Abstract
Kidney injury disrupts the intricate renal architecture and triggers limited regeneration, together with injury-invoked inflammation and fibrosis. Deciphering the molecular pathways and cellular interactions driving these processes is challenging due to the complex tissue structure. Here, we apply single cell spatial transcriptomics to examine ischemia-reperfusion injury in the mouse kidney. Spatial transcriptomics reveals injury-specific and spatially-dependent gene expression patterns in distinct cellular microenvironments within the kidney and predicts Clcf1-Crfl1 in a molecular interplay between persistently injured proximal tubule cells and their neighboring fibroblasts. Immune cell types play a critical role in organ repair. Spatial analysis identifies cellular microenvironments resembling early tertiary lymphoid structures and associated molecular pathways. Collectively, this study supports a focus on molecular interactions in cellular microenvironments to enhance understanding of injury, repair and disease.
Similar content being viewed by others
Introduction
The mammalian kidney removes waste products from the blood, maintains fluid homeostasis, and releases hormones that control blood pressure1. These and other renal functions hinge on a stereotypic organization of diverse renal cell types along a cortico-medullary axis2. Nephrons, the primary filtering units of the kidney, contain over twenty distinct and spatially organized cell types, including species-specific sexual diversity in proximal tubule segments, to ensure recovery of key molecules from the primary glomerular filtrate1,3. Proximal tubule cells are highly active metabolically and are, therefore, particularly susceptible to acute kidney injury (AKI), an abrupt loss of excretory kidney function that can be caused by multiple insults, including ischemia, sepsis, and nephrotoxic drugs4. AKI affects approximately 20 − 25% of all hospitalized patients in the western world and is associated with increased morbidity and mortality5, including the development of Chronic Kidney Disease (CKD)6. CKD is characterized by renal inflammation and fibrosis7,8 and is predicted to become the fifth most common cause of death by 20409. Despite its high prevalence and medical burden, therapeutic strategies for treating AKI and preventing the AKI-to-CKD transition are lacking.
The heightened risk of CKD following AKI has been linked to the limited regenerative capacity of the mammalian kidney10. Murine models of ischemic AKI predominantly result in pronounced cell death of the proximal tubule, particularly within the S3 segment9,10,11,12,13,14. The injury also invokes compensatory de-differentiation and cell division of surviving proximal tubule cells, which can lead to at least partial restoration of kidney structure and function11,12. However, even mild injury is associated with epithelial scarring and the persistence of maladaptive epithelial cells expressing inflammation-inducing cytokines and fibrosis-associated secretory factors. Inflammation, fibrosis, and vascular rarefaction are key features of the pathological niche surrounding maladaptive epithelial cells13,14,15,16,17,18,19,20,21,22,23,24. Several molecular and cellular features of the AKI response are conserved between mice and humans25,26,27,28 and persistent pro-inflammatory and pro-fibrotic signaling are believed to promote the AKI-to-CKD progression29,30,31.
Single-cell studies have generated new insight into AKI through the identification of cell types associated with pathology and underlying transcriptional programs8,13,15,28,32,33. However, tissue dissociation in these studies removes the spatial context of interacting cell types, and so precludes a deeper understanding of the complex local cellular interactions following renal injury. In contrast, spatial transcriptomics with single-cell resolution provides a readout of cell type-specific transcriptional activity while preserving information about cellular organization in tissues34,35,36,37,38. As such, this approach is well-suited to provide new insight into the pathophysiological mechanisms driving the progression of kidney disease and identifying cell interactions for potential therapeutic intervention.
Sequential Fluorescence In Situ Hybridization (seqFISH)35,39,40, a spatial transcriptomics approach, can detect and quantify thousands of mRNA transcripts at single-cell resolution in intact tissues. seqFISH’s high capture efficiency and sensitivity, coupled with the capability of examining a large number of genes, facilitates the identification of cellular subtypes and enables the prediction of specific patterns of cell-cell interactions35,41. In this work, we focused on the mammalian kidneys response to AKI at the single-cell level, leveraging seqFISH to investigate changes in cell composition, gene expression, and cell-cell interactions. This approach uncovered AKI-specific cellular neighborhoods and cell-cell interactions relevant to inflammation and fibrosis and identified target molecules and pathways potentially driving AKI progression.
Results
seqFISH profiling of acute kidney injury
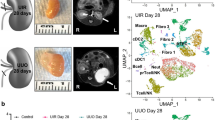
To examine AKI outcomes after the initial phase of injury-invoked renal repair27, we subjected C57BL/6 J male mice (8–13 weeks of age) to a mild ischemia-reperfusion injury (IRI; serum creatinine levels at 48 h 0.3–1.7 mg/dL), waited four weeks and then collected and analyzed the kidneys. We selected 1300 target genes for seqFISH profiling based on key cell-type enriched markers from single-cell studies of normal and IRI kidney3,13,15 and a long-term study of injury-associated expression during the AKI to CKD transition in mice (27; Fig. 1A). We used Cellpose 2.042 and DAPI based segmentation to achieve maximal accuracy of our analysis and minimize segmentation errors. Integration of control (n = 3) and post AKI (n = 3) samples resulted in a data set comprising 245,171 single cells, with an average of 185 ± 153 (mean ± SD) total transcripts and 87 ± 53 individual genes detected per cell, with a strong concordance between samples (Supplementary Fig. 1 and Supplementary Fig 4). Following rigorous quality filtering, we retained 220,753 cells for subsequent analysis. By clustering on single-cell gene expression, we identified all major kidney cell types, highlighting the segmental cell diversity along the conjoined epithelial networks of the nephron and collecting system (Fig. 1B and Supplementary Data 1), as well as divergent vascular endothelial sub-sets, interstitial fibroblasts and distinct immune cell types (Fig. 1B, C). The cell types identified by seqFISH were concordant with cell types previously identified using single-cell RNA sequencing (scRNAseq) (Supplementary Fig. 2A, B). Further, their spatial localization conformed to well-documented kidney anatomy (Fig. 1C, D, F and Supplementary Figs. 3, 5). However, the seqFISH analysis captured a notably larger fraction of several kidney-resident cell types relative to that previously reported in scRNA-seq studies43,44, where the tissue dissociation procedures cause certain cell types to be over- or under-represented43,45,46. For instance, in past work, fibroblasts were reported to comprise ~ 4–10% and macrophages less than 5% in kidney single cell- and single nucleus-RNAseq datasets, whereas our data showed that fibroblasts comprise close to 20% and macrophages ~ 10% of all cells (Supplementary Fig. 2C). These results are consistent with the idea that detecting cell types in kidney tissue by seqFISH will be less biased compared with contemporary single cell-RNAseq protocols.
A Experimental overview: mice were subject to IRI at day 0 to induce AKI. Control and AKI kidneys were collected on day 28, and seqFISH was performed on frozen kidney sections. B Umap depicting all cell types identified using seqFISH. Normal kidney-specific cell types are highlighted and their locations within the nephron are illustrated. DC – dendritic cells; Macroph – macrophages; DCT-CNT – distal convoluted and connecting tubule; Fib – fibroblasts; IC – intercalated cells; Injured PT – injured proximal tubule cells; LOH-TL-C – thin limb of the loop of Henle (comprising cells of the thin descending limb of the loop of Henle of cortical nephrons and the thin ascending limb of the loop of Henle of juxtamedullary nephrons); LOH-TL-JM – thin limb of the loop of Henle of juxtamedullary nephrons; PC – principal cells; Per – pericytes; Podo – podocytes; PT – proximal tubule; PTS1/2/3 – proximal tubule segment 1/2/3; T – T cells; TAL – thick ascending limb of the loop of Henle; Uro – urothelium; Vasc – endothelial cells. C Spatial location of all cell types in one representative control sample. Left - all cell types, right - 10 cell types out of the total 22 that were identified when plotted individually. These cell types span the cortex and medulla, showing that seqFISH analysis captures all major cell types in different areas of the kidney. D Representative field of view outlined in panel (C) showing the cell masks color-coded by cell type (top) and a zoomed-in image showing RNA expression of representative marker genes (bottom). The image is comprised of the sum of the background and DAPI image to illustrate the underlying tissue morphology. E Violin plot showing the normalized expression of marker genes for each cell type. The bar plot shows the relative abundance of each cell type within the control and AKI samples. F Spatial locations of PTS3, Vasc_2, Injured PT, fibroblasts, and macrophages within one representative control (top) and one AKI (bottom) sample.
We next compared AKI with control samples to identify changes in the abundance and spatial locations of specific cell types. At the cortico-medullary boundary, which is known to be most sensitive to AKI47, the proximal tubule segment 3 (PTS3) and vascular endothelium (Vasc_2) were strongly reduced (Fig. 1E, F and Supplementary Figs. 2D, 5). Conversely, a marked increase was observed in injured proximal tubule cells, as identified by the expression of Vcam1 (13,15; Injured PT). In addition, fibroblast, macrophage, T cell, and dendritic cell populations were all elevated in proportion in AKI samples relative to control (Fig. 1E, F and Supplementary Fig. 2D, 5). In contrast to T cells and dendritic cells, the increase in fibroblasts and macrophages was associated with a cortical expansion in injured kidney samples (Fig. 1F and Supplementary Fig. 5); fibroblasts and macrophages predominated within the medulla of the uninjured kidney. In addition, we observed an over-representation of a Slc12a1+/Ptger3+ subset of thick ascending limb (TAL)-subtypes in the distal medullary loop-of-Henle (Fig. 1E48;). Thus, AKI induces global changes in both cell type composition and location.
