resistance induced by PD-L1 in cancer cells.
IR induces nuclear translocation of PD-L1 in a manner that is dependent on deglycosylation of PD-L1 at N219
PD-L1 is an immune checkpoint type I transmembrane glycoprotein, which is mainly localized on the plasma membrane of cancer cells44. Since PD-L1 accelerates the repair of IR-induced DSBs (Fig. 2g), PD-L1 should be translocated into the nucleus before engagement in DSB repair. To test this possibility, H460 cells expressing exogenous GFP-tagged human PD-L1 were treated with IR (4 Gy), followed by observation of time-lapse images of living cells under fluorescence microscope. After IR exposure, PD-L1 was gradually entered the intracellular area (i.e., cytoplasm and nucleus) from the cell membrane in a time-dependent manner (Fig. 3a).
a H460 cells expressing GFP-tagged PD-L1 were treated with 4 Gy of IR at 2 Gy/min dose rate, followed by time-lapse imaging of GFP-PD-L1 in living cells. The pictures were captured every 5 min. b H460 cells were treated with 4 Gy of IR at 2 Gy/min dose rate, followed by isolation of cytoplasmic and nuclear fractions. PD-L1 was analyzed by western blot. α-Tubulin or PCNA was used as cytoplasmic or nuclear marker, respectively. **PD-L1: glycosylated form, *PD-L1: nonglycosylated form. c H460 cells expressing Flag-tagged PD-L1 were treated with glycosylation inhibitor tunicamycin (5 µg/ml) for various times, followed by immunofluorescence staining with anti-Flag antibody. d Schematic representation of the N → Q mutation at single- or multi-glycosylation site(s) in PD-L1. e, f Flag-tagged WT and various PD-L1 mutants were transfected into H460 cells, followed by western blot or immunofluorescence staining using anti-Flag antibody. **PD-L1: glycosylated form, *PD-L1: nonglycosylated form. Source data are provided as a Source Data file.
PD-L1 has been demonstrated to be an N-linked glycosylated protein, and its N-glycosylation shifts its molecular weight from 33 to 45 kDa15. Here we confirmed that PD-L1 is N-linked glycosylated since peptide-N-glycosidase F (PNGase F) can remove the N-glycans from PD-L1 leading to a change in molecular weight of PD-L1 from 45 to 33 kDa in both H460 and U2OS cells (Supplementary Fig. 4). Intriguingly, IR exposure also induced a change in PD-L1 molecular weight from 45 to 33 kDa (Fig. 3b). After IR exposure, levels of the 33 kDa form of PD-L1 in the nuclear and cytoplasmic fraction were significantly increased (Fig. 3b). These findings suggest that IR promotes PD-L1 deglycosylation, and the deglycosylated form (33 kDa) of PD-L1 mainly resides in the nucleus with a small portion in the cytoplasm. α-Tubulin was used as a cytoplasmic marker45, and PCNA was used as a nuclear marker46, respectively, to verify the purity of each fraction. It was recently reported that a heavily glycosylated PD-L1 (150 kDa) was observed in the nucleus in breast cancer MDA231 and colon carcinoma RKO cells45. To test whether IR induces the heavily glycosylated 150 kDa PD-L1 in human lung cancer cells, H460 and H358 cells were treated with IR (4 Gy), followed by analysis of PD-L1 via western blot. We did not observe the heavily glycosylated 150 kDa PD-L1 in the nucleus or cytoplasm in the presence or absence of IR exposure. However, IR significantly enhanced the 33 kDa deglycosylated PD-L1 in the nucleus and cytoplasm in both H460 and H358 cells (Supplementary Fig. 5). Tunicamycin is an N-linked glycosylation inhibitor that potently induces deglycosylation of PD-L1 leading to a reduction in molecular weight from 45 to 33 kDa15. To further test whether deglycosylation promotes PD-L1 nuclear localization, H460 or H460 PD-L1 knockout cells that express exogenous Flag-tagged PD-L1 were treated with tunicamycin (5μg/ml) for various times. Intriguingly, tunicamycin-induced deglycosylation led to translocation of PD-L1 from the membrane into the cytoplasm and nucleus (Fig. 3c and Supplementary Fig. 6). 2-deoxyglucose (2-DG) is a glucose analog that has recently been reported to decrease PD-L1 glycosylation47. It is interesting to test whether 2-DG promotes PD-L1 cytoplasm and/or nuclear localization. H460 cells were treated with 2-DG (5 mM) for 72 h, followed by immunostaining using PD-L antibody. Intriguingly, treatment of H460 cells with 2-DG also led to significant accumulation of PD-L1 in the nucleus and cytoplasm (Supplementary Fig. 7). Our findings indicate that the subcellular localization of PD-L1 mainly depends on its glycosylation/deglycosylation status. Four physiological glycosylation sites (i.e., N35, N192, N200 and N219) in PD-L1 have been recently identified by Dr. Hung’s research group15. To further test which glycosylation site(s) dictates PD-L1’s nuclear and cytoplasmic localizations, a series of asparagine (N) → glutamine (Q) PD-L1 mutants including N35Q, N192Q, N200Q, N219Q, N35Q/N192Q/N200Q (3NQ), N35Q/N192Q/N200Q/N219Q (4NQ) were created to abrogate either single-site or multisite glycosylation as shown in Fig. 3d. Flag-tagged PD-L1 WT and all mutants were exogenously expressed in H460 cells. A slight molecular weight decrease was observed in single-site mutants N35Q, N192Q, N200Q and N219Q as compared to WT. Significant molecular weight reductions were observed in multisite mutants 3NQ and 4NQ (Fig. 3e). The Flag-tagged WT and all mutants were exogenously expressed in H460 cells, followed by immunofluorescence using Flag antibody. Compared with WT PD-L1, mutation at single site N35 (N35Q), N192 (N192Q) or N200 (N200Q) or the compound mutations at these three sites (3NQ) did not change PD-L1’s membrane location (Fig. 3f). However, mutation at single site N219 (N219Q) or compound mutations at all four sites (4NQ) resulted in translocation of PD-L1 from the membrane into the nucleus and cytoplasm around the nucleus (Fig. 3f), indicating that glycosylation at N219 is essential for PD-L1’s membrane localization, and that deglycosylation at N219 leads to PD-L1 cytoplasmic and nuclear localizations. Similar experiments were performed in H460 PD-L1 knockout cells and HeLa cells expressing Flag-tagged WT or mutant PD-L1 and similar results observed (Supplementary Fig. 8). To further confirm the intracellular localization of WT and various PD-L1 mutants, co-immunofluorescent staining with anti-Flag, anti-HSP90B1 and DAPI was conducted. HSP90B1 was used as an endoplasmic reticulum (ER) marker as previously reported48. Consistently, WT, N35Q, N192Q, N200Q and 3NQ PD-L1 mutants were mainly localized on the membrane. The N219Q and 4NQ PD-L1 mutants were found to be co-localized with DAPI and HSP90B1 (Supplementary Fig. 9), indicating that the nonglycosylated N219Q and 4NQ mutant PD-L1 proteins are localized in the nucleus and ER. Thus, our data support and extend the findings from a previous report48. We also analyzed cell surface PD-L1 by FACS in H460 cells expressing various PD-L1 mutant(s) compared with WT or vector-only as described49. Consistently, only N219Q and 4NQ showed significantly less cell surface PD-L1 signal compared to WT (Supplementary Fig. 10).
The smaller band (33 kDa) of PD-L1 has been identified as deglycosylated PD-L1 and well characterized in a previous publication15. To further confirm whether the IR-induced smaller band is indeed deglycosylated PD-L1, H460 PD-L1 knockout cells expressing exogenous PD-L1 WT or nonglycosylated 4NQ mutant were treated with IR (4 Gy) or tunicamycin (TM, 5 μg/ml) for 24 h, followed by analysis of PD-L1. Intriguingly, when cells expressing WT PD-L1 were treated with IR or TM, the protein bands corresponding to WT PD-L1 shifted down to the same size as the nonglycosylated form (4NQ) while in cells expressing 4NQ mutant treated with IR or TM, the protein bands had no size change (Supplementary Fig. 11). These results indicate that both IR- and TM-induced smaller bands from WT-PD-L1 are indeed deglycosylated PD-L1.
