Abstract
Pentraxin 3 (PTX3), a long pentraxin and a humoral pattern recognition molecule (PRM), has been demonstrated to be protective against Aspergillus fumigatus, an airborne human fungal pathogen. We explored its mode of interaction with A. fumigatus, and the resulting implications in the host immune response. Here, we demonstrate that PTX3 interacts with A. fumigatus in a morphotype-dependent manner: (a) it recognizes germinating conidia through galactosaminogalactan, a surface exposed cell wall polysaccharide of A. fumigatus, (b) in dormant conidia, surface proteins serve as weak PTX3 ligands, and (c) surfactant protein D (SP-D) and the complement proteins C1q and C3b, the other humoral PRMs, enhance the interaction of PTX3 with dormant conidia. SP-D, C3b or C1q opsonized conidia stimulated human primary immune cells to release pro-inflammatory cytokines and chemokines. However, subsequent binding of PTX3 to SP-D, C1q or C3b opsonized conidia significantly decreased the production of pro-inflammatory cytokines/chemokines. PTX3 opsonized germinating conidia also significantly lowered the production of pro-inflammatory cytokines/chemokines while increasing IL-10 (an anti-inflammatory cytokine) released by immune cells when compared to the unopsonized counterpart. Overall, our study demonstrates that PTX3 recognizes A. fumigatus either directly or by interplaying with other humoral PRMs, thereby restraining detrimental inflammation. Moreover, PTX3 levels were significantly higher in the serum of patients with invasive pulmonary aspergillosis (IPA) and COVID-19-associated pulmonary aspergillosis (CAPA), supporting previous observations in IPA patients, and suggesting that it could be a potential panel-biomarker for these pathological conditions caused by A. fumigatus.
Similar content being viewed by others
Introduction
Humoral immune components are an integral part of the host immune system. Animal studies and human genetic analyses have provided evidence that several of the humoral immune components play essential roles against invading fungal pathogens1: (a) deficiency of C3, a central component of the complement system of hosts, significantly decreased survival of mice infected with Candida albicans, Candida glabrata and Aspergillus fumigatus2,3, (b) mice genetically deficient in C1q, a key component of the classical complement pathway (CP), have increased susceptibility to A. fumigatus infection4, (c) C1q knockout mice succumbed sooner to Cryptococcus neoformans infection5, (d) polymorphisms in the genes coding for mannose binding lectin (MBL) and MBL associated serine proteinase-2 (MASP-2), a PRM and a transducer, respectively, of the lectin complement pathway (LP) are associated with increased susceptibility of the host to Sporothrix infection6, and (e) gene variation in serum amyloid-P component (SAP) affects the risk of invasive pulmonary aspergillosis (IPA) in hematopoietic stem cell transplant recipients7. Despite these studies, host humoral immune components have been underexplored in the context of fungal infections.
In line with those reports, pentraxin-3 (PTX3) knockout mice are also highly susceptible to IPA4, and genetic variations in the human PTX3 gene have been associated with increased risk for IPA in a number of clinical settings at high-risk, including solid-organ transplantation (particularly, lung), chronic obstructive pulmonary disease, reactivation of latent human cytomegalovirus in hematopoietic stem-cell transplant patients, and hematological malignancies4,8,9,10,11,12,13,14. PTX3 is a member of the pentraxin superfamily with pleiotropic functional properties in innate immunity, where it acts both as a soluble PRM and a regulator of inflammation, and plays additional roles in tissue remodeling, female fertility, and cancer4. PTX3 is stored in neutrophil granules and released in response to microbial recognition and/or inflammatory stimulation15 or, de novo synthesized and secreted by both phagocytes and non-immune cells at sites of infection (and inflammation)16. The locally made PTX3 promotes killing of A. fumigatus conidia by immune cells via complement and Fc-γ receptors, suggesting that PTX3 acts at the crossroad between other soluble PRMs17.