Distinct cellular microenvironments are specific for AKI and normal kidneys
We next sought to understand whether changes in cell populations also lead to the re-organization of the local kidney architecture. Specifically, we asked whether we could detect localized microenvironments (MEs) with distinct cell type compositions within the kidney and whether those environments changed following AKI. To this end, we calculated the frequencies of each cell type within a 30 µm radius of each of the cells in the kidney tissue. We reasoned that this radius is of physiological relevance as it represents ~ 2 cell layers around each cell, which is a distance scale that allows for short-range paracrine and juxtacrine signaling49. We then used the resulting cell-by-neighbor matrix to cluster individual cells using Leiden clustering (Fig. 2A and Supplementary Fig. 6). To determine the sensitivity of ME clustering to different radii, we varied the radii and repeated the neighboring calculation maintaining the same clustering parameters (Supplementary Fig. 6). Interestingly, this clustering approach detected MEs with similar compositions within a radius range of 10–50 µm (Supplementary Fig. 6A). However, smaller (10, 20 µm) and larger (50, 100 µm) radii resulted in multiple redundant MEs with highly similar cellular compositions (Supplementary Fig. 6B). In addition, larger radii produced multiple MEs that were sample specific reflecting slight differences in the orientation of sections, and potentially, biases caused by individual imaging Fields Of View (FOVs) (Supplementary Fig. 6C). In summary, decreasing or increasing the radius for neighborhood analysis from 30 µm does not improve the capture of biologically relevant cellular interactions and can introduce redundancy or bias.
A Zoomed in image of one field of view in the AKI sample: The cellular composition within a 30um radius around each cell is calculated, and these compositions are then clustered to create 17 distinct MEs. B Cellular composition within each ME is calculated as the relative mean abundance of each cell type. Barplot showing the relative frequency of each ME within the Control and AKI samples. Gray asterisk represents enrichment in control and blue enrichment in AKI (*p < 0.05; **p < 0.01 of a one-sided t test). Source data are provided as a Source Data file. C Spatial locations of five MEs in one representative control and one AKI sample. D Spatial locations of all injured PT cells in one AKI sample plotted in color over the locations of all cells belonging to ME-5 plotted in gray. E Zoomed-in image of the box plot in D showing an overlay of cell masks on Vcam1 antibody and DAPI signal. The cell masks are colored according to the ME assignment of each cell. The Vcam1 signal is shown in white over the DAPI gray signal. F Expression of top injury marker genes in injured PT cells within ME-5 and the Injured PT cells outside ME-5 in Control samples (left) and AKI (right). (*p < 0.05; **p < 0.01 of a paired two-sided t test). Source data are provided as a Source Data file. G Normalized expression of injury markers in Injured PT cells within ME-5, sorted by the eccentricity of each cell. Cells were sorted according to eccentricity values, and the gene expression was averaged using a moving window with a window size of 10% total number of cells. Expression values were normalized for each gene in the heatmap.
Using a 30 µm radius for neighborhood analysis, we detected 17 MEs with a distinct cell type composition and spatial location (Fig. 2B, C and Supplementary Fig. 7). Eight MEs were not markedly different between individual control and AKI samples, five were enriched in the control kidneys and four were specific to AKI samples (Fig. 2B and Supplementary Fig. 7). To limit sampling bias, we focused our analysis on MEs that were equally enriched or depleted in all control and AKI replicates (indicated by an asterisk in Fig. 2B, gray – enriched in control and blue-enriched in AKI). In Fig. 2C, we show examples of MEs that are similar between control and AKI (ME-0), absent from AKI (ME-3), or enriched in AKI (ME-5, ME-15, ME-16). AKI-specific MEs were largely a combination of injured epithelium, fibroblasts, and immune cells in varying proportions, and were mostly depleted of the primary tissue cells. Interestingly, dendritic cells, macrophages, T cells, and fibroblasts were distributed across different MEs in the control samples, but concentrated within specific MEs in AKI samples (Supplementary Fig. 7B). Therefore, the emergence of these cells, whether by differentiation, proliferation, or recruitment from the blood, results in redistribution into distinct local environments upon AKI.
Characterization of the pathogenic niche in AKI
Recent studies in mouse and humans have drawn attention to the tissue micro-environment around injured proximal tubule cells, which have been associated with renal pathology13,15,28. Injured proximal tubule cells display the cell adhesion molecule Vcam1 and adopt a senescence-associated secretory phenotype (SASP)13,15,28,33. We identified a similar population of Vcam1-positive epithelial cells in our data set by unsupervised clustering (Cluster Injured PT, Fig. 1B, E). The majority of these injured epithelial cells reside within a single ME (ME-5, Fig. 2B, D). As a means of internal validation, we stained with anti-Vcam1 antibody concurrently with seqFISH analysis and found that Vcam1 protein is mainly present in cells within ME-5 (Fig. 2E).
Figure 2F plots the mean expression of top injured PT marker genes in control (left) and AKI (right), comparing the expression of these genes in injured PT cells outside ME-5 to the expression of their counterparts within ME-5. Interestingly, while the expression of injury genes was elevated in injured PT cells in AKI samples compared to control, the expression within ME-5 is significantly higher than in cells outside ME-5 for both AKI and control samples. Injured PT cells have been detected in normal samples and are thought to be naturally occurring senescent cells13,15,17. Our analysis suggests that the cellular neighborhood of injured cells correlates with a more severe injury phenotype even within AKI and that cellular interactions within this neighborhood could be a driver of injury progression.
Proximal tubule cells undergo a de-differentiation in response to AKI that manifests in several morphological changes, including a loss of the apical brush border, and a flattening of the epithelium, and likely with this, the nucleus of the cell50. To assess whether nuclear morphology might be an informative parameter, we correlated changes in the expression of injury genes to the eccentricity of the nucleus in Injured PT cell masks. An eccentricity value of 0 represents a perfect circle, and values between 0 and 1 represent an ellipse. We found that eccentricity analysis could sub-divide Injured PT cell expression profiles. With increasing eccentricity values (corresponding to a flattening of the nucleus), Vcam1 expression increased, while expression of other injury-related genes did not correlate with high eccentricity (Fig. 2G). Thus, morphological criteria can have a predictive value of the injury state of the cell. Taken together, our findings show that ME-5 represents an injured and likely pathogenic niche. The majority of injured PTs and injury-associated fibroblasts, vasculature, and immune cells reside within this niche, and morphological changes within the injured epithelium are associated with elevated expression of injury-response genes.
Clcf1 - Crlf1 interactions between injured PT and fibroblasts shape the injured niche
We sought to identify signaling between Injured PT and the other cell types present within ME-5 (fibroblasts, macrophages, T cells, DCs, Vasc_1, few normal PT cells), that would be potential drivers organizing the injured niche. We first identified predicted signaling initiated by the Injured PTs using Nichenet51 and found that Clcf1 – Crlf1 and Jag1-Notch3 signaling were upregulated between Injured PTs and fibroblasts within the AKI samples (Fig. 3A and Supplementary Fig. 9A). Upregulation of Crlf1 and Clcf1 in kidney injury and disease has been indicated in several prior studies27,52,53,54, and Crlf1 upregulation has been reported in the fibrotic lung55. Single-cell RNAseq data analyzing IRI in the mouse kidney has predicted a Clcf1-Crlf1 signaling axis between injured PT cells and fibroblasts15. Clcf1 encodes a member of the Il-6 cytokine family, which is thought to engage Crlf1 as a chaperone in signaling through the receptor Cntfr56,57. While Clcf1 was specifically expressed within injured PTs (Fig. 3A and Supplementary Figs. 9A, 10), Crlf1 was predominantly expressed in a distinct subset of ME-5 associated fibroblasts (Fig. 3A and Supplementary Figs. 9A, 11).
A Mean expression of two top ligands and their receptors identified by NicheNet calculated for the cell types comprising ME-5. B Illustration showing hypothesized cellular behaviors under distance-dependent and distance-independent signals. Cell A expresses a ligand while the expression of the receptor on cell B can be constant over the distance between Cell A and Cell B (distant independent) or changing over distance (distant dependent). C Fraction of Fibroblasts expressing Crlf1 and Notch3 over distance from the nearest Injured PT cell. Cells were divided into 20um distance bins and the fraction of expressing fibroblasts was calculated out of all fibroblasts in each bin. D Spatial expression of Clcf1 on Injured PT (gold) and Crlf1 (blue) on fibroblasts. E Zoomed-in image on the inset in (D) showing Clcf1 on Injured PT and Crlf1 on fibroblasts. F Zoomed-in image showing mRNA dots of the corresponding genes. G Antibody-staining of the Injured PT marker Vcam1 coupled with RNAscope evaluation of Clcf1, Crlf1, and Cntfr, the receptor for the Clcf1-Crfl1 complex. Staining was done on a control sample. The image is of one representative control sample out of two. A larger kidney area from this experiment is presented in Supplementary Fig. 13. H Same as in (G), measured on AKI sample. The image is of one representative AKI sample out of three. A larger kidney area from this experiment is presented in Supplementary Fig. 13.