We further performed fractionation analysis for PD-L1 from H460 PD-L1 knockout cells expressing exogenous WT or the glycosylation-deficient 4NQ mutant, with or without IR exposure. Results reveal that IR not only promoted WT PD-L1 downshift to the smaller band (33 kDa) like the nonglycosylated form (4NQ), but also increased localization of the smaller PD-L1 band in the cytoplasmic and nuclear fractions (Supplementary Fig. 12a), indicating that IR can induce WT PD-L1 deglycosylation and promote localization of the deglycosylated PD-L1 in the cytoplasm and nucleus. Intriguingly, the 4NQ mutant PD-L1 was localized in both cytoplasmic and nuclear fractions, but significantly higher levels of 4NQ were observed in the nuclear fraction than in the cytoplasm. IR had no significant effect on 4NQ mutant PD-L1 (Supplementary Fig. 12a). Co-immunofluorescence staining demonstrated that IR promoted co-localization of WT PD-L1 with ER marker HSP90B1 and nuclear marker DAPI (i.e., ER and nuclear localizations) (Supplementary Fig. 12b). The 4NQ mutant PD-L1 had ER and nuclear localizations in the absence or presence of IR (Supplementary Fig. 12b). To compare the half-life of 4NQ versus WT PD-L1, Flag-tagged WT PD-L1 and 4NQ mutant PD-L1 were exogenously expressed in H460 PD-L1 knockout cells, followed by cycloheximide (CHX) chase assay. The half-life of nonglycosylated 4NQ mutant PD-L1 (t1/2: 9.28 h) was significantly shorter than that of WT PD-L1 (t1/2:18.2 h) in human lung cancer cells (Supplementary Fig. 13). These findings indicate that deglycosylation reduces PD-L1 stability, which supports previous findings15.
In addition to IR, we also tested the effects of chemotherapeutic agents cisplatin, gemcitabine, and etoposide on PD-L1 nuclear translocation. H460 or H358 cells were treated with cisplatin (2 μM), gemcitabine (2 μM), or etoposide (50 μM) for 24 h, followed by immunofluorescence staining with anti-PD-L1. Results indicate that exposure of H460 cells to cisplatin, gemcitabine or etoposide also increased localization of PD-L1 in the nucleus (Supplementary Fig.14).
Radiation promotes interaction between PD-L1 and NGLY1 leading to PD-L1 deglycosylation
We and others discovered that PNGase can directly remove the N-glycans from PD-L1 leading to a change in molecular weight of PD-L1 from 45 to 33 kDa in cell lysates isolated from human lung cancer H460 and human osteosarcoma U2OS cells (Supplementary Fig. 4) or from breast cancer MDA-MB-231 and BT-549 cells15. NGLY1 (PNGase) is a de-N-glycosylating enzyme that plays an essential role in the release of intact N-glycans from N-glycosylated proteins50,51. To test how IR induces deglycosylation of PD-L1, H460 cells were exposed to IR (4 Gy), followed by co-IP using PD-L1 antibody at various time points. NGLY1 had a weak interaction with PD-L1 without IR exposure. IR significantly enhanced NGLY1/PD-L1 interaction after 5 min and maintained high levels of interaction for 30 min, which returned to a weak interaction by 60 min (Supplementary Fig. 15). Intriguingly, IR induced enhancement of NGLY1/PD-L1 interaction in association with downregulation of the glycosylated form (45 kDa) of PD-L1 and upregulation of deglycosylated form (33 kDa) of PD-L1 in a time-dependent manner (Supplementary Fig. 15). These findings suggest that IR-induced PD-L1 deglycosylation may occur through NGLY1 (PNGase).
Radiation-induced PD-L1 nuclear translocation occurs through CMTM6-mediated transportation
Radiation-induced accumulation of PD-L1 in the nucleus may have two potential sources, translocation from cell plasma membrane or new protein synthesis. To test these possibilities, H460 cells were treated with IR (4 Gy) in the absence or presence of cycloheximide (CHX, 20 µg/mL) for various times. PD-L1 in cytoplasmic and nuclear fractions was analyzed at various time points, and cytoplasmic PD-L1 was found to be the glycosylated form (45 kDa) while nuclear PD-L1 was the deglycosylated form (33 kDa) (Supplementary Fig. 16). Radiation downregulated the glycosylated form of PD-L1 in the cytoplasm and upregulated the deglycosylated form in the nucleus in a time-dependent manner (Supplementary Fig. 16). Levels of PD-L1 in the nuclear fractions had no significant change in the absence or presence of CHX (Supplementary Fig. 16). These results indicate that IR-induced nuclear PD-L1 is translocated from the cell membrane rather than newly synthesized.
CMTM6 has recently been found to directly interact with PD-L1 at the plasma membrane and in recycling endosomes to prevent PD-L1 degradation49,52. To test whether CMTM6 interacts with and transports PD-L1 from the cytoplasm into the nucleus following radiation exposure, we employed a Duolink® proximity ligation assay (PLA) system to detect PD-L1/CMTM6 interactions in H460 cells according to the manufacturer’s instructions as we recently described53. Two primary antibodies raised in different species (i.e., PD-L1 antibody from rabbit and CMTM6 antibody from mouse) are used to detect two unique protein targets. A pair of oligonucleotide-labeled secondary antibodies (PLA probes) then binds to the primary antibodies. After PLA, we stained samples using α-tubulin antibody as contrast imaging and quantified PLA signals. IgG or single PD-L1 or CMTM6 antibody alone were used as negative controls and no discrete spots were observed (Fig. 4a, upper panel). IR significantly enhanced discrete spots (PLA signals) in H460 cells (Fig. 4a, lower panel), indicating IR promotes PD-L1/CMTM6 interactions. Small number of PD-L1/CMTM6 interactions occurred only in the cytoplasm but not in the nucleus in untreated cells. However, PD-L1/CMTM6 interactions gradually increased and moved into the nucleus in a time-dependent manner following IR exposure (Fig. 4a, lower panel), indicating that CMTM6 may transport PD-L1 from the cytoplasm into the nucleus through direct interaction following IR. Co-IP experiments further demonstrated that IR promoted PD-L1/CMTM6 interactions in H460 cells, and CMTM6 can interact with both glycosylated (45 kDa) and deglycosylated (33 kDa) forms of PD-L1 (Fig. 4b). To further test the effect of CMTM6 on PD-L1 nuclear translocation, CMTM6 was depleted from H460 cells using CMTM6 siRNA (Fig. 4c), followed by IR treatment. Silence of CMTM6 not only reduced nuclear localization of PD-L1 in untreated cells, but also significantly blocked IR-induced nuclear translocation of PD-L1 (Fig. 4d), suggesting that CMTM6 is an essential transporter for PD-L1 nuclear translocation.
a PD-L1/CMTM6 interactions were analyzed by PLA at various times following IR exposure in H460 cells. PLA signals were detected by fluorescence microscopy as discrete spots. IgG, single PD-L1 antibody alone or single CMTM6 antibody alone was used as negative control. After PLA, immunofluorescence staining using α-tubulin antibody was performed as phase contrast imaging. Error bars represent ± s.d., n = 10 per group. ****P < 0.0001, by two-tailed t test. b PD-L1/CMTM6 interactions were analyzed by co-IP using PD-L1 antibody following IR exposure in H460 cells. **PD-L1: glycosylated form, *PD-L1: nonglycosylated form. c Ctrl siRNA or CMTM6 siRNA was transfected into H460 cells, followed by western blot using CMTM6 antibody. d H460 cells expressing Ctrl siRNA or CMTM6 siRNA were treated with IR (4 Gy), followed by immunofluorescence staining with PD-L1 antibody. Source data are provided as a Source Data file.