Based on competition experiments, galactomannan (GM), a polysaccharide present in the cell wall of A. fumigatus, has been proposed as a PTX3 ligand4. However, GM is concealed within the rodlets and melanin pigment layers of dormant conidia, obscuring the fungal ligands interacting with PTX318. Indirect recognition of microbes by PTX3 has been reported: (i) PTX3 was described to bind another soluble PRM, ficolin-1, activating the LP; however, the ficolin-1/PTX3 complex could not form on A. fumigatus conidia19, (ii) ficolin-2 and PTX3 recruit each other on conidial surfaces20,21,22 amplifying the LP that, however, was appreciated only during C1q and MBL deficiencies, the major recognition units of the CP and LP, respectively, and (iii) although ficolin-3 is known to recognize PTX322, the functional consequence of this interaction in antifungal immunity is unknown. PTX3 was shown to interact with MBL, and the resulting complex promoted an LP-CP cross-activation on C. albicans23; however, it remains unknown whether this is also the case for A. fumigatus. Moreover, no evidence is available regarding the potential interplay of this long pentraxin with other humoral PRMs that are known to play fundamental roles in lung pathophysiology. On the other hand, defective recognition of A. fumigatus conidia by antigen-presenting cells resulting in an altered type-2 immune response was the attributed reason for PTX3-/- mice susceptibility to IPA4. However, neither B cell deficiency nor passive administration of specific antibodies in mice altered their susceptibility to aspergillosis24, suggesting an alternative protective antifungal mechanism associated with PTX3.
Here, we aimed at the identification of A. fumigatus ligands of PTX3. Moreover, we assessed if other soluble PRMs that are known to interact with A. fumigatus and PTX3 could promote the recruitment of PTX3 on conidia and the functional implications of these tri-partite interactions. Finally, as a way to assess the clinical significance of our observations, we measured serum levels of PTX3 in a range of human fungal infections, including CAPA, candidemia and mucormycosis, in addition to IPA.
Results
Pentraxin-3 (PTX3) directly binds to swollen and germinating conidia of A. fumigatus
In previous studies, PTX3 was shown to bind to the dormant conidia of A. fumigatus, with no binding to the hyphal morphotype of this fungus4,17. Moreover, it was observed that the PTX3 interaction with conidia was abolished in the presence of galactomannan (GM), one of the cell wall constituents of A. fumigatus, suggesting that GM is the fungal ligand for PTX3. However, (i) in dormant conidia, GM is concealed by surface rodlet/melanin layers18, and (ii) the recombinant PTX3 concentrations used in this study were 10–200 µg/mL (0.24–23.5 mM, based on the molecular weight of PTX3 protomer subunit, that is ∼42.5 kDa), whereby the median levels of PTX3 in the bronchoalveolar lavage fluid (BALF) and plasma of the patients with IPA are in the range of 2.50–6.97 ng/mL and 5.00–7.10 ng/mL, respectively12,25,26. Therefore, we re-visited these interaction studies by incubating different morphotypes of A. fumigatus (2 × 106 dormant, swollen, or germinating conidia) with 50 ng of PTX3 [i.e., 1 µg/mL, which is a close-to-physiology (pathology) concentration of the protein and up to 200-times lower compared to that used in the previous studies], followed by immunolabelling and microscopy. At this concentration of PTX3, immunolabelling of swollen and germinating conidia was observed, with no labeling of the dormant conidial morphotype (Fig. 1), suggesting that the A. fumigatus ligand(s) for PTX3 is/are exposed on the surfaces of swollen and germinating conidial morphotypes, and are either absent or concealed within that of dormant conidia.
Dormant, swollen and germinating conidia were blocked for 1 h with PBS containing 1% BSA, incubated with PTX3 [1 μg/mL in HEPES buffer (pH 7.4) supplemented with CaCl2 and MgCl2] followed by sequential incubations with primary anti-human PTX3 polyclonal antibodies (raised in rabbit, 1:500 dilution) and secondary Alexa Fluor-647 labeled anti-rabbit IgG (with PBS washing between these two incubations) followed by confocal microscopy. Only swollen and germinating conidia showed PTX3 binding, but not dormant conidia of A. fumigatus (controls: treated like other samples, but without incubation with PTX3). Immunolabelling was performed with three different cultures of A. fumigatus (biological replicates), and every time at least five images were taken for each assay condition.