Our analysis also identified upregulation of Cxcr6 in T cells and Itgax in DCs within AKI samples suggestive of Injured PT signaling to T cells and to dendritic cells through Cxcl16 - Cxcr6 and Icam1 – Itgax signaling axes, respectively, recruiting immune cells within AKI (Supplementary Fig. 9A). In the context of liver fibrosis, Cxcr6-dependent recruitment of CD8 T cells and NKT cells is known to contribute to disease progression58,59,60 and a similar role has been suggested for a Cxcl16 - Cxcr6 axis in kidney fibrosis61,62,63. Macrophages in AKI also express Cxcl16, consistent with a contribution to T-cell recruitment (Supplementary Fig. 9A). Csf1 - Csf1r signaling axis between injured PTs and macrophages was also upregulated within AKI samples (Supplementary Fig. 9A), in line with previous studies that link injured epithelial Csf1 production to macrophage-mediated recovery following AKI64. Analysis of two published scRNAseq and snRNAseq datasets of dissociated human and mouse kidney samples showed that the elevated expression of the top ligands identified here, including Clcf1 and Csf1 as well as Cxcl16, Icam1, and Tgfb2, was conserved across species (Supplementary Fig. 9B).
We hypothesized that spatially localized ligand-receptor interactions (distance dependent; Fig. 3B) will contribute specifically to the structuring of the microenvironment around and via injured PTs as opposed to signaling with no spatial preference (distance independent; Fig. 3B). To assess the spatial localization of the signals, we quantified the fraction of receptor-expressing target cells as a function of the distance of the target cells from the nearest Injured PT, regardless of ME assignment. Figure 3C plots the fraction of Crlf1 + (left) and Notch3 + (right) fibroblasts over distance from injured PTs. While there is a relatively constant fraction of Notch3 + fibroblasts at varying distances from injured PT cells, the fraction of Crlf1 + fibroblasts decreases as the distance from the injured cells increases. Similar to Notch3, little to no spatial preference was detected for other receptors associated with Injured PTs signaling to fibroblasts, vasculature, and different immune cell types (Supplementary Fig. 12). To validate the predicted expression and spatial distribution of the Clcf1-Crlf1 pairings, we combined anti-Vcam1 immunodetection with serial RNA-FISH on control and AKI samples. Consistent with seqFISH analysis, Crlf1 + cells were specifically concentrated adjacent to Clcf1 + Vcam1 + Injured PTs (Fig. 3G, H and Supplementary Fig. 13). Contrary to the specific localization of Crlf1 around injured cells, Acta2, a well-documented marker of inflammatory myofibroblasts15, exhibited heterogeneous expression (Supplementary Fig. 14). Clcf1, the predicted target of Crfl1 activity, signals through Cntfr. Cntfr expression colocalized with Crlf1 in peri-epithelial myofibroblasts but was not limited to this population (Fig. 3G, H).
Taken together, our findings implicate injured PT signals in the remodeling of vasculature, recruitment of immune cells, and reshaping of the fibroblast population within the AKI samples. We show that while several signaling events are upregulated within AKI samples, only Clcf1-Crlf1 interactions are highly spatially localized. Signaling which is upregulated but does not show spatial localization could present global changes following AKI or past events leading to the formation of the injured niche. Our combined spatial data and validation suggest that Crlf1 is a specific identifier of fibrotic processes closely coupled to Injured PTs and that Clcf1 - Clcf1 interaction with fibroblast cells is a constitutive defining feature of the injured niche.
Fibroblasts show distinct expression patterns between different MEs
To determine whether gene expression changes could be identified more precisely in different MEs, we analyzed fibroblasts within the four AKI-specific MEs (ME-5, ME-9, ME-15, and ME-16). Both the composition of cell types and location in the tissue were distinct for each ME (Fig. 4A – left, middle). ME-5 localized to the cortex and corticomedullary boundary, ME-15 extended from the corticomedullary boundary to the medulla, ME-9 was concentrated around TAL_2 cells in the medulla, and ME-16, comprising predominantly immune cell types, was scattered throughout the kidney (Fig. 4A and Supplementary Fig. 8).
A Left - coordinates plot showing the composition of four MEs enriched in AKI samples. Middle-spatial locations of each of the MEs in one AKI sample. Right - expression of marker genes of each ME in all fibroblasts in the sample. B Differentially expressed genes within fibroblasts between the different MEs. The heatmap shows the mean expression of each of the genes, calculated for fibroblasts within each ME as well as for all fibroblasts in control and AKI. The depicted MEs represent four enriched in AKI (ME-5,15,9,16) and five enriched in control (ME-1,3,7,8,12). C Top – sub-clusters of Fibroblasts based on gene expression presented in Umap space. Clusters were calculated using Leiden clustering. Bottom – same ME and marker genes as in (A), presented on Umap space. D Mean expression score for the DE genes shown in panel (B) calculated within fibroblast populations in a time course snRNAseq data following injury in a mouse model of AKI. The scores were calculated for individual fibroblasts using Seurat and averaged for each mouse. The data presented in the heatmap shows the average of all mice in each condition. The minimum values were subtracted for each score such that minimum score = 0. N = 3 mice per time point. E The same calculation as in (D) was performed on a dataset of samples collected from patients who have undergone AKI (n = 11), exhibit CKD (n = 13), and are compared to normal kidney biopsies (n = 14). F Vcam1 antibody-staining with Clcf1, Crlf1 and Npr3 expression measured using RNAscope on control sample. The image is of one representative control sample out of two. G Same as in (F) on the AKI sample. The image is of one representative AKI sample out of three.
The top enriched fibroblast gene sets for each ME, shown in Fig. 4A, B, revealed specific ME-associated gene expression (see Supplementary Data 1 for a full list of differentially expressed genes). Figure 4A shows that the expression of the top ME-associated genes is highly restricted to the location of each ME in the physical space. Interestingly, we found that many of these genes were not detected when clustering fibroblasts based on gene expression alone without considering spatial ME information (Fig. 4C). Out of the four ME-specific fibroblast maker genes Crlf1, Actg2, Igfbp5 and Iigp1, only Igfbp5 was enriched in the gene expression-based cluster (cluster 3 in Fig. 4C), while the other genes did not show cluster-specific expression. These data underscore the importance of considering the spatial environment of fibroblasts to identify genes with a potential functional significance.
We found that Crlf1, Npr3 (which encodes the natriuretic peptide receptor 3), and Timp1 (which encodes TIMP metallopeptidase inhibitor 1) were markedly enriched in the ME-5 fibroblasts (Fig. 4A, B). In previous work, Npr3, a known blood pressure regulator, reduced disease severity upon inhibition in a model of cardiac fibrosis65, whereas upregulation of Timp1 was reported to increase kidney scarring66,67. Here, we validated the co-expression of Crlf1 and Npr3 in association with injured PTs using immunostaining and serial RNA-FISH (Fig. 4F, G). In ME-15, which comprises fibroblasts and macrophages, Actg2, encoding a member of the smooth muscle actin family, was the top gene identifier of ME-15 fibroblasts (Fig. 4A). Actg2 has not previously been associated with kidney disease, although a recent study identified Actg2 as a candidate in long term kidney impairment following acute decompensated heart failure68. ME-15 fibroblasts also elevate the expression of Apoe and C1qa, key inflammatory genes highly expressed by macrophages, suggesting both macrophages and fibroblasts contribute to the inflammatory environment. Fibroblasts in ME-16 were distinguished by the expression of Ligp1, which encodes the interferon-induced GTPase1, and the expression of Bcl3, an inhibitor of apoptosis, and the caspase Casp4. In contrast to ME-5, -15, and -16 fibroblasts, ME-9 fibroblast marker genes were not highly expressed within the total AKI fibroblast population, and several genes were higher in control fibroblasts. One of the most strongly enriched fibroblast genes in this ME was the insulin growth factor binding protein (IGFBP), encoded by the Igfbp5 gene. IGFBPs have been linked to CKD progression69 and disease progression in a mouse model of diabetic kidney disease70.
Spatially dependent stromal gene expression in the mouse and human kidney
Given the highly distinct gene signatures of fibroblasts across the different MEs, we next asked if the expression of the identified ME-specific fibroblast marker genes changes along the AKI-to-CKD transition. To answer this question, we analyzed published mouse snRNA-seq15 collected at five-time points post IRI-invoked AKI (4 h, 12 h, 2 days, 14 days, and 6 weeks; Fig. 4D). We calculated a ME score for fibroblasts, defined as the combined expression of all the ME-specific genes for each of the AKI-enriched MEs (ME-5, ME-15, ME-9, and ME-16). We also calculated scores for fibroblast marker genes in control and AKI samples regardless of ME assignment (Control-Fib and AKI-fib). Figure 4D presents the average score for all the samples in each condition, normalized such that the minimum score for each gene set = 0.
As expected, following AKI, the control-fibroblast score is reduced immediately and remains relatively low until 6 weeks post AKI, while the AKI-fibroblast score is low in control samples, but increases and peaks at day 2 after AKI, remaining elevated at 6 weeks. This timeframe suggests that the injury induces fibroblast differentiation and recruitment, as captured in the AKI-fibroblast score, and that AKI-induced fibroblasts persist 6 weeks after injury. We observed divergent patterns in the ME-specific fibroblast scores, where ME-5 and ME-15 scores were similar to the score of AKI-fibroblast, peaking at day 2 post AKI, with ME-15 score remaining high also at later time points. However, the ME-9 score was significantly delayed and peaked at 14 days post-AKI, suggesting a later appearance for this fibroblast population following AKI. Although ME-16 genes were highly specific to AKI samples in our data (Fig. 4B), the ME-16 score was low throughout the time course. As this population of fibroblasts is relatively small, the signal may be diluted within other populations within the sequencing data.
We repeated this analysis with a scRNA-seq data set from human patients with AKI, CKD, or normal kidneys as control28(Fig. 4E). Similarly to the mouse data, we find that here, too, ME-5 and ME-15 scores are high in AKI relative to control and CKD. ME-16 scores were elevated in both AKI and CKD, although at lower levels. Assuming that the sequencing data is capturing cells from different sites of the tissue, our analysis suggests that the abundance of the different fibroblast subsets detected in each condition reflect changes along the AKI-to-CKD transition and that these populations are common to the mouse and human kidney. Thus, beyond spatial mapping, seqFISH analysis provides additional molecular granularity as to how cell populations acquire distinct properties during a biological process.