PD-L1 enhances NHEJ and inhibits HR
Our findings reveal that PD-L1 promotes the repair of IR-induced DSBs, and IR-induced deglycosylation facilitates nuclear translocation of PD-L1 (Figs. 2 and 3). It is known that the repair of IR-induced DSBs occurs mainly through the NHEJ pathway in nucleus. It is possible that PD-L1 may be involved in the NHEJ-mediated DSB repair. To test this hypothesis, first, the EJ5-GFP-H460 NHEJ reporter cell system was used for analysis of NHEJ efficiency54. Depletion of PD-L1 using PD-L1 siRNA significantly reduced NHEJ efficiency compared to control siRNA (Fig. 5a–c). It is known that NHEJ repair needs the binding between DSBs and 53BP1, RIF1 or FAM35A (a component of shieldin complex) to inhibit end resection20,26. As expected, silence of 53BP1 or RIF1 resulted in decreased NHEJ activity (Fig. 5a–c). Intriguingly, IR-induced foci of 53BP1, RIF1 and FAM35A in PD-L1KO H460 cells were significantly reduced compared to H460 parental cells (Fig. 5d). Western blot analysis revealed that knockout of PD-L1 without IR exposure slightly decreased FAM35A but did not significantly affect expression levels of 53BP1 (Supplementary Fig. 17). Second, the effect of PD-L1 on HR was also assessed using DR-GFP-H460 reporter cells55. Depletion of PD-L1 using PD-L1 siRNA enhanced HR activity, which was similar to the effect of 53BP1 silencing on HR (Fig. 5e, f). Intriguingly, the enhanced HR activity by depletion of PD-L1 or 53BP1 could be abrogated by BRCA1 siRNA (Fig. 5e, f). To assess the effect of PD-L1 loss on early stage of HR (i.e., DNA resection), PD-L1 was knocked out from H460 cells by CRISPR/Cas9. H460 PD-L1 knockout (KO) cells were treated with IR, followed by immunofluorescence analysis of pRPA2 or RPA70 foci. Knockout of PD-L1 significantly increased IR-induced foci of both pRPA2 and RPA70 (Fig. 5g). Western analysis shows that knockout of PD-L1 without IR exposure slightly increased pPRA2 without significant effect on RPA70 (Supplementary Fig. 17). These results indicate that depletion of PD-L1 promotes DNA resection and HR. To further confirm our findings, the effects of PD-L1 on NHEJ and HR were tested in more cell lines. First, PD-L1 was overexpressed in U2OS, H1299 and A549 cells that express low or undetectable levels of endogenous PD-L1, followed by analysis of NHEJ and HR. Results reveal that overexpression of exogenous PD-L1 resulted in significant upregulation of NHEJ and downregulation of HR (Supplementary Fig. 18). Consistently, depletion of endogenous PD-L1 from H460 or H358 cells led to decreased NHEJ and increased HR (Supplementary Fig. 19). Based on our findings above, we propose that, PD-L1 may dictate DSB repair pathway choice to be favorable to NHEJ and unfavorable to HR.
a H460 cells were transfected with Ctrl siRNA, PD-L1 siRNA, 53BP1 siRNA, RIF1 siRNA or BRCA1 siRNA, followed by western blot. b, c NHEJ activity was analyzed by FACS using BDTM FACSCanto II in H460 cells expressing Ctrl siRNA, PD-L1 siRNA, 53BP1 siRNA, or RIF1 siRNA. Y axis represents the recovery rate of GFP from I-SceI-induced DSBs in EJ5-GFP reporter. X axis represents 7-AAD (a nucleic acid dye) staining, which was used for the exclusion of nonviable cells for quality control in flow cytometric analysis. Error bars represent ± s.d., n = 3 per group. ***P < 0.001, by two-tailed t test. d H460 cells or H460 PD-L-1 knockout (KO) cells were treated with IR (4 Gy), followed by immunofluorescence co-staining of γH2AX with anti-53BP1, RIF1 or FAM35A, respectively. e, f HR activity was analyzed by FACS using BDTM FACSCanto II in H460 cells expressing Ctrl siRNA, PD-L1 siRNA, 53BP1 siRNA, BRCA1 siRNA+ PD-L1 siRNA or BRCA1 + 53BP1 siRNA. Y axis represents the recovery rate of GFP from I-SceI-induced DSBs in DR-GFP reporter. X axis represents 7-AAD (a nucleic acid dye) staining, which was used for the exclusion of nonviable cells for quality control in flow cytometric analysis. Error bars represent ± s.d., n = 3 per group. *P < 0.05, **P < 0.01, by two-tailed t test. g H460 cells or H460 PD-L-1 knockout (KO) cells were treated with 4 Gy of IR at 2 Gy/min dose rate, followed by immunofluorescence co-staining of γH2AX with pRPA2 or RPA70, respectively. Source data are provided as a Source Data file.
IR induces recruitment of PD-L1 to DSB sites and interaction with Ku in nucleus
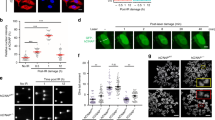
Our findings have demonstrated that IR induces PD-L1 nuclear translocation and promotes DSB repair by enhancing the NHEJ pathway (Figs. 2, 3 and 5). It is known that IR-induced DSBs are recognized by Ku to initiate NHEJ56. To test the mechanism by which PD-L1 promotes DSB repair via NHEJ, first, we tested whether IR-induced nuclear PD-L1 co-localizes with the major NHEJ factor Ku. Ku is the main DSB sensor but it is very difficult to detect Ku foci because Ku presents as a hidden repair complex57. Thus, a pre-extraction technology for high-resolution imaging of Ku foci using cytoskeleton buffer plus RNase A (CSK + R) was employed as described57. Without CSK extraction, IR induced nuclear translocation of PD-L1 but no PD-L1 and Ku80 foci were observed (Fig. 6a, left panel). In contrast, IR induced clear PD-L1 foci as well as Ku80 foci after CSK + R extraction, which were co-localized in the nucleus (Fig. 6a, left lower panel). Furthermore, IR-induced PD-L1 foci were partially co-localized with γH2AX foci after CSK + R extraction (Fig. 6a, right lower panel). These results suggest that IR exposure may facilitate co-localization of PD-L1 with Ku at DSB sites.
a Co-localizations of PD-L1 with Ku80 or γH2AX were analyzed in H460 cells following IR exposure by immunofluorescence with pre-extraction using CSK plus RNase A (CSK + R). b H460 cells were treated with IR, followed by isolation of cytoplasmic and nuclear fractions. Co-IP experiments using PD-L1 antibody were performed in cytoplasmic and nuclear fractions, followed by western blot. IgG and lysate were used as controls. **PD-L1: glycosylated form, *PD-L1: nonglycosylated form. α-tubulin as cytoplasmic marker, and PCNA as nuclear marker, for purity of each fraction. c PD-L1/Ku80 interactions were analyzed by PLA following IR (4 Gy) exposure in H460 cells. PLA signals were detected by fluorescence microscopy as discrete spots. IgG, single PD-L1 antibody alone or single Ku80 antibody alone was used as negative control. After PLA, immunofluorescence staining using α-tubulin antibody was performed as phase contrast imaging. Error bars represent ± s.d., n = 10 per group. ***P < 0.001, by two-tailed t test. Source data are provided as a Source Data file.
To test whether PD-L1 interacts with Ku, co-IP experiments using PD-L1 antibody were carried out in cytoplasmic and nuclear fractions isolated from H460 cells with or without IR exposure. PD-L1 did not interact with Ku70 or Ku80 in cytoplasm in the absence or presence of IR (Fig. 6b, lane 2 vs. lane 3, and lane 8 vs. lane 9). PD-L1 slightly interacted with Ku80 but not Ku70 in the nuclear fraction in the absence of IR (Fig. 6b, lane 5). IR exposure induced a significant PD-L1/ Ku80 interaction and a weak PD-L1/ Ku70 interaction in the nuclear fraction (Fig. 6b, lane 5 vs. lane 11). Additionally, co-IP experiments were performed in the mixture of purified recombinant Ku70/Ku80 with recombinant Flag-tagged WT PD-L1 or nonglycosylated 4NQ mutant PD-L1 proteins. Both WT and mutant 4NQ PD-L1 proteins interacted with recombinant Ku proteins in the cell-free system (Supplementary Fig. 20). To identify where PD-L1/Ku interactions occur in cells, PLA was employed to detect intracellular PD-L1/Ku interaction as described in Fig. 4a. PLA can amplify protein signals by 1000x to directly visualize specific protein-protein interactions. IgG, single PD-L1 antibody alone, and single Ku80 antibody alone were used as negative controls and no discrete spots were observed (Fig. 6c). In PD-L1/Ku80 PLA experiments, only weak discrete spots were detected in the nucleus without IR treatment. Significantly enhanced discrete spots (i.e., PLA signals) were observed in the nucleus after IR exposure (Fig. 6c), indicating that IR-induced PD-L1/Ku interactions occur exclusively in the nucleus.