Galactosaminogalactan (GAG), a germination-specific A. fumigatus cell wall polysaccharide, is the PTX3 ligand exposed on swollen and germinating conidial surfaces
Fungi are endowed with a cell wall that is the first structure to interact with the host immune system. To identify components of the A. fumigatus cell wall that are recognized by PTX3, different cell wall fractions were extracted from dormant, swollen and germinating conidial morphotypes and tested for their interaction with PTX3 by indirect ELISA. The cell wall alkali-insoluble (AI) fractions from all three morphotypes and the alkali-soluble (AS) fraction from dormant conidia failed to interact with PTX3, while AS fractions from swollen and germinating conidia were clearly bound to this protein (Fig. 2A). Among purified cell wall components from all the three morphotypes of A. fumigatus, PTX3 only bound to galactosaminogalactan (GAG; Fig. 2B) in a concentration-dependent manner (Figs. 2C, D). Moreover, swollen and germinating conidia of the A. fumigatus ∆gt4bc mutant (that lacks GAG biosynthetic genes) gave no PTX3-specific signal in immunolabelling experiments (Supplementary Fig. S1), suggesting that GAG, a cell wall polysaccharide synthesized during the course of germination27, is a ligand for PTX3 on swollen and germinating conidial morphotypes. Furthermore, PTX3 did not recognize the hydrophobin RodA and melanin pigments (Fig. 2B) that form superficial rodlet and pigment layers on dormant conidial surface.
ELISA was performed by coating A. fumigatus cell wall components into ELISA plate wells, adding biotinylated PTX3 (1 μg/mL) followed by horseradish peroxidase (HRP) conjugated to streptavidin, and revealing by O-phenylenediamine method. A PTX3 interacts with alkali-soluble (AS) fractions from swollen (SC) and germinating (GC) conidia, but not dormant conidia (DC). B PTX3 binds to GC-AS fraction and GAG, but not other cell wall components (10 μg/mL) or DC specific RodAp and melanin pigments. C PTX3 binds to GC-AS fraction in a concentration dependent manner. D Irrespective of the batches of preparations, PTX3 shows better affinity towards insoluble galactosaminogalactan (GAG) (10 μg/mL). E PTX3 binds to native and completely deacetylated GAGs, but not with acetylated GAG. F Germinating conidia were incubated with PTX3, followed by immunolabelling for GAG with mouse monoclonal anti-GAG antibodies and anti-mouse IgG-TRITC and for PTX3 with monoclonal anti-PTX3 antibodies raised in rat and Alexa Fluor 405 conjugated rat IgG; merged image (in pink) suggests the colocalization of GAG and PTX3. AS and AI fractions extracted from two independent batches of dormant, swollen and germinating conidia were used in the assays, with technical replicates (2A, B, C). A recombinant RodA-protein, melanin (the DC components), β−1,3-glucan, chitin, α−1,3-glucan, galactomannan (GM) and GAG extracted from two independent mycelial cultures of A. fumigatus, two batches of native GAG, as well as acetylated and deacetylated GAGs derivatized from native GAGs were used for the binding assays (2B, D, E). Data are presented as mean values ± SD. PTX3-GAG double immunolabelling was performed with two independent cultures of A. fumigatus (biological replicates); each time at least five images were captured for a condition presented (2F). Source data (2A–E) are provided as a Source data file.
GAG is a heteropolysaccharide composed of N-acetyl-galactosamine, galactosamine and galactose, and can be extracted as urea-soluble and insoluble fractions depending on the ratio of hexoses and hexosamines27. Urea-insoluble GAG showed concentration-dependent and batch-independent interaction with PTX3, whereas urea-soluble GAG showed batch variations (Fig. 2D). Further, native urea-insoluble GAG was completely acetylated or deacetylated prior to interaction studies. Deacetylation increased PTX3 binding, while acetylation almost completely abolished recognition of GAG by PTX3 (Fig. 2E), suggesting that the degree of acetylation plays an essential role in the interaction of GAG with PTX3. Immunolabelling of germinating conidia for GAG and PTX3 indicated their co-localization (Fig. 2F). We then assessed the minimal length of GAG that retains binding to PTX3 by means of an inhibition assay. For that, oligosaccharides obtained from partial hydrolysis of GAG were pre-incubated with PTX3 prior adding to native GAG-coated ELISA plates and residual binding of the protein was determined. In this competitive assay, the GAG oligosaccharide mixture with 9-14 monosaccharide units could effectively inhibit the binding of PTX3 to GAG, whereas the oligosaccharide mixture with 6-10 monosaccharide units did not show any inhibition. Also, a mixture of the building blocks of GAG (N-acetyl-galactosamine, galactosamine, galactose) failed to inhibit PTX3 interaction with native GAG. This assay suggested that the oligosaccharides composed of N-acetyl-galactosamine, galactosamine and galactose constituting more than 10 monosaccharide units define the minimal chain-length required for GAG to interact with PTX3 (Supplementary Fig. S2).