Spatial distribution of immune cells correlates with inflammatory potential
Next, we focused on immune cells as drivers of inflammatory fibrosis. To identify T cell subtypes present in our data, we mapped T cell populations onto a reference data set of mouse T cells71, using SEURAT. We identified known T cell populations, including CD4 and Tregs (high Cd83 and Ctla4 expression), naïve, effector (high Cxcr3 expression and expression of cytotoxic genes such as Nkg7), and exhausted (no Cxcr3 expression but expression of the inhibitory molecule Pdcd1) CD8 T cells (Fig. 5A). When we quantified the fraction of each T cell subtype within MEs − 5, − 15 and − 16 in the AKI samples, we found a difference in the fraction of CD4 T cells amongst all T-cells: ~ 40% in ME-5, 50% in ME-15 and 55% in ME-16 (Fig. 5B). We noticed a similar trend for Tregs, although, as expected, Tregs represented a smaller subset of the T cell population (Fig. 5B). The inverse trend was apparent for effector CD8 T cells, and no clear trend was evident for naïve and exhausted CD8 cells (Fig. 5B).
A All T cells in our dataset were mapped to a tumor-infiltrating lymphocyte dataset. The T cells were mapped to six known cell populations - helper T cells (CD4T), cytotoxic T cells (CD8T Eff_mem), naive and exhausted CD8 (CD8_Naive, CD8_exhauseted), and regulatory T cells (Treg). The violin plot shows marker gene expression for each of the T cell subtypes in our data. B Fraction of T cells of each subtype in AKI enriched MEs (5, 15, 16) for n = 3 AKI mice. Each dot represents one mouse. The fraction is the total fraction of each subtype out of all T cells within the specified ME. The boxplot middle line represents the median value, and box boundaries show the 25th and 75th percentiles. Source data are provided as a Source Data file. C Average expression of three M1 (blue) and four M2(red) markers within Macrophages in the AKI enriched MEs. D Fractions of Mrc + (left) and Ccr7 + (right) Macrophages within the AKI enriched MEs boxplots showing the distribution for n = 3 AKI mice as in (B). Dots represent individual mice. The boxplot middle line represents the median value, and box boundaries show the 25th and 75th percentiles. Source data are provided as a Source Data file. E Spatial distribution of the same genes as in (D). Left - ME-15 is indicated and Mrc+ cells are colored in red. Right - ME-16 is indicated and Ccr7+ cells are colored in blue. F. Spatial locations of the T cell subtypes within one AKI sample. ME-16 is depicted in color. G Vcam1 antibody staining, as well as the markers Cd4, Cd8, and the DC marker Clec9a, are detected with RNAscope on a control sample. The image is of one representative control sample out of two. A larger kidney area from this experiment is presented in Supplementary Fig. 15. H Same as in G for an AKI sample. The image is of one representative AKI sample out of three. A larger kidney area from this experiment is presented in Supplementary Fig. 15.
Since macrophages represent a large fraction of the immune cells in AKI-specific MEs, we measured marker genes linked to differential activation states of M1 and M2 macrophages. In general, M1 macrophages promote disruptive, disease-associated inflammatory responses, whereas M2 macrophages promote constructive, inflammatory-associated tissue repair72. We found that the average expression of the M2-related genes Mrc1, Cd163 and Arg1 was increased in macrophages within the ME-5 and ME-15 groupings, while M1-related genes Cxcl10, Ccr7, Cd40 and Cd86, were expressed at higher levels on the average in ME-16 macrophages (Fig. 5C). In agreement with these data, Mrc1+ macrophages were more prevalent in ME-5 and ME-15, and Ccr7 + macrophages were more prevalent in ME-16 (Fig. 5D, E).
Tertiary Lymphoid Structures (TLS), increasingly recognized as contributors to chronic inflammation22, are organized lymphoid aggregates that form ectopically in response to a disturbance in tissue homeostasis and then act as localized hubs that enable signal exchange between immune cells, which in turn promotes the development of adaptive immunity within a tissue. Unlike secondary lymphoid structures (such as the lymph nodes and the spleen), TLS is unencapsulated, exposing cells within to multiple signals from the environment. In the context of autoimmune and chronic inflammation, the presence of TLS is correlated with severe disease, and in the context of chronic kidney disease, with more severe inflammation and fibrosis20. Mechanistically, the emergence of TLS in a long-term model of the AKI to CKD transition was linked to the maturation of B cell-directed autoimmunity against the target kidney tissue27,73.
We found that the cellular composition and appearance of ME-16 show characteristics of immune aggregates reminiscent of nascent TLS. ME-16 is composed of T cells, DCs, and macrophages with a small number of fibroblasts (Fig. 4A). ME-16 clusters also appear scattered without a preference for the cortex or the medulla (Figs. 2C,4A). When overlaid on the locations of ME-16 (Fig. 5F), the spatial distribution of CD4 and CD8 T cells were dispersed within and outside of ME-16, with CD4 T cells clearly aggregated in ME-16. Thus, ME-16 has the expected properties of early-forming immune aggregates in which CD4 T cells and Tregs are providing instructive signals to CD8 and other immune cells. Further, employing RNA-FISH detection of Cd4, Cd8a, and the DC marker Clec9, we detect early aggregation of DCs, CD4, and CD8 T cells 4 weeks following AKI (Fig. 5G, H and Supplementary Fig. 15). CD8 T cells trend towards cortical prevalence and CD4 towards medullary enrichment. Moreover, elevated Cxcr6 expression in CD8 T cells (Supplementary Fig. 16) suggests interaction with Cxcl16-expressing Injured PTs (Supplementary Fig. 9A).
Although the clear enrichment of T cells and DCs within ME-16 is consistent with localized antigen presentation, we did not detect any B cells, in line with kidney profiling data showing a later engagement of the B cell response26,73. Because M1 macrophages are preferentially enriched within ME-16, we propose that ME-16 represents an early lymphoid aggregate that propagates inflammation. In contrast, the presence of M2 macrophages in ME-15 and ME-5 suggests that these MEs are undergoing a combination of inflammatory and fibrotic processes where the inflammatory response is sequestered and replaced by fibrotic processes74.
Discussion
Cell identity is largely defined by gene expression profiles. However, unique microenvironmental interactions within tissues also represent important determinants of both gene expression and cell identity. To gain a more granular understanding of how cell identity and function are shaped by such micro-level interactions, we need to incorporate information about gene expression with cellular locations within tissues. Transcriptionally heterogeneous cellular populations characterized by scRNA-seq analysis are typically clustered into sub-populations. However, without a specific reference to a corresponding purified cellular subtype, this approach is prone to over- or under-clustering, and may even fail to identify important cellular populations41. Spatial transcriptomics overcomes this limitation as we first cluster cells globally into well-characterized cell types, and then integrate this information with the cellular locations to identify cellular microenvironments (MEs) for each individual cell. Thus, each cell is given two identifiers – cell type, based on its gene expression, and a cellular ME, based on the composition of its close neighbors.
We leveraged seqFISH to study cellular and structural changes following kidney injury, which dramatically disrupts the highly complex kidney architecture and triggers two parallel responses: regeneration of the damaged epithelia and fibrosis leading to the development of CKD. Based on our analysis, we have created a comprehensive map of the cellular, molecular, and structural changes following AKI, which can be leveraged to enhance our understanding of pathophysiologic processes underlying the AKI-to-CKD transition. To ease data accessibility for the scientific community, we have created an interactive website through which the data can be explored (https://woldlab.caltech.edu/ci2-celltiles/Mouse-Kidney-Fibrosis/).
Our approach allowed us to gain a deeper understanding of an injured ME undergoing responses specific for the AKI-insult; here referred to as ME-5. Within ME-5, we identified injured PTs, fibroblasts, macrophages, as well as immune cells (T, DC), and normal kidney cell types (PTS1, PTS2, PTS3, and vasculature). Injured cells within the ME expressed specific ligands, which were conserved in human-injured PTs. When we used spatial coordinates to identify specific ligand-receptor interactions that were highly spatially localized, we found that Clcf1-Crlf1 interactions were upregulated within the injured ME, and spatially localized between injured PTs and fibroblasts. This finding suggests that Crlf1 expression within fibroblasts is a highly specific determinant for the fibrotic processes surrounding the injured cells. At a broader level, our approach illustrates that analyzing localized interactions at a microenvironmental level can help us gain a multi-faceted view of how cell-cell communication contributes to tissue organization under both normal and injury settings.
Following injury, global gene expression and cellular populations differed across the emerging MEs. Importantly, we detected gene markers delineating ME-specific fibroblast subpopulations that we did not detect by our initial gene expression-based clustering. Specifically, we found that Crlf1, together with Timp1 and Npr3, are specific markers for fibroblasts localized around injured cells, possibly contributing to fibrosis. Conversely, Actg2, Apoe, and C1q labeled fibroblasts, which were more medullary and distant from the injured cells, suggesting that these propagate inflammation. Interestingly, we identified ME-16, a specific microenvironment that resembled a tertiary lymphoid structure both in terms of morphology and cell type composition. In this ME, we found preferentially CD4 T cells and Tregs, as well as macrophages with a pro-inflammatory gene expression. Thus, our spatial analysis was able to identify discrete cellular subsets not identified by gene expression alone and revealed a morphological structure with potential functional importance.