PD-L1/Ku interaction and PD-L1 deglycosylation occur in a cell cycle-dependent manner
To test the effect of cell cycle on deglycosylation of PD-L1 and its interaction with Ku, H460 cells were synchronized at the G1/S boundary by double thymidine blockade, followed by release into the medium containing nocodazole for cell cycling. Co-IP experiments using anti-PD-L1 were performed at various cell cycle phases (G1, S, G2, M). We found that deglycosylated PD-L1 (33 kDa) interacted with Ku80 mainly in the nucleus at G1 and S phases (Supplementary Fig. 21a). PD-L1/Ku80 interactions in G1, S and G2 phases were also measured by PLA assay. Consistent with co-IP analysis, G1 and S phases contained significantly greater PLA signals (i.e., PD-L1/Ku interactions) than G2 phase in the nucleus (Supplementary Fig. 21b). It is known that NHEJ is active throughout all phases of the cell cycle but predominates in G1 and early S phases21,55. Consistent with NHEJ activity, the deglycosylated form of PD-L1 (i.e., 33 kDa) was observed mainly in G1 and S phases (Supplementary Fig. 21c). When the cell cycle progressed into G2 and M phases, which are known to have relatively low NHEJ activities, PD-L1 became a fully glycosylated form (i.e., 45 kDa) (Supplementary Fig. 21c). These findings indicate that PD-L1 is deglycosylated and interacts with Ku in the nucleus to positively regulate NHEJ activity in G1 and S phases.
PD-L1 facilitates Ku to bind DNA bearing DSBs and promotes interactions between NHEJ factors
To further test how PD-L1/Ku interactions support NHEJ activity, we tested the effect of PD-L1 on Ku/DNA binding in EJ5-GFP H460 cells by ChIP assay using Ku80 antibody. I-Scel was transfected into cells to generate DSBs. A strong Ku80/DNA binding was observed in EJ5-GFP H460 cells after transfection of I-Scel (Supplementary Fig. 22a, lane 3 vs. lane 1). Knockout of PD-L1 abrogated Ku80/DNA binding (Supplementary Fig. 22a, lane 4 vs. lane 3). After transfection of exogenous PD-L1 cDNA into EJ5-GFP H460 PD-L-1 knockout cells, Ku/DNA binding was restored in a dose-dependent manner (Supplementary Fig. 22a, lanes 4–7). To directly test the effect of PD-L1 on Ku/DNA binding in a cell-free system, electrophoretic mobility shift assay (EMSA) using dsDNA was also performed as described previously26. Intriguingly, addition of purified PD-L1 protein significantly enhanced dsDNA/Ku binding (Supplementary Fig. 22b). These findings uncover the mechanism by which PD-L1 positively regulates NHEJ, which occurs through promotion of Ku/dsDNA binding.
It is known that DNA-PKcs is recruited to DNA whose end is bound to Ku, which forms the active DNA-PK complex, and DNA-PK continues to recruit additional NHEJ factors, such as DNA ligase IV/XRCC458. To test whether PD-L1 affects interactions between NHEJ factors, co-IP experiments were carried out using Ku70 antibody or Ku80 antibody, respectively, in H460 vs. H460 PD-L1 knockout (PD-L1KO) cells. IR exposure promoted Ku70 or Ku80 to associate with DNA-PKcs and XRCC4 in H460 cells (Supplementary Fig. 22c). In contrast, knockout of PD-L1 significantly suppressed Ku70 or Ku80 to associate with DNA-PKcs and XRCC4 (Supplementary Fig. 22c), which could negatively affect the NHEJ-mediated DSB repair. These findings suggest that PD-L1 plays a critical role in supporting the interactions of Ku70/80 with other NHEJ factors for DSB repair.
PD-L1 via its IgC domain interacts with Ku at the core domain
PD-L1 is composed of four domains: IgV-like, IgC-like, transmembrane (TM), and cytoplasmic domain (CD)59. Ku80 contains three domains: vWA, core, and c-terminal domain (C)20. To identify PD-L1/Ku interaction sites on PD-L1 or Ku80, a series of PD-L1 deletion mutants (ΔIgV, ΔIgC, ΔTM and ΔCD) and Ku80 deletion mutants (ΔvWA, Δcore and ΔC) were created as illustrated in Fig. 7a. HA-tagged WT Ku80 was co-transfected with Flag-tagged WT-PD-L1 or PD-L1 deletion mutants (ΔIgV, ΔTM and ΔCD) in 293 T cells, followed by co-IP using anti-HA or anti-Flag antibody, respectively. In anti-HA co-IP experiments, HA-WT Ku80 can pull-down WT, ΔIgV, ΔTM and ΔCD mutant PD-L1 but not the ΔIgC mutant PD-L1 (Fig. 7b). In anti-Flag co-IP experiments, HA-WT Ku80 was pulled-down by Flag-WT, ΔIgV, ΔTM and ΔCD mutant PD-L1 but not by the ΔIgC mutant PD-L1 (Fig. 7c). These findings reveal that Ku80 interacts with PD-L1 at the IgC domain. Similar co-IP experiments were carried out in 293T cells following co-transfection using Flag-tagged WT PD-L1 with HA-Ku80 deletion mutants (ΔvWA, Δcore and ΔC). Intriguingly, deletion of the core domain (Δcore) from Ku80 resulted in Ku failure to interact with PD-L1 (Fig. 7d, e), indicating that PD-L1 interacts with Ku80 at the core domain, which is also the DNA binding domain in Ku80.
a Schematic representation of structural WT and various deletion mutants of PD-L1 and Ku80 proteins. b, c HA-tagged WT Ku80 was co-transfected with Flag-tagged WT PD-L1 or various PD-L1 deletion mutant(s) into 293T cells, followed by co-IP using anti-HA (b) or anti-Flag antibody (c), respectively. The Ku-associated PD-L1 (b, upper) or PD-L1-associated Ku (c, upper) was analyzed by western blot. IgG or 1/10 input was used as negative or positive control, respectively. d, e Flag-tagged WT PD-L1 was co-transfected with HA-tagged WT Ku80 or various Ku80 deletion mutant(s) into 293T cells, followed by co-IP using anti-Flag (d) or anti-HA antibody (e), respectively. The PD-L1-associated Ku (d, upper) or Ku-associated PD-L1 (e, upper) was analyzed by western blot. IgG or 1/10 input was used as negative or positive control, respectively. Source data are provided as a Source Data file.
Since the Ku binding site on PD-L1 (i.e., IgC domain) also contains the N192, N200 and N219 glycosylation sites, it is very interesting to test the localization of IgC deletion mutant PD-L1 (ΔIgC). H460 PD-L1 KO cells were transfected with Flag-tagged WT or ΔIgC mutant PD-L1, followed by immunofluorescence. We observed that WT PD-L1 was mainly localized on the plasma membrane while the ΔIgC mutant PD-L1 was mainly localized in the nucleus with a small amount in the cytoplasm around the nucleus (Supplementary Fig. 23). Since N219 site is also removed in ΔIgC mutant PD-L1, it is understandable that ΔIgC is mainly localized in nucleus.