PTX3 interacts with A. fumigatus dormant conidial surface-associated proteins
In our microscopic analyses and ELISA, PTX3 showed negative immunolabelling to dormant conidia and failed to interact with the RodA protein (that forms surface rodlet layer on dormant conidia28) and melanin pigments. However, several studies have reported a direct interaction of PTX3 with dormant conidia of A. fumigatus4,7,17. We revisited these observations by flow cytometry; nonetheless, the interaction between dormant conidia and PTX3 was weak. Only when a supra-physiological concentration of PTX3 (i.e., 50 µg/mL) was applied, ~19.5% of dormant conidia showed positive PTX3 labeling (Supplementary Fig. S3A). To account for strain-dependent effects, dormant conidia from another clinical isolate, Af293, and a laboratory parental strain of A. fumigatus, Δku80 pyrG+, were assessed for PTX3 binding. Respectively, only ~19.7% of Af293 conidia and ~12.5% of Δku80 pyrG+ conidia were recognized by PTX3 at 1 µg/mL. Also, the binding could not be saturated by supra-physiological concentrations of PTX3 (45.9% and 49.1% Af293 and 34.4% and 51.9% Δku80 pyrG+ conidia were positive for PTX3 labeling at 5 and 20 μg/mL, respectively) (Supplementary Figs. S3B & C).
We then attempted to identify the weakly interacting dormant conidial surface associated ligand(s) of PTX3. As PTX3 has been shown to interact with bacterial outer membrane glycoproteins, vesicular membrane glycoproteins, and assumed to bind to viral surface glycoproteins29,30,31, we hypothesized that conidial surface proteins could be the PTX3 ligands. To prove this, we extracted conidial surface proteins and analyzed their interaction with PTX3 by ELISA. Indeed, this extracted protein mixture showed interaction with PTX3 in a concentration-dependent manner (Supplementary Fig. S4A). On the other hand, genetic deletion of RodA (that forms a rodlet layer on dormant conidial surface28) strengthened recognition of respective mutant dormant conidia by PTX3, compared to dormant conidia from the parental strain, Δku80 pyrG+, at PTX3 concentrations of 1 µg/mL (Supplementary Fig. S4B). Earlier, we showed that ΔrodA mutant dormant conidia harbor ~4-fold more extractable surface proteins compared to its parental strain Δku80 pyrG+32. This increase in the number and amount of dormant conidial surface proteins in ΔrodA mutant could explain the increased binding of PTX3, and also suggests that conidial surface proteins are indeed potential but weak ligands of PTX3 on dormant conidia.
To identify potential conidial protein ligand(s) of PTX3, we subjected the conidial surface extract to proteomic analysis. From three biological replicates, a total number of 329 proteins were identified and assigned to unique UniProt IDs. This initial list of candidates was shortened based on negative selection of proteins with Gene Ontology (GO) terms associated with intracellular localization (i.e., cytoplasm, mitochondrion, nucleus, and endoplasmic reticulum), which led to a panel of 74 putative membrane and cell wall proteins (Table S1). Of them, probable glycosidase Crf1 (Q8J0P4), catalase B (CatB; Q92405), alkaline protease 1 (Alp1; P28296), exo-β-1,3-glucanase (Q4WLJ9), endo-1,3-β-glucanase (Q4WJZ7) and aspergillopepsin (Q4WAG2), available in our recombinant A. fumigatus protein collection, were used in solid-phase binding experiments. None of them was bound by PTX3, suggesting that other and yet not tested membrane/cell wall proteins are dormant conidial ligands of this long pentraxin or that these dormant conidial surface-associated proteins (including those tested) must form functionally active complexes to be recognized by PTX3.