Although our extensive gene panel allowed us to detect all major kidney cell types as well as location-specific gene expression patterns and intercellular interactions, our panel is still limited. To comprehensively capture the consequences of intercellular interactions, there is a need to identify additional molecular pathways and, therefore, scale up the number of measured genes in future studies. In addition, since intercellular signaling was inferred from RNA expression of ligands and receptors, identified molecular interactions will have to be validated using additional approaches in future studies. We have demonstrated that the identification of MEs captures the kidney morphology and is highly coherent between the different samples. However, similarly to cell type clustering, the process of ME identification required thresholding the data (30 µm neighborhoods) and manual curation, which could potentially introduce biases. Furthermore, our analysis suggests that, upon injury, specific fibroblast populations appear in earlier time points, and TLS-like MEs, which can develop into larger TLS, form in later timepoints. To verify these hypotheses, performing a time-course experiment ranging from very early timepoints following injury (2 days) and up to several months would be required. This can be addressed in future work.
In conclusion, this study employed high-resolution spatial information to characterize injury-invoked changes in the kidney, including the formation of injured microenvironments and lymphoid aggregates, localized AKI-specific inter-cellular interactions, and fibroblasts subtypes with potential functional roles in injury progression. Our data highlight the importance of spatial context and cellular interactions in shaping cellular responses to kidney injury. We envision that this kind of data can be leveraged beyond constructing spatial microenvironments where correlations between cell types and gene expression within their neighbors can be used to find continuous changes and flow of information between cells within the tissues.
Methods
Mice
Mouse handling, husbandry, and surgical procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee at the University of Southern California (protocol number 11911). Mice were kept on a 14/10 light/dark cycle, with an average humidity range of 45–50% and temperatures of 21–23 °C.
Ischemia-reperfusion injury model and tissue collection
Warm bilateral renal ischemia-reperfusion injury (IRI) was performed as previously described on male C57BL/6 J (weight 26–27 g, age 8–13 weeks)27. Male non-surgery C57BL/6 J mice were used as controls. Kidneys were collected 28 days post-IRI. After organ perfusion with ice-cold RNase-free phosphate-buffered saline (PBS), the kidney capsule was removed and the kidneys were fixed in RNase-free 4% paraformaldehyde-PBS overnight at 4 °C. Kidneys were then equilibrated overnight in RNase-free 30% sucrose-PBS, embedded in O.C.T. compound (Tissue-Tek) in a dry-ice ethanol bath and stored at − 80 °C. Ten-micrometer cryostat sections were cut from embedded kidneys and placed on the functionalized coverslips on dry ice for subsequent seqFISH experiments. Secondary validation experiments used sections adjacent to those analyzed by seqFISH.
Readout probe design and synthesis
Readout probes 15 nt in length were designed as previously described75. In brief, a set of probe sequences was randomly generated with combinations of A, T, G, or C nucleotides. Readout-probe sequences within a GC-content range of 40–60% were selected. We performed a BLAST search against the mouse transcriptome to ensure the specificity of the readout probes. To minimize cross-hybridization of the readout probes, any probes with ten contiguously matching sequences between readout probes were removed. The reverse complements of these readout-probe sequences were included in the primary probes according to the designed barcodes. The fluorophore-coupled 15-nt readout probes (Alexa 488, 647 (Thermo Fisher Scientific) and Cy3B (GE Healthcare).
Primary probe design
Primary probes were designed as previously described35,76 with some modifications. The probe length was set to 35nt. Any gene with less than 16 probes was discarded. Housekeeping genes and highly expressed genes were discarded to avoid optical crowding of the fluorescence probes. The probes were divided into two pool designs: the first pool comprised 131 cell marker genes; this pool was barcoded with 4 pseudocolors to be decoded over 16 hybridization rounds in total. The second pool contained the rest of the genes and was barcoded with 9 pseudocolors to be decoded over 36 hybridization rounds in total. Each pool was read in three channels, however we used the seqFISH+35 design scheme where a set of genes was only decoded over one channel to avoid chromatic aberration. The first cell marker pool was designed such that there were 44, 43, and 44 genes barcoded in the first, second, and third fluorescent channels respectively. The second pool contained 390, 389, and 390 genes in the first, second, and third channels, respectively. The codebooks and probe sequences are available as Supplementary Data 2 and 3.
Serial probe design and hybridization
To ensure detection of the injury specific markers Vcam1 and Havcr1, we created primary probes which were not barcoded for these two genes. Vcam1 was targeted with 30 probes and Havcr1 with 27 probes. The probes were designed such that each one carried two binding sites for the same readout (without splitting into 4 readout barcodes). The probes were obtained from IDT and no further synthesis was performed. Probes were hybridized on the samples together with the barcoded probe pool at a concentration of 10 nM per probe. The probe sequences are available as Supplementary Data 4.
Primary probe synthesis
Primary probes were generated from oligoarray pools (Twist Bioscience) as previously described35. In brief, probe sequences were amplified from the oligonucleotide pools with limited two-step PCR cycles, and PCR products were purified using QIAquick PCR Purification Kit (Qiagen 28104). Then, in vitro transcription (NEB E2040S) was performed, followed by reverse transcription (Thermo Fisher EP0751). After reverse transcription, the single-stranded DNA (ssDNA) probes were alkaline hydrolyzed with 1 M NaOH at 65 °C for 15 min to degrade the RNA templates and neutralized with 1 M acetic acid. Finally, probes were ethanol precipitated, and eluted in nuclease-free water.
Coverslip functionalization
Coverslips were cleaned with a plasma cleaner on a high setting (PDC-001, Harrick Plasma) for 5 min. Then coverslips were rinsed with 100% ethanol three times and heat-dried in an oven at > 90 °C for 30 min. Next, the coverslips were treated with 100 μg μl−1 of poly-D-lysine (P6407; Sigma) in water for > 3 h at room temperature, followed by three rinses with water. The coverslips were then air-dried and kept at 4 °C for no longer than 2 weeks.
Probe hybridization
Tissue sections were permeabilized in 70% ethanol at − 20 °C for > 1 h, cleared with 8% SDS (AM9822; Invitrogen) in 1 × PBS for 45 min at room temperature, then washed in PBS and left to dry. Once dry, a house-made flow cell was attached to the coverslip to allow the flow of hybridization reagents. A hybridization mix with 1 nM of each oligo in 50% Hybridization Buffer (50% HB: 2 × SSC, 50% Formamide (v/v) (Invitrogen AM9344), 10% Dextran Sulfate (Sigma D8906) in Ultrapure water) was applied to the tissue section. Sections were then incubated for 30 h at 37 °C, washed in 55% Wash Buffer (55% WB: 2 × SSC, 55% Formamide (v/v), 0.1% Triton X-100 (Sigma 93443)) for 30 min at 37 °C, and then washed twice in 2× SSC at room temperature.
Microscope setup and seqFISH imaging
Setup, sequential hybridizations, and imaging were performed as previously described35,77 with some modifications. Data was captured on a Leica DMi8 microscope equipped with a confocal scanner unit (Yokogawa CSU-W1), fiber-coupled lasers (643, 561, 488 and 405 nm) from CNI, Shanghai Dream Lasers Technology and filter sets from Semrock, a sCMOS camera (Andor Zyla 4.2 Plus), 63 × oil objective lens (Leica 1.40 NA), and a motorized stage (ASI MS2000). A custom-made automated sampler moved designated readout probes in the hybridization buffer from a 2.0- ml 96-well plate through a multichannel fluidic valve (IDEX Health & Science EZ1213-820-4) to the custom-made flow cell with a syringe pump (Hamilton Company 63133-01). The syringe pump was also used to move other buffers through the multichannel fluidic valve to the custom-made flow cell. Integration of the imaging and the automated fluidics delivery system was controlled by custom-written scripts in μManager.
For hybridization and imaging, the sample within the custom-made flow cell was first connected to the automated fluidics system on the motorized stage of the microscope. The fields of view (FOVs) were registered using nuclear signals revealed by preincubation in 5 μg ml−1 DAPI (Sigma D8417) in 4 × SSC. Imaging was performed with sequential hybridization and imaging routines. The serial hybridization buffer contained three unique readout probes (12.5 nM each) with different fluorophores (Alexa Fluor 647, Cy3B, or Alexa Fluor 488) in 10% EC buffer (10% ethylene carbonate (Sigma E26258), 10% dextran sulfate (Sigma D4911) and 4 × SSC) and was picked up from a 96-well plate and distributed into the flow cell for a 20 min incubation. After the serial hybridization, the sample was washed with 1 ml of 4 × SSCT (4 × SSC and 0.1% Triton-X), followed by a washing step with 500 μl of the 12.5% wash buffer. Then, the samples were rinsed with 500 μl of 4 × SSC, and stained with 200 μl of the DAPI solution for 60 s to visualize nuclei. Next, the anti-bleaching buffer was flown through the sample for imaging. The anti-bleaching buffer was made of 50 mM Tris-HCl pH 8.0 (Invitrogen 15568025), 300 mM NaCl (Invitrogen AM9759), 2 × SSC, 3 mM trolox (Sigma 238813), 0.8% D-glucose (Sigma G7528), 1000-fold diluted catalase (Sigma C3155) and 0.5 mg ml−1 glucose oxidase (Sigma G2133).
Snapshots were acquired with 643, 561, 488, and 405- nm fluorescent channels per field of view (FOV). After image acquisition, 1 ml of the 55% wash buffer was flown for 1 min to strip off readout probes, followed by a 5 min incubation before rinsing with 4 × SSC. The serial hybridization, imaging and signal extinguishing steps were repeated until all rounds were completed. Blank images displaying only cellular autofluorescence were imaged at the beginning and end of the routines.