The PD-L1/Ku interaction is required for PD-L1 to reduce radiosensitivity by accelerating NHEJ-mediated DSB repair
To further test whether PD-L1/Ku binding is essential for PD-L1 promotion of DSB repair via the NHEJ pathway, WT PD-L1 and the Ku binding deficient ΔIgC mutant PD-L1 were exogenously expressed in H460 PD-L1 knockout cells (PD-L1KO) (Fig. 8a), followed by IR exposure. DSBs were analyzed by immunofluorescence staining for γH2AX. Results reveal that IR potently induced γH2AX foci (i.e., DSBs) in all cell lines (Fig. 8b, left half images). Knockout of PD-L1 significantly retarded DSB repair at 2 h time point compared to H460 parental cells. Importantly, expression of exogenous WT PD-L1 but not the Ku binding deficient ΔIgC mutant PD-L1 restored PD-L1’s acceleration of DSB repair (Fig. 8b, right half images). These findings were also confirmed by Comet assay (Supplementary Fig. 24). In support of these findings, knockout of PD-L1 from H460 cells significantly reduced NHEJ efficiency (Fig. 8c), and expression of exogenous WT PD-L1 but not the Ku binding deficient ΔIgC mutant PD-L1 restored NHEJ activity (Fig. 8c). Importantly, knockout of PD-L1 led to significant sensitization of H460 lung cancer cells or xenografts to radiation treatment, which could be reversed by expression of exogenous WT PD-L1 but not the Ku binding deficient ΔIgC mutant PD-L1 in vitro and in vivo (Fig. 8d, e). Additionally, WT Ku80 or the PD-L1 binding deficient mutant Ku80 (Δcore) were exogenously expressed in the Ku80 silenced H460 cells. As expected, expression of exogenous WT Ku80 but not the PD-L1 binding deficient Δcore mutant Ku80 restored DSB repair capacity and NHEJ activity from the Ku80 silenced H460 cells (Supplementary Fig. 25). Based on our results, we conclude that interaction of PD-L1 via its IgC domain with Ku at the core domain is required for PD-L1 to accelerate NHEJ-mediated DSB repair.
a Western blot analysis of PD-L1 using PD-L1 or Flag antibody, respectively, in H460, H460 PD-L1 KO, H460 PD-L1 KO cells expressing exogenous WT PD-L1 or PD-L1 ΔIgC mutant. b Cells were treated with 4 Gy of IR at 2 Gy/min dose rate, followed by immunofluorescence analysis of γH2AX foci and DAPI at different time points. Foci ≥5/cell were counted as foci-positive cells. Error bars represent ± s.d., n = 3 per group. ***P < 0.001, ****P < 0.0001, by two-tailed t test. c NHEJ activity was analyzed by FACS using CytekTM Aurora in various cells as indicated. Y axis represents the recovery rate of GFP from I-SceI-induced DSBs in EJ5-GFP reporter. X axis represents 7-AAD (a nucleic acid dye) staining, which was used for the exclusion of nonviable cells for quality control in flow cytometric analysis. Error bars represent ± s.d. n = 3 per group. **P < 0.01, ***P < 0.001, by two-tailed t test. d Cells were treated with IR (4 Gy), followed by colony formation assay. Error bars represent ± s.d., n = 3 per group. ***P < 0.001, by two-tailed t test. e Nu/Nu mice with various xenografts were treated with IR (2 Gy every other day, and 5 times in total). Tumor volume was measured once every 2 days. Mice were sacrificed at day 20, and tumors were removed and analyzed. Error bars represent ± s.d., n = 6 mice each group. **P < 0.01, by two-tailed t test. Source data are provided as a Source Data file.
Discussion
PD-L1 renders cancer cells resistant to T cell-mediated cytotoxicity34. Using genome-scale CRISPR/Cas9 knockout screening, we identified that PD-L1, in addition to its well-known immune inhibitory function, also confers on cancer cells resistance to radiotherapy through acceleration of the NHEJ-mediated DNA double-strand-break repair. Accumulation of endogenous PD-L1 was observed in radioresistant human lung cancer cells. Depletion of endogenous PD-L1 sensitized various human lung cancer cells to IR. Other investigators also reported that PD-L1 expression is associated with radioresistance43,60,61 and/or positively regulates DNA damage repair in various cancer types such as breast cancer61,62,63,64, colon cancer61, bladder62, lung cancer43,61 and head and neck squamous cell carcinoma60. Our findings are consistent with previous reports and further uncovered the mechanisms by which PD-L1 promotes DSB repair contributing to radioresistance.
Although an early report from Zhang et al showed only one form of PD-L1 and did not consider PD-L1 glycosylation/deglycosylation65, several recent reports have demonstrated that PD-L1 can be N-glycosylated. Two forms of PD-L1, including glycosylated (45 kDa) and unglycosylated (33 kDa) PD-L1, were observed in various cancer cells15,47,66. Intriguingly, PD-L1 glycosylation was reported to stabilize PD-L1 protein15 as well as be required for PD-L1 to interact with PD-166. Here, we discovered that IR-induced deglycosylation of PD-L1 promotes PD-L1 translocation from the membrane into the cytoplasm (i.e., ER) and nucleus. N35, N192, N200, and N219 in PD-L1 protein have recently been identified as physiological N-glycosylation sites15. Since the non-glycosylatable N219Q or 4NQ mutant PD-L1 is localized in both nucleus and ER while the N35Q, N192Q and N200Q PD-L1 mutants remain localized on the membrane like WT PD-L1 (Fig. 3 and Supplementary Fig. 9), glycosylation at N219 may be required for PD-L1’s membrane localization, and deglycosylation at N219 could lead to PD-L1 nuclear and ER localizations. Thus, glycosylation/deglycosylation of PD-L1 at N219 may act as a switch to functionally control the subcellular localization of PD-L1. Cha et al reported that the fully non-glycosylatable 4NQ mutant PD-L1 protein is predominantly localized in the ER48 while we found that 4NQ mutant PD-L1 was localized in both nucleus and ER (Supplementary Figs. 9 and 12). The reason for this difference remains unclear. It is known that deglycosylation-mediated ER localization mainly contributes to PD-L1 degradation via enhanced ubiquitination15,48. Based our findings, we propose that deglycosylation of PD-L1 not only facilitates its degradation via ER localization, but also regulates DSB repair through its nuclear localization. Shao et al recently reported that 2-deoxyglucose (2-DG) acts as a glucose analog to inhibit PD-L1 glycosylation47. Studying images in Fig. 3d from their article, in addition to ER localization, we also observed significantly increased nuclear localization of PD-L1 after treatment with 2-DG or the N-linked glycosylation inhibitor tunicamycin (TM) compared to no treatment control. Unfortunately, these interesting findings were not described in their article. Our data confirm that 2-DG and TM promote PD-L1 nuclear and ER localizations in human lung cancer cells (Fig. 3c and Supplementary Fig. 7). Thus, in addition to ER localization, 2-DG and TM-induced inhibition of PD-L1 glycosylation (i.e., deglycosylation) also promotes PD-L1 nuclear localization.
CMTM6 contains the MARVEL domain which is implicated in regulating the trafficking of transmembrane and secretory proteins67. Importantly, CMTM6 has recently been identified as a critical regulator of PD-L1, which is required for efficient endocytic recycling of PD-L149. Following IR exposure, CMTM6/PD-L1 interactions gradually move from the cytoplasm into the nucleus in a time-dependent manner, and that silencing of CMTM6 significantly blocks IR-induced PD-L1 nuclear translocation (Fig. 4). Based on these findings, we propose that CMTM6 is an essential transporter for PD-L1 translocation from the membrane/cytoplasm into the nucleus.