Anti-Vcam1 antibody staining was performed as the last step of the hybridizations. Prior to antibody labeling, the tissue was washed multiple times with 1X PBS and a blocking buffer containing 1x PBS (Ambion, AM9625) ultra-pure BSA 1% (Ambion, AM2616) TritonX-100 0.3% (Sigma-Aldrich, 93443) Dextran Sulfate Low MW 0.1% (Sigma-Aldrich, D4911-10G) Salmon Sperm DNA 0.5 mg/ml (Invitrogen, am9680) was applied and incubated with the sample for 15 min at room temperature. Then, primary rabbit anti-Vcam1 antibody (Abcam, ab134047) was introduced at a 1:100 dilution in the blocking buffer and the sample was incubated at room temperature for 1 hr, washed three times in 1X PBS and a secondary anti-rabbit 647 antibody (Invitrogen, A32733) applied at 1:500 dilution for 1 hr at room temperature. Finally, the sample was washed extensively in > 2 ml of PBS and incubated for 30 min with PBS at room temperature to remove excess antibodies. Finally, DAPI and anti-bleach buffer were applied before imaging. The same FOVs were then imaged with the antibody label.
Immunofluorescence and RNAscope
Immunofluorescence and RNAscope experiments were performed as previously described19. using the following antibodies and probes: mouse IgG2a anti-alpha Smooth Muscle Actin (aSMA, 1:2000, Sigma-Aldrich, A5228), goat anti-Vcam1 (1:200, R&D Systems, AF643), rabbit anti-Vcam1 (1:200, Abcam, ab134047), donkey anti-rabbit AlexaFluor 594 (1:500, Molecular probes, A21207), donkey anti-goat AlexaFlour 594 (1:500, Life Technologies, A11058), chick anti-rat AlexaFluor 647 (1:500, Molecular Probes, A21472). RNAscope Mm-Acta2 (ACD, 319531), RNAscope Mm-Cd4-C3 (ACD, 406841-C3), RNAscope Mm-Cd8a-C2 (ACD, 401681-C2), RNAscope Probe - Mm-Clcf1-C1 (ACD, 457971), RNAscope Mm-Clec9a-O1 (ACD, 537731), RNAscope Probe - Mm-Cntfr-C3 (ACD, 457981-C3), RNAscope Probe - Mm-Crlf1-C2 (ACD, 446891-C2), RNAscope Probe - Mm-Npr3-C3 (ACD, 502991-C3). Images were acquired on Leica Sp8.
Image analysis of seqFISH data
Image registration
Image registration was performed using phase cross-correlation on DAPI-stained images for each FOV. The initial hybridization round was used as a reference for estimating required translational shifts.
Image preprocessing
Aligned images underwent background subtraction using dilated blank images to further remove unwanted background noise. Post background subtraction, further background removal was performed using a 5 × 5 high pass Gaussian filter, followed by a 3 × 3 low pass Gaussian filter to mitigate hot pixels and make spots more Gaussian-like for improved 2D Gaussian fitting. Image intensities across channels and serial hybridizations were normalized by 80-99.999% percentile clipping and rescaling between 0-1.
Spot calls and spot feature generation
Sub-pixel centroids of identified spots were obtained using DAOStarFinder, a spot-picking algorithm from Astropy that performs fast 2D Gaussian fits. The full-width half-max (FWHM) was numerically optimized to find the best parameter for spot calling. Features such as flux, peak amplitude, sharpness, bilateral to four-fold symmetry, and symmetry of Gaussian fits were recorded and stored for each spot from the DAOStarFinder algorithm. To obtain additional features such as total spot area, a 7 × 7 bounding box was used to isolate each spot, and then a local adaptive threshold using a Gaussian kernel was used to obtain the area of the spot.
Mask generation and spot mapping
Cell masks were generated using Cellpose 2.0. As cell borders are hard to discern unambiguously, to maximize the accuracy of single-cell detection, we segmented based on proximity to the nucleus (DAPI visualization), not the cell membrane. This resulted in lower RNA counts per cell but increased the confidence of single-cell analysis. Masks touching the edges of the image were removed to prevent edge bias or undercounting of spots in cells where a cell will be truncated because it touches an edge. In addition, 2 pixels were deleted between two or more masks that touch to prevent spot mixing between cell masks. Spots were mapped to each cell mask to assign cell IDs.
Decoding
Super-resolved, mapped spots were decoded using a support vector machine (SVM) embedded, feature-based symmetrical nearest neighbor algorithm. First, the decoder removed unwanted noise through an SVM model with a radial-basis function or polynomial kernel to filter false spots based on their spot characteristics. Post-filtering, each spot was assigned a probability score on the likelihood that a given spot corresponded to a true barcode based on its spot features. Then, the algorithm performed a nearest neighbor radial search for each spot at a distance of 1-2 pixels across barcoding rounds (ignoring spots within the same barcoding round). Searches are performed comparing each round to the other individual rounds in a parallel fashion. When multiple spots fell within the search radius of a given reference spot, each spot was assigned a score based on distance, intensity, and size. The highest scoring spot was chosen, and an overall codeword score was assigned after picking the best spot for each barcoding round. These scores (including the overall codeword score) are influenced by the number of total neighbors encountered. Each codeword was also assigned an ambiguity score, which is the total number of additional neighbors found over the expected for each barcoding reference round. The probability score (obtained from individual spot probabilities) and the codeword score were used to generate the overall score for each decoded barcode. Each completed barcode also underwent a parity check to distinguish true signal from noise, while also allowing one drop in spot calls. Once the best set of spots was chosen, they were filtered based on the number of times the same set of spots was picked when changing the barcoding round reference, keeping spot sets that appear >= 3 times. If there are any codewords that have overlapping spots, then their overall codeword score was used to pick the best one. If they had the same score, then the codeword with the smallest total distance between spots was used. After the 1st round of decoding, unused spots were resubmitted for additional rounds of decoding (up to 2) to try and assign leftover spots. Decoded spots, including “trues” and “fakes”, were combined to generate a final decoded spots table. These decoded spots were sorted based on their overall codeword score and subsampled to calculate false positive rates (FPR). Generally, the codeword score had a direct relationship with fake spots with low scores corresponding more with fake spots and high scores corresponding to true spots. Once an FPR of 15% was reached, only those subsets of true and fake codes were used for analysis. Once the desired barcodes are obtained, a gene-by-cell matrix is generated for downstream analysis.
SVM training
Called spots underwent a quick pass decoding to obtain labels for true and fake spots. The number of fake spots must be 500–500,000 for the SVM classifier to become active. The number of true spots is down-sampled to match the number of fake spots for balanced training. Eighty percent of the data was used for training and 20% for validation. Spot features were normalized using a MinMax Scaler and stored, then the SVM model underwent hyper-parameter tuning of C, gamma, and degree parameters using GridSearchCV with 8-fold cross-validation using either a polynomial or radial-basis function kernel. Once the optimal parameters were obtained, the test set underwent the same feature scaling parameters that were used for the training data to gauge out-of-sample performance.
False Positive Rate
The false positive rate was calculated as follows:
The frequency of off-targets (\({\nu }_{{off}}\)) was estimated as the number of observed off-target codes \(({N}_{{off}})\) divided by the number of off-target codes in the codebook (\({M}_{{off}}\)). Assuming that on-target codes have the same frequency of error as off-target codes, we can multiply the \({\nu }_{{off}}\) by the number of on-targets (\({M}_{{on}}\)) in the codebook to obtain estimated false positive counts (\({{FP}}_{{counts}}\)). \({{FP}}_{{counts}}\) were then normalized by the number of observed on-targets \(({N}_{{on}})\) to obtain false positive rate (FPR).
Image analysis of serial probes
Images of the serial probes were subjected to the same background subtraction and processing as described above. The RNA spots were then manually threshold based on visual inspection of the images. The same threshold was used for all experiments.
Gene expression preprocessing and clustering
The R package Seurat (version 4.1.0)78 was used to process the data. Low-quality cells (RNA count ≥ 25 and/or genes/cell ≤ 2) were removed, and the data was normalized and scaled using the NormalizeData() and ScaleData() functions. Principal component analysis was performed using RunPCA(), considering all genes in the dataset. Batch effects were removed using the Harmony algorithm within the Seurat wrapper RunHarmony()79. The functions RunUMAP(), FindNeighbors(), and FindClusters() (resolution 0.8) were used for dimensionality reduction and cluster identification, resulting in a total of 31 clusters. The distribution of clusters across samples and imaged positions was examined, and six clusters with a strong position bias caused by technical issues such as imaging in a different focal plane (9452 cells in total) were excluded from downstream analysis, resulting in a final data set of 220,970. Differentially expressed genes between clusters were identified using the FindAllMarkers() function with a Wilcoxon Rank Sum test.
ME detection
Detecting the Microenvironments (MEs) was done in the following steps: first, the Euclidean distance between all cells in each sample was calculated. For each cell in the sample, a 30μm radius was defined, and the number of cells within this radius that belong to each cell type was counted. Then, the cell-by-neighbor matrix was centered to have a mean of 0, PCA was performed, followed by Leiden clustering using Scanpy with sc.pp.neighbors() with n_neighbors = 20 and sc.tl.Leiden() with resolution = 0.5. This process resulted in 19 clusters. The results were filtered to remove clusters with < 500 cells total - this resulted in the removal of one cluster. Then the cell type composition in each cluster was evaluated by calculating the mean counts of each cell type within the clusters as in Fig. 2D. Two clusters that shared a highly similar cell type composition were manually merged to create a total of 17 clusters (Fig. 2D) with 220,753 total cells.
NicheNet analysis
NicheNet51 was used to detect signaling between injured cells to the other cell types within the injured niche (ME-5, Fig. 3A and Supplementary Fig. 9A). The Seurat implementation NicheNet was used. Only cell types which had ≥ 500 total cells in ME-5 were considered for the analysis. The calculation of the top ligand-receptor was run for each of the cell types within ME-5 using nichenet_seuratobj_cluster_de() where the sender was set to be Injured PT, the receiver_reference = cell type in Control and receiver_effected = cell type in AKI. The top 10 ligand-receptor pairs were chosen as pairs with the highest ‘weight’ term out of all pairs that NicheNet identified. Supplementary Fig. 9A plots the mean expression of the top ligand-receptor pairs for all cell types in ME-5.
Fibroblasts differential expression analysis
To reduce noise, we first calculated the average expression of all genes across all cell types and z-scored average expression values. All genes with a z-score < 0.1 within fibroblasts were removed from differential expression analysis. We then identified the differentially expressed genes between fibroblasts within the different MEs using the FindAllMarkers() function in Seurat with the following parameters: min.pct = 0.1,logfc.threshold = 0.25,only.pos = T. Figure 4B shows the average expression of the genes detected in each of the AKI enriched or AKI depleted MEs - as calculated by the enrichment of each ME in the control or AKI using a one sided students’ t-test (significance values shown in Fig. 2B). To calculate the score of each gene set on RNAseq data presented in Fig. 4D, E the function AddModuleScore() was used with the differentially expressed genes as features, to apply a score to fibroblasts in the sequencing data as identified by the authors. Score values were averaged for all fibroblasts in each mouse/patient in the data, and then averaged for all mice/patients in each condition. Number of mouse samples n = 3 per time point. Number of patients: n = 11 for AKI, n = 13 for CKD, and n = 14 for normal controls. The minimum values were subtracted for each score such that minimum score = 0.
Reference mapping
For reference mapping of kidney cell types presented in Supplementary Fig. 2B, we mapped our data onto two mouse kidney sequencing datasets from Kirita et al.15 and Ransick et al.3. We compared the un-injured kidney cell types from our control to those of male control samples from both datasets. RunUMAP() was run on the reference object to return the UMAP model and then reference mapping was done using the common genes present in both datasets. Anchors were found with the FindTransferAnchors() functions with normalization.method = “LogNormalize” and reference.reduction = “pca”. Then, MapQuery() was performed with the detected anchors with reference.reduction = “pca” and reduction.model = “umap”. We then enumerate the fraction of cells with the original label that received each of the new labels in each comparison. This data is presented in Supplementary Fig. 2B.
For T cell reference mapping A reference atlas of T cell subsets was used71 reference RDS object can be found here: https://figshare.com/articles/dataset/ProjecTILs_murine_reference_atlas_of_tumor-infiltrating_T_cells_version_1/12478571/2. RunUMAP() was run on the reference object to return the UMAP model and then reference mapping was done using 1121 genes present in both datasets. Anchors were found with the FindTransferAnchors() functions with normalization.method = “LogNormalize” and reference.reduction = “pca”. Then, MapQuery() was done with the detected anchors with reference.reduction = “pca” and reduction.model = “umap”. Reference cell types that were mapped with ≤ 100 cells in total were removed.
Statistics and reproducibility
No statistical method was used to determine sample size. The two serial probe sets against Vcam1 and Havcr1 were not used on one AKI sample (AKI1). Therefore, this sample was excluded from the analysis presented in Fig. 1E and main Fig. 2F, G, which include these genes. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw and processed data generated in this study were deposited in the Dryad database (https://datadryad.org/stash/dataset/doi:10.5061/dryad.bnzs7h4hj) and Zenodo (https://doi.org/10.5281/zenodo.12709329). The codebooks and probe sequences used to generate seqFISH probes used in this study are available as Supplementary Data 2 and 3. The probe sequences used to generate serial probes are available as Supplementary Data 4. Processed data can be browsed interactively at https://woldlab.caltech.edu/ci2-celltiles/Mouse-Kidney-Fibrosis/. Mouse sequencing data generated by Ransick et al. and by Kirita et al. and used in this study are available on GEO with accessions GSE129798 and GSE139107. Mouse T cell expression profiles were obtained from figshare (https://figshare.com/articles/dataset/ProjecTILs_murine_reference_atlas_of_tumor-infiltrating_T_cells_version_1/12478571/2). Human sequencing data was obtained from cellxgene (https://cellxgene.cziscience.com/collections/bcb61471-2a44-4d00-a0af-ff085512674c). Source data are provided with this paper.
Code availability
Scripts used for processing seqFISH images can be found in GitHub (https://github.com/CaiGroup/pyfish_tools) and Zenodo (https://doi.org/10.5281/zenodo.12192195).
References
Su, W., Cao, R., Zhang, X.-Y. & Guan, Y. Aquaporins in the kidney: physiology and pathophysiology. Am. J. Physiol. Ren. Physiol. 318, F193–F203 (2020).
Yu, A. S. L. et al. Brenner & Rector’s the Kidney 11th edition (2020).
Ransick, A. et al. Single-cell profiling reveals sex, lineage, and regional diversity in the mouse kidney. Dev. Cell 51, 399–413 (2019).
Ronco, C., Bellomo, R. & Kellum, J. A. Acute kidney injury. Lancet 394, 1949–1964 (2019).
Mehta, R. L. et al. International Society of Nephrology’s 0by25 initiative for acute kidney injury (zero preventable deaths by 2025): a human rights case for nephrology. Lancet 385, 2616–2643 (2015).
Kellum, J. A. et al. Acute kidney injury. Nat. Rev. Dis. Prim. 7, 1–17 (2021).
Romagnani, P. et al. Chronic kidney disease. Nat. Rev. Dis. Prim. 3, 1–24 (2017).
Kuppe, C. et al. Decoding myofibroblast origins in human kidney fibrosis. Nature 589, 281–286 (2020).
Foreman, K. J. et al. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet 392, 2052–2090 (2018).
Chawla, L. S. & Kimmel, P. L. Acute kidney injury and chronic kidney disease: an integrated clinical syndrome. Kidney Int 82, 516–524 (2012).
Witzgall, R., Brown, D., Schwarz, C. & Bonventre, J. V. Localization of proliferating cell nuclear antigen, vimentin, c-Fos, and clusterin in the postischemic kidney. Evidence for a heterogenous genetic response among nephron segments, and a large pool of mitotically active and dedifferentiated cells. J. Clin. Invest. 93, 2175–2188 (1994).
Kusaba, T., Lalli, M., Kramann, R., Kobayashi, A. & Humphreys, B. D. Differentiated kidney epithelial cells repair injured proximal tubule. Proc. Natl. Acad. Sci. USA 111, 1527–1532 (2014).
Gerhardt, L. M. S., Liu, J., Koppitch, K., Cippà, P. E. & McMahon, A. P. Single-nuclear transcriptomics reveals diversity of proximal tubule cell states in a dynamic response to acute kidney injury. Proc. Natl. Acad. Sci. USA 118, e2026684118 (2021).
Ide, S. et al. Ferroptotic stress promotes the accumulation of pro-inflammatory proximal tubular cells in maladaptive renal repair. Elife 10, e68603 (2021).
Kirita, Y., Wu, H., Uchimura, K., Wilson, P. C. & Humphreys, B. D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl. Acad. Sci. USA 117, 15874–15883 (2020).
Kramann, R., Tanaka, M. & Humphreys, B. D. Fluorescence microangiography for quantitative assessment of peritubular capillary changes after AKI in mice. J. Am. Soc. Nephrol. 25, 1924–1931 (2014).
Dixon, E. E., Wu, H., Muto, Y., Wilson, P. C. & Humphreys, B. D. Spatially resolved transcriptomic analysis of acute kidney injury in a female murine model. J. Am. Soc. Nephrol. 33, 279–289 (2022).
Ó hAinmhire, E. & Humphreys, B. D. Fibrotic changes mediating acute kidney injury to chronic kidney disease transition. Nephron 137, 264–267 (2017).
Gerhardt, L. M. S. et al. Lineage tracing and single-nucleus multiomics reveal novel features of adaptive and maladaptive repair after acute kidney injury. J. Am. Soc. Nephrol. 34, 554–571 (2023).
Sato, Y. et al. Developmental stages of tertiary lymphoid tissue reflect local injury and inflammation in mouse and human kidneys. Kidney Int 98, 448–463 (2020).
Conway, B. R. et al. Kidney Single-Cell Atlas Reveals Myeloid Heterogeneity in Progression and Regression of Kidney Disease. J. Am. Soc. Nephrol. 31, 2833–2854 (2020).
Sato, Y., Silina, K., van den Broek, M., Hirahara, K. & Yanagita, M. The roles of tertiary lymphoid structures in chronic diseases. Nat. Rev. Nephrol. 19, 525–537 (2023).
Huang, R., Fu, P. & Ma, L. Kidney fibrosis: from mechanisms to therapeutic medicines. Signal Transduct. Target Ther. 8, 129 (2023).
Li, L., Fu, H. & Liu, Y. The fibrogenic niche in kidney fibrosis: components and mechanisms. Nat. Rev. Nephrol. 18, 545–557 (2022).
Hinze, C. et al. Single-cell transcriptomics reveals common epithelial response patterns in human acute kidney injury. Genome Med 14, 103 (2022).