A pre-extraction technology was used to detect PD-L1 foci following IR exposure. IR not only promotes PD-L1 nuclear translocation but also induces PD-L1 foci which co-localize with Ku foci as well as γH2AX foci, indicating that PD-L1 could be recruited to DSB sites to associate with major NHEJ factor Ku protein in the nucleus upon IR exposure (Fig. 6). Functionally, depletion of endogenous PD-L1 downregulates NHEJ while overexpression of exogenous PD-L1 upregulates NHEJ (Fig. 5 and Supplementary Fig. 18 ). These findings support the notion that PD-L1 acceleration of DSB repair occurs through positive regulation of the NHEJ pathway. Mechanistically, PD-L1 not only promotes Ku to bind dsDNA but also facilitates Ku to associate with other NHEJ factors, including DNA-PKcs and XRCC4 (Supplementary Fig. 22). Thus, PD-L1 may function as a critical recruiter of NHEJ factors to DSB sites for DSB repair. These findings uncover the mechanism by which PD-L1 enhances NHEJ activity to accelerate DNA double-strand-break repair. However, it was recently reported that depletion of endogenous PD-L1 by shRNA decreased HR and increased NHEJ in U2OS cells62, which seems to oppose our findings. To experimentally address this contradiction, we repeated the NHEJ and HR experiments in multiple different cell lines. Results consistently indicate that overexpression of PD-L1 led to increased NHEJ and decreased HR in H1299, A549 and U2OS cells (Supplementary Fig. 18). Knockout of endogenous PD-L1 from H460 and H358 reduced NHEJ and enhanced HR (Supplementary Fig. 19). Our consistent findings from repeated experiments in multiple cell lines are convincing, and thus may resolve the discordance with previous report62 to avoid the potential misleading. Importantly, our and others’ findings demonstrated that PD-L1 contributes to radioresistance43,60,62,64 (Figs. 1 and 2 and Supplementary Fig. 2). It is well known that the repair of IR-induced DSBs occurs mainly through NHEJ68,69, which further supports our findings that PD-L1-mediated radioresistance occurs through positive regulation of NHEJ.
Double thymidine block release experiments reveal that the deglycosylated form of PD-L1 (33 KDa) was detected in G1 and S phases while the glycosylated form (45 kDa) was observed in G2 and M phases. Importantly, PD-L1/Ku interactions occur mainly in G1 and S phases which are known to have a relatively high NHEJ activity (Supplementary Fig. 21). Therefore, deglycosylation regulates PD-L1/Ku interaction and NHEJ activity in a cell cycle-dependent manner. Domain-mapping studies reveal that PD-L1 via its IgC domain interacts with Ku at the core domain (Fig. 7). The IgC domain, as the Ku binding site on PD-L1, is essential for PD-L1 to accelerate NHEJ-mediated DSB repair. The IgC domain is also required for PD-L1 to reduce lung cancer radiosensitivity in vitro and in vivo (Fig. 8), suggesting that the IgC domain of PD-L1 may represent an ideal target for overcoming radioresistance in lung cancer therapy.
In summary, our findings have demonstrated that deglycosylation of PD-L1 promotes not only its ER localization and degradation, but also its nuclear localization and interaction with Ku at DSB sites, leading to acceleration of NHEJ-mediated DSB repair, which eventually contributes to radioresistance of human lung cancer. CMTM6 is a required transporter for translocation of PD-L1 into the nucleus from the membrane. PD-L1 promotes Ku/DNA binding and recruitment of major NHEJ factors to DSB sites. Physiological deglycosylation of PD-L1 and its interaction with Ku occur predominantly in G1 and S phases, which could confer efficient NHEJ activity in a cell cycle-dependent manner. Direct interaction between the IgC domain of PD-L1 and the core domain of Ku is required for PD-L1 to accelerate NHEJ-mediated DSB repair and produce radioresistance in human lung cancer cells. Based on our findings, we propose that PD-L1, in addition to its inhibitory effect on T cell-mediated immune activity, appears to be a strong driver of the mechanism utilized for DSB repair in cancer. Specifically targeting this novel function of PD-L1 may represent a potentially effective strategy for cancer therapy.
Methods
Materials
Cell lines, cell culture and transfection
NSCLC cell lines (i.e., A549, H292, H358, H460, HCC827, H1299, and H1975), U2OS, HeLa and HEK-293T cells were purchased from the American Type Culture Collection (ATCC). Two months after receipt, these cell lines were employed for the described experiments without further authentication by authors. A549 cells were cultured in F-12K DMEM media with 5% fetal bovine serum (FBS), 5% bovine serum (BS) and 1% penicillin-streptomycin. H292, H358, H460, HCC827, H1299, and H1975 cells were maintained in RPMI-1640 media with 5% FBS, 5% BS and 1% penicillin-streptomycin. Human osteosarcoma U2OS cells were cultured in McCOY’s-5A media with 5% FBS, 5% BS and 1% penicillin-streptomycin. HeLa and HEK-293T cells were cultured in DMEM media with 5% FBS, 5% BS and 1% penicillin-streptomycin. Lipofectamine 3000 was used to transfect plasmid, siRNA and sgRNA into various cells according to the manufacturer’s instructions. For lentivirus infection, HEK-293T as packaging cells were plated into six-well plates and transfected with lentiCas9-Blast or LVX-ZsGreen-PD-L1, psPAX2 and VSVG (4:3:2). After 72 h, we harvested the culture media by spinning down and removal of the pellets. The medium with lentivirus was mixed with 5 µg/mL polybrene and used to infect H460 cells for 8 h, followed by culture in normal medium. After 3 days, 1 µg/mL puromycin was added for selection of the infected cells. The surviving cells were separated into single cells, followed by expansion and identification of Cas9 and PD-L1 expression.
Genome-wide CRISPR/Cas9 knockout screening
CRISPR/Cas9 knockout screening was carried out as described in Nature protocols from Dr. Zhang’s laboratory28. We used human CRISPR knockout pooled library (GeCKO v2) which is a pooled gRNA library in a 1 vector lentiCRISPRv2 containing Cas9 from Addgene. The entire library was delivered as two half-libraries (A and B) which contain 6 sgRNAs per gene. To amplify the library, lentiCRISPRv2 A or B was separately electroporated into Endura™ electroCompetent cells. After recovery on ice for 1 h, Endura E. coli cells were coated on Petri dish with LB containing ampicillin and incubated at 32 °C overnight, followed by purification of library plasmids using Qiagen plasmid kit. Purified lentiCRISPRv2 A and B plasmids were co-transfected into HEK-293T cells with the packaging plasmids pVSVg and psPAX2 using Lipofectamine 2000 for 8 h. The media was then changed to DMEM supplemented with 1% BSA for 60 h. The medium was collected by centrifugation at 3000 rpm for 10 min, followed by filtering with 0.45 µm membrane. The resulting medium with lentivirus bearing library was used for cell infection. H460 cells (5 × 106) were seeded in 24.5 cm cell culture dishes (Corning) for cell growth to 30% confluence. The amount of virus in the medium was optimized as a multiplicity of infection (MOI) of 0.4. The appropriate volume of virus at 0.4 MOI and 8 µg/ml polybrene were added to the cell culture dish for 8 h, then replaced with 10% FBS RPMI-1640 and cultured for 72 h. Puromycin (1 µg/ml) was applied to the cell culture dish (24.5 cm) for a 7-day selection. H460 cells were then divided into two groups (no treatment control and IR treatment), triplicate in each group. H460 cells in the IR group were treated with 4 Gy of IR at 2 Gy/min dose rate, followed by normal cell culture for 7 days until the cells covered almost the entire dish (i.e., 95% confluence). The cells were harvested, and the genome DNA from each sample was extracted using blood & cell culture DNA maxi kit (QIAGEN). Purified genome DNA samples (triplicate in each group, DNA amount per sample >272 μg) were sent to Admera Health (South Plainfield, NJ) for sequencing the distribution of sgRNAs. Raw sequencing reads were mapped to Human GeCKO sgRNA v2 library with no mismatches tolerated using Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK) method70. The number of sequencing reads, mapped reads, sgRNAs with zero read count, and the Gini index of read count distribution were evaluated for each sample. The read counts derived from MAGeCK ‘count’ were median-normalized to adjust for the effect of library sizes and read count distributions. MAGeCK ‘test’ was done to compare samples between two treatments using Robust Rank Aggregation algorithm (‘MAGeC-RRA’). In addition, MAGeCK ‘mle’ was done to compare samples between two treatments using Maximum Likelihood Estimation algorithm (‘MAGeCK-MLE’). The P values were calculated from the Negative Binomial model using a modified robust ranking aggregation algorithm. The false discovery rate (FDR) was computed from the empirical permutation P values using the Benjamini–Hochberg procedure.