Balzer, M. S. et al. Single-cell analysis highlights differences in druggable pathways underlying adaptive or fibrotic kidney regeneration. Nat. Commun. 13, 4018 (2022).
Liu, J. et al. Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight 2, e94716 (2017).
Lake, B. B. et al. An atlas of healthy and injured cell states and niches in the human kidney. Nature 619, 585–594 (2023).
Zuk, A. & Bonventre, J. V. Acute kidney injury. Annu. Rev. Med. 67, 293–307 (2016).
Ferenbach, D. A. & Bonventre, J. V. Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nat. Rev. Nephrol. 11, 264–276 (2015).
Canaud, G. & Bonventre, J. V. Cell cycle arrest and the evolution of chronic kidney disease from acute kidney injury. Nephrol. Dial. Transpl. 30, 575–583 (2015).
Hansen, J. et al. A reference tissue atlas for the human kidney. Sci. Adv. 8, eabn4965 (2022).
Schreibing, F. & Kramann, R. Mapping the human kidney using single-cell genomics. Nat. Rev. Nephrol. 18, 347–360 (2022).
Shah, S., Lubeck, E., Zhou, W. & Cai, L. seqFISH Accurately detects transcripts in single cells and reveals robust spatial organization in the hippocampus. Neuron 94, 752–758.e1 (2017).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Alon, S. et al. Expansion sequencing: Spatially precise in situ transcriptomics in intact biological systems. Science 371, eaax2656 (2021).
Shah, S., Lubeck, E., Zhou, W. & Cai, L. In situ transcription trofiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92, 342–357 (2016).
Lohoff, T. et al. Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis. Nat. Biotechnol. 40, 74–85 (2022).
Zhu, Q., Shah, S., Dries, R., Cai, L. & Yuan, G.-C. Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat. Biotechnol. 36, 1183–1190 (2018).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Gaedcke, S. et al. Single cell versus single nucleus: transcriptome differences in the murine kidney after ischemia-reperfusion injury. Am. J. Physiol. Ren. Physiol. 323, F171–F181 (2022).
Dixon, E. E., Wu, H., Sulvarán-Guel, E., Guo, J. & Humphreys, B. D. Spatially resolved transcriptomics and the kidney: many opportunities. Kidney Int. 102, 482–491 (2022).
Denisenko, E. et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 21, 130 (2020).
Wu, H., Kirita, Y., Donnelly, E. L. & Humphreys, B. D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 30, 23–32 (2019).
Scholz, H. et al. Kidney physiology and susceptibility to acute kidney injury: implications for renoprotection. Nat. Rev. Nephrol. 17, 335–349 (2021).
Chen, L., Chou, C.-L. & Knepper, M. A. Targeted single-cell RNA-seq identifies minority cell types of kidney distal nephron. J. Am. Soc. Nephrol. 32, 886–896 (2021).
Oyler-Yaniv, A. et al. A tunable diffusion-consumption mechanism of cytokine propagation enables plasticity in cell-to-cell communication in the immune system. Immunity 46, 609–620 (2017).
Gerhardt, L. M. S. & McMahon, A. P. Multi-omic approaches to acute kidney injury and repair. Curr. Opin. Biomed. Eng. 20, 100344 (2021).
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020).
Attanasio, M. et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat. Genet. 39, 1018–1024 (2007).
Xu, S. et al. Leukemia inhibitory factor is a therapeutic target for renal interstitial fibrosis. EBioMedicine 86, 104312 (2022).
Wang, S., Hu, X., Ma, L., Zhang, L. & Tian, Y. CLCF1 is up-regulated in renal ischemia reperfusion injury and may associate with FOXO3. Ann. Transl. Med. 10, 399 (2022).
Kass, D. J. et al. Cytokine-like factor 1 gene expression is enriched in idiopathic pulmonary fibrosis and drives the accumulation of CD4+ T cells in murine lungs: evidence for an antifibrotic role in bleomycin injury. Am. J. Pathol. 180, 1963–1978 (2012).
Crisponi, L., Buers, I. & Rutsch, F. CRLF1 and CLCF1 in Development, health and disease. Int. J. Mol. Sci. 23, 992 (2022).
Murakami, M., Kamimura, D. & Hirano, T. Pleiotropy and specificity: insights from the interleukin 6 family of cytokines. Immunity 50, 812–831 (2019).
Wehr, A. & Tacke, F. The roles of CXCL16 and CXCR6 in liver inflammation and fibrosis. Curr. Pathobiol. Rep. 3, 283–290 (2015).
Wehr, A. et al. Chemokine receptor CXCR6-dependent hepatic NK T Cell accumulation promotes inflammation and liver fibrosis. J. Immunol. 190, 5226–5236 (2013).
Dudek, M. et al. Auto-aggressive CXCR6 CD8 T cells cause liver immune pathology in NASH. Nature 592, 444–449 (2021).
Wu, Y., An, C., Jin, X., Hu, Z. & Wang, Y. Disruption of CXCR6 ameliorates kidney inflammation and fibrosis in deoxycorticosterone acetate/salt hypertension. Sci. Rep. 10, 133 (2020).
Ma, Z., Jin, X., He, L. & Wang, Y. CXCL16 regulates renal injury and fibrosis in experimental renal artery stenosis. Am. J. Physiol. Heart Circ. Physiol. 311, H815–H821 (2016).
Zhang, M. & Zhang, S. T cells in fibrosis and fibrotic diseases. Front. Immunol. 11, 1142 (2020).
Wang, Y. et al. Proximal tubule-derived colony stimulating factor-1 mediates polarization of renal macrophages and dendritic cells, and recovery in acute kidney injury. Kidney Int. 88, 1274–1282 (2015).
Venkatesan, B., Tumala, A., Subramanian, V. & Vellaichamy, E. Transient silencing of Npr3 gene expression improved the circulatory levels of atrial natriuretic peptides and attenuated β-adrenoceptor activation- induced cardiac hypertrophic growth in experimental rats. Eur. J. Pharmacol. 782, 44–58 (2016).
Chromek, M. et al. Matrix metalloproteinase-9 and tissue inhibitor of metalloproteinases-1 in acute pyelonephritis and renal scarring. Pediatr. Res. 53, 698–705 (2003).
Kökény, G. et al. Susceptibility to kidney fibrosis in mice is associated with early growth response-2 protein and tissue inhibitor of metalloproteinase-1 expression. Kidney Int. 102, 337–354 (2022).
Rademaker, M. T. et al. Acute decompensated heart failure and the kidney: physiological, histological and transcriptomic responses to development and recovery. J. Am. Heart Assoc. 10, e021312 (2021).
Wang, S., Chi, K., Wu, D. & Hong, Q. Insulin-like growth factor binding proteins in kidney disease. Front. Pharmacol. 12, 807119 (2021).
Song, C. et al. IGFBP5 promotes diabetic kidney disease progression by enhancing PFKFB3-mediated endothelial glycolysis. Cell Death Dis. 13, 340 (2022).
Andreatta, M. et al. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 12, 2965 (2021).
Wang, X. et al. The role of macrophages in kidney fibrosis. Front. Physiol. 12, 705838 (2021).
Cippà, P. E. et al. A late B lymphocyte action in dysfunctional tissue repair following kidney injury and transplantation. Nat. Commun. 10, 1157 (2019).
Adler, M. et al. Principles of cell circuits for tissue repair and fibrosis. iScience 23, 100841 (2020).
Shah, S. et al. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174, 363–376.e16 (2018).
Eng, C.-H. L., Shah, S., Thomassie, J. & Cai, L. Profiling the transcriptome with RNA SPOTs. Nat. Methods 14, 1153–1155 (2017).
Takei, Y. et al. Integrated spatial genomics reveals global architecture of single nuclei. Nature 590, 344–350 (2021).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Acknowledgements
Work in APM’s and LC’s laboratory was supported by a Broad Innovation Grant from the Eli and Edythe Broad Center for Regenerative Medicine and Stem Cell Research at the University of Southern California and a grant from the NIH to APM (NIDDK UC2DK126024). Additional support from the German Research Foundation grant GE 3179/1-1 (L.M.S.G.) and German Society of Internal Medicine (DGIM) Clinician Scientist grant (L.M.S.G.). The authors thank Inna-Marie Strazhnik for help with figure illustration and design.
Author information
Authors and Affiliations
Contributions
M.P., L.M.S.G., A.P.M., and L.C. conceptualized the study. J.Y. and M.P. designed the probes with input from L.M.S.G. L.M.S.G. and K.K. performed surgeries, tissue collection, and validation experiments. J.Y. performed seqFISH experiments. K.L.C. and M.P. wrote image processing scripts. M.P., L.M.S.G., and S.Z. analyzed data with input from G.C.Y., M.T., A.P.M., L.C. M.P., L.M.S.G., and H.A. were responsible for visualization with input from B.W. M.P. and L.M.S.G. wrote the initial draft of the manuscript. M.P., L.M.S.G., A.P.M., and L.C. edited and reviewed the manuscript. A.P.M. and L.C. supervised all aspects of the project.
Corresponding authors
Ethics declarations
Competing interests
LC is a cofounder of Spatial Genomics, Inc. APM is a consultant to eGENESIS, Trestle Biotherapeutics, IVVA Medical and GentiBIO. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Benjamin Humphreys, Arjun Raj, and Katalin Susztak for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Polonsky, M., Gerhardt, L.M.S., Yun, J. et al. Spatial transcriptomics defines injury specific microenvironments and cellular interactions in kidney regeneration and disease. Nat Commun 15, 7010 (2024). https://doi.org/10.1038/s41467-024-51186-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51186-z
- Springer Nature Limited