Immunofluorescence staining and high-resolution imaging of Ku and PD-L1 foci
Cells were grown on chamber slides (BD Falcon, MA), washed with cold 1 × PBS, fixed with 4.0% paraformaldehyde in 1 × PBS for 15 min at room temperature, and permeabilized with 0.5% Triton X-100 in 1 × PBS for 10 min. After blocking with 10% normal goat serum (Life Technologies, Carlsbad, CA) for 1 h, samples were incubated with the indicated primary antibody at 4 °C overnight. After washing, samples were incubated with goat anti-mouse IgG Alexa Fluor 488 (green) (1:1000) or goat anti-rabbit IgG Alexa Fluor 568 (red) (1:1000) for 45 min at room temperature in the dark. After washing, samples were mounted with Prolong Gold antifade reagent containing DAPI. Images were captured using a Leica SP8 confocal microscope (Wetzlar, Germany). Because Ku or PD-L1 presents as a hidden repair complex, Ku foci or PD-L1 foci are very difficult to detect. A pre-extraction for high-resolution imaging of Ku foci or PD-L1 foci using cytoskeleton buffer (CSK) plus RNase A (CSK + R) was performed as described previously57. Cells were washed with 1 × PBS and then incubated twice for 3 min at room temperature with CSK buffer (10 mM Pipes, pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2 and 0.7% Triton X-100) plus 0.3 mg/ml RNase A (CSK + R). After pre-extraction, cells were washed with 1 × PBS and fixed with 2% PFA, followed by standard immunofluorescence staining using Ku80 (1:400) or PD-L1 (1:50, Novus) as primary antibody as above.
NHEJ and HR assays
The EJ5-GFP or DR-GFP reporter plasmid was applied to measure NHEJ or HR activity, respectively, as previously described54,55,71. Cells stably expressing EJ5-GFP or DR-GFP reporter were seeded in six-well plates. When cell growth reached 50–80% confluence, cells were transfected with 2 µg of pCBA-SceI per well and incubation for 48 h. Then, cells were harvested, washed, resuspended in 1 mL 1 × PBS with 0.5% BSA and 1 μg/ml 7-aminoactinomycin D (7-AAD)71. 7-AAD, a nucleic acid dye, was used for the exclusion of nonviable cells for quality control in flow cytometric analysis. The GFP positive cells (GFP restoration) were counted by flow cytometry (BDTM FACSCanto II or CytekTM Aurora). All data were analyzed by FlowJo software. Each experiment was repeated three times.
Proximity ligation assay (PLA)
PLA was performed according to the manufacturer’s instructions (Sigma-Aldrich). Cells were grown on chamber slides, washed with cold 1 × PBS, fixed with 4.0% paraformaldehyde in 1 × PBS for 15 min at room temperature, and permeabilized with 0.5% Triton X-100 in 1 × PBS for 10 min. After incubation with blocking solution at 37 °C for 1 h, cells were incubated with two primary antibodies from different species at 4 °C overnight. After washing, cells were incubated with a mixture of the corresponding secondary antibodies with Duolink in situ PLA probe at 37 °C for 1 h. Cells were incubated with the ligation solution at 37 °C for 30 min, followed by incubation with the amplification solution for 100 min at 37 °C. After washing, slides were mounted with a coverslip using Duolink PLA mounting medium with DAPI. After 15 min, PLA signals were captured by Leica SP8 confocal microscope.
Lung cancer xenograft and treatment
Six-week-old male and female (half/half) Nu/Nu nude mice were purchased from Envigo (Indianapolis, IN) and housed under pathogen-free conditions with a 14 h light/10 h dark cycle, 18–23 °C temperature and 40–60% humidity in microisolator cages. All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of Emory University. In total, 3 × 106 lung cancer cells were injected into subcutaneous tissue in the flank region of nude mice. After 6–9 days, when the tumor volume increased to 100 mm3, mice were irradiated with 2 Gy every other day for 5 treatments in total using an X-RAD 320 irradiator (Precision X-ray) to deliver whole-body irradiation to the mice. Tumor volume was measured every two days by caliper measurement after the first therapy and continued to 20 days using formula (V = π/6 (L × W × H)). Mice were sacrificed by inhaled CO2 at the end of treatment, and tumor tissues were collected for further analysis.
Statistics and reproducibility
Each experiment was repeated at least three times independently with similar results. All data are presented as mean ± standard deviation (s.d.) from at least three independent experiments. The statistical significance of differences between groups was analyzed by two-tailed unpaired Student’s t test. We chose the sample size to detect a minimum effect size of 1.5 with at least 80% power and a type I error of 0.05 for each comparison. P value less than 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The CRISPR screen genome DNA sequencing data generated in this study has been deposited in NCBI’s Gene Expression Omnibus (GEO) database under GEO series accession code GSE218225. Source data are provided with this paper.
References
Chen, L. & Han, X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Investig. 125, 3384–3391 (2015).
Nishino, M., Ramaiya, N. H., Hatabu, H. & Hodi, F. S. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat. Rev. Clin. Oncol. 14, 655–668 (2017).
Ghahremanloo, A., Soltani, A., Modaresi, S. M. S. & Hashemy, S. I. Recent advances in the clinical development of immune checkpoint blockade therapy. Cell Oncol. 42, 609–626 (2019).
George, J. et al. Genomic amplification of CD274 (PD-L1) in small-cell lung cancer. Clin. Cancer Res. 23, 1220–1226 (2017).
Zak, K. M. et al. Structure of the complex of human programmed death 1, PD-1, and its ligand PD-L1. Structure 23, 2341–2348 (2015).
Liu, J. et al. Extracellular vesicle PD-L1 in reshaping tumor immune microenvironment: biological function and potential therapy strategies. Cell Commun. Signal 20, 14 (2022).
Dong, H. et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 8, 793–800 (2002).
Cha, J. H., Chan, L. C., Li, C. W., Hsu, J. L. & Hung, M. C. Mechanisms controlling PD-L1 expression in cancer. Mol. Cell 76, 359–370 (2019).
Thompson, R. H. et al. Costimulatory B7-H1 in renal cell carcinoma patients: Indicator of tumor aggressiveness and potential therapeutic target. Proc. Natl. Acad. Sci. USA 101, 17174–17179 (2004).
Sperduto, P. W. et al. The graded prognostic assessment (GPA) for lung cancer patients with brain metastases: initial report of the small cell lung cancer GPA and update of the non-small cell lung cancer GPA including the effect of programmed death ligand-1 (PD-L1) and other prognostic factors. Int. J. Radiat. Oncol. Biol. Phys. 114, 60–74 (2022).
Kim, J. H., Jeong, S. Y., Lee, J. J., Park, S. T. & Kim, H. S. A Bayesian network meta-analysis of first-line treatments for non-small cell lung cancer with high programmed death ligand-1 expression. J. Clin. Med. 11, 1492 (2022).
Ohaegbulam, K. C., Assal, A., Lazar-Molnar, E., Yao, Y. & Zang, X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol. Med. 21, 24–33 (2015).
Yu, X., Li, W., Young, K. H. & Li, Y. Posttranslational modifications in PD-L1 turnover and function: from cradle to grave. Biomedicines 9, 1702 (2021).
Gao, Y. et al. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat. Cell Biol. 22, 1064–1075 (2020).
Li, C. W. et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat. Commun. 7, 12632 (2016).
Mouw, K. W., Goldberg, M. S., Konstantinopoulos, P. A. & D’Andrea, A. D. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 7, 675–693 (2017).
Sato, H. et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 8, 1751 (2017).
Deng, L. et al. Irradiation and anti-PD-L1 treatment synergistically promote antitumor immunity in mice. J. Clin. Investig. 124, 687–695 (2014).
Dovedi, S. J. et al. Acquired resistance to fractionated radiotherapy can be overcome by concurrent PD-L1 blockade. Cancer Res. 74, 5458–5468 (2014).
Chang, H. H. Y., Pannunzio, N. R., Adachi, N. & Lieber, M. R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 (2017).
Chapman, J. R., Taylor, M. R. & Boulton, S. J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 (2012).
Burma, S., Chen, B. P. & Chen, D. J. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair 5, 1042–1048 (2006).
Hefferin, M. L. & Tomkinson, A. E. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair 4, 639–648 (2005).
Ceccaldi, R., Rondinelli, B. & D’Andrea, A. D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64 (2016).
Gupta, R. et al. DNA Repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 173, 972–988 e923 (2018).
Dev, H. et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 20, 954–965 (2018).
Ozawa, N. et al. PD-L1 upregulation is associated with activation of the DNA double-strand break repair pathway in patients with colitic cancer. Sci. Rep. 11, 13077 (2021).
Joung, J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Lander, E. S. The heroes of CRISPR. Cell 164, 18–28 (2016).
Lan, B. et al. CRISPR-Cas9 screen identifies DYRK1A as a target for radiotherapy sensitization in pancreatic cancer. Cancers 14, 326 (2022).
Saber, A., Liu, B., Ebrahimi, P. & Haisma, H. J. CRISPR/Cas9 for overcoming drug resistance in solid tumors. DARU 28, 295–304 (2020).
Chen, X. et al. Targeting mitochondrial structure sensitizes acute myeloid leukemia to venetoclax treatment. Cancer Discov. 9, 890–909 (2019).
Joung, J. et al. CRISPR activation screen identifies BCL-2 proteins and B3GNT2 as drivers of cancer resistance to T cell-mediated cytotoxicity. Nat. Commun. 13, 1606 (2022).
Bishara, L. A., Machour, F. E., Awwad, S. W. & Ayoub, N. NELF complex fosters BRCA1 and RAD51 recruitment to DNA damage sites and modulates sensitivity to PARP inhibition. DNA Repair 97, 103025 (2021).
Jegadesan, N. K. & Branzei, D. DDX11 loss causes replication stress and pharmacologically exploitable DNA repair defects. Proc. Natl. Acad. Sci. USA 118, e2024258118 (2021).
Martino, J. et al. The human Shu complex functions with PDS5B and SPIDR to promote homologous recombination. Nucleic Acids Res. 47, 10151–10165 (2019).
Hopkins, J. et al. Meiosis-specific cohesin component, Stag3 is essential for maintaining centromere chromatid cohesion, and required for DNA repair and synapsis between homologous chromosomes. PLoS Genet. 10, e1004413 (2014).
Sekhar, K. R. et al. Targeting nucleophosmin 1 represents a rational strategy for radiation sensitization. Int. J. Radiat. Oncol. Biol. Phys. 89, 1106–1114 (2014).
Shao, B. et al. Effects of tumor-derived exosome programmed death ligand 1 on tumor immunity and clinical applications. Front. Cell Dev. Biol. 9, 760211 (2021).
Park, D. et al. Novel small-molecule inhibitors of Bcl-XL to treat lung cancer. Cancer Res. 73, 5485–5496 (2013).
Tu, X. et al. PD-L1 (B7-H1) competes with the RNA exosome to regulate the DNA damage response and can be targeted to sensitize to radiation or chemotherapy. Mol. Cell 74, 1215–1226 e1214 (2019).
De, S., Holvey-Bates, E. G., Mahen, K., Willard, B. & Stark, G. R. The ubiquitin E3 ligase FBXO22 degrades PD-L1 and sensitizes cancer cells to DNA damage. Proc. Natl. Acad. Sci. USA 118, e2112674118 (2021).
Polioudaki, H. et al. Nuclear localization of PD-L1: artifact or reality? Cell Oncol. 42, 237–242 (2019).
Yu, J. et al. Regulation of sister chromatid cohesion by nuclear PD-L1. Cell Res. 30, 590–601 (2020).
Deng, X., Gao, F., Flagg, T., Anderson, J. & May, W. S. Bcl2’s flexible loop domain regulates p53 binding and survival. Mol. Cell Biol. 26, 4421–4434 (2006).
Shao, B. et al. Deglycosylation of PD-L1 by 2-deoxyglucose reverses PARP inhibitor-induced immunosuppression in triple-negative breast cancer. Am. J. Cancer Res. 8, 1837–1846 (2018).
Cha, J. H. et al. Metformin promotes antitumor immunity via endoplasmic-reticulum-associated degradation of PD-L1. Mol. Cell 71, 606–620 e607 (2018).
Burr, M. L. et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105 (2017).
Suzuki, T., Huang, C. & Fujihira, H. The cytoplasmic peptide:N-glycanase (NGLY1)—structure, expression and cellular functions. Gene 577, 1–7 (2016).
Miao, X., Wu, J., Chen, H. & Lu, G. Comprehensive analysis of the structure and function of peptide:N-glycanase 1 and relationship with congenital disorder of deglycosylation. Nutrients 14, 1690 (2022).
Mezzadra, R. et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549, 106–110 (2017).
Chen, G. et al. Mcl-1 interacts with Akt to promote lung cancer progression. Cancer Res. 79, 6126–6138 (2019).
Bennardo, N., Cheng, A., Huang, N. & Stark, J. M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008).
Chen, G. et al. Targeting Mcl-1 enhances DNA replication stress sensitivity to cancer therapy. J. Clin. Investig. 128, 500–516 (2018).
Walker, J. R., Corpina, R. A. & Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 (2001).
Britton, S., Coates, J. & Jackson, S. P. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 202, 579–595 (2013).
Ochi, T., Wu, Q. & Blundell, T. L. The spatial organization of non-homologous end joining: from bridging to end joining. DNA Repair 17, 98–109 (2014).
Bailly, C., Thuru, X. & Quesnel, B. Soluble programmed death ligand-1 (sPD-L1): a pool of circulating proteins implicated in health and diseases. Cancers 13, 3034 (2021).
Schulz, D. et al. Increased PD-L1 expression in radioresistant HNSCC cell lines after irradiation affects cell proliferation due to inactivation of GSK-3beta. Oncotarget 10, 573–583 (2019).
Cheon, H., Holvey-Bates, E. G., McGrail, D. J. & Stark, G. R. PD-L1 sustains chronic, cancer cell-intrinsic responses to type I interferon, enhancing resistance to DNA damage. Proc. Natl. Acad. Sci. USA 118, e2112258118 (2021).
Kornepati, A. V. R. et al. Tumor intrinsic PD-L1 promotes DNA repair in distinct cancers and suppresses PARP inhibitor-induced synthetic lethality. Cancer Res. 82, 2156–2170 (2022).
Xue, Z. et al. PD-L1 deficiency sensitizes tumor cells to DNA-PK inhibition and enhances cGAS-STING activation. Am. J. Cancer Res. 12, 2363–2375 (2022).
Zhang, R. et al. D-mannose facilitates immunotherapy and radiotherapy of triple-negative breast cancer via degradation of PD-L1. Proc. Natl. Acad. Sci. USA 119, e2114851119 (2022).
Zhang, J. et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018).
Li, C. W. et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell 33, 187–201 e110 (2018).
Mamessier, E., Birnbaum, D. J., Finetti, P., Birnbaum, D. & Bertucci, F. CMTM6 stabilizes PD-L1 expression and refines its prognostic value in tumors. Ann. Transl. Med. 6, 54 (2018).
Mahaney, B. L., Meek, K. & Lees-Miller, S. P. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417, 639–650 (2009).
Wang, Q. et al. Bcl2 negatively regulates DNA double-strand-break repair through a nonhomologous end-joining pathway. Mol. Cell 29, 488–498 (2008).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Budke, B. et al. Noncanonical NF-kappaB factor p100/p52 regulates homologous recombination and modulates sensitivity to DNA-damaging therapy. Nucleic Acids Res. 50, 6251–6263 (2022).
Acknowledgements
This work was supported by NIH/NCI grants R01CA255257and R01CA200905 (to X. Deng). We are grateful to Dr. William Dynan (Emory University) for kindly providing purified Ku70/80 proteins. We thank Anthea Hammond for editing of the manuscript.
Author information
Authors and Affiliations
Contributions
X.D. conceived the project. Z.S. performed experiments. B.D. and J.M.S. analyzed CRISPR/Cas9 screening sequence data. X.D. and Z.S. wrote the manuscript. D.S.Y. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shu, Z., Dwivedi, B., Switchenko, J.M. et al. PD-L1 deglycosylation promotes its nuclear translocation and accelerates DNA double-strand-break repair in cancer. Nat Commun 15, 6830 (2024). https://doi.org/10.1038/s41467-024-51242-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51242-8
- Springer Nature Limited