The interaction of PTX3 with dormant conidia is facilitated by complex formation with other humoral pattern recognition molecules (PRMs)
A consolidated body of evidence indicates that PTX3 interacts with several other soluble PRMs and modulate their functions33,34,35. We therefore addressed this property in the context of recognition of A. fumigatus dormant conidia. When conidia were opsonized with normal human serum (NHS) or heat-inactivated NHS followed by sequential incubation with primary anti-PTX3 antibodies (human) and secondary human IgG conjugated with a fluorochrome, all conidia showed immunolabelling for PTX3 (Fig. 3A). This suggests that heat-resistant factors in the serum might complex with PTX3, and thereby facilitate its interaction with dormant conidia. Indeed, PTX3 has been demonstrated to engage in molecular complexes with several serum humoral factors22,34,36. We focused on those humoral PRMs that have been reported to bind A. fumigatus dormant conidia and assessed their interaction with PTX3. ELISA indicated that surfactant protein-D (SP-D) and, to a lesser extent, the complement proteins C1q and C3b are the major binders of PTX3 (Fig. 3B). Of the investigated PRMs, C1q and C3b are known interactomes of PTX3, whereas, to our knowledge, this is the first report of a direct interaction between a pulmonary surfactant protein SP-D and PTX3. We further confirmed the interaction between SP-D, C1q or C3b and PTX3 by immunolabelling and followed by confocal microscopy (Supplementary Fig. S5).
A Upon opsonization with pooled normal human serum (NHS) or heat-inactivated (HI) NHS followed by incubation with primary human anti-PTX3 polyclonal antibodies (rabbit, at 1:500 dilution) and FITC-conjugated anti-rabbit IgG, dormant conidia showed positive labeling, suggesting that serum factor(s) mediate PTX3-conidia interaction. Positive labeling with inactivated NHS suggested that the mediating serum factor(s) are heat stable. B Humoral pattern-recognition molecules (PRMs) reported to interact with A. fumigatus conidia were coated on 96-well ELISA plate, followed by sequential incubations with biotinylated PTX3 (1 μg/mL), streptavidin-horseradish peroxidase (HRP), and 3,3’,5,5’-tetramethylbenzidine (TMB; HRP substrate) followed by reading the optical densities at 450 nm (with a reference at 530 nm). Among these humoral PRMs, surfactant protein D (SP-D), and complement proteins C1q, C3b showed PTX3 interaction. Control has been subtracted to plot the graph. SP-A: Surfactant protein-A, C4b: complement protein C4b, MBL: mannose binding lectin, FCN: Ficolin. Immunolabelling was performed two times with independent cultures of A. fumigatus (biological replicates); each time, at least five images were captures for an assay condition (3 A). PTX3 and other PRM interaction study (3B) was performed in triplicate (technical replicate); data are presented as mean values ± SD. Source data are provided as a Source data file.
We then set out to investigate potential complexes between PTX3 and SP-D, C1q or C3b in situ, i.e., on the surface of dormant conidia of A. fumigatus. Opsonization of dormant conidia individually with SP-D, C1q or C3b showed ~98% of labeling for each of them (Fig. 4A) and with PTX3 was ~19.5% (Supplementary Fig. S3A). On the other hand, pre-opsonization with SP-D, C1q or C3b enhanced the interaction of PTX3 with dormant conidia; flow cytometry indicated a mean increase (in percentage) of dormant conidial labeling by PTX3 of 13.9 ± 7.5, 32.0 ± 11.4 and 14.0 ± 7.4% upon pre-opsonization with SP-D, C1q or C3b, respectively (p < 0.05) compared to direct opsonization with PTX3 (Fig. 4B). These findings indicate that SP-D, C1q or C3b form complexes, thereby facilitate the interaction of PTX3, indirectly, with dormant conidia of A. fumigatus.
A Bar-graph representing the percentage of conidia positive for SP-D, C1q or C3b binding, as determined by flow-cytometry. B Bar-graph showing conidia positive for PTX3 binding alone or upon its complexing with SP-D, C1q or C3b (flow-cytometry). PTX3 binding was detected by anti-human PTX3-FITC. Median values are presented. Statistical analysis was performed with two-sided paired t-test (*p < 0.05). C Histograms (flow-cytometry data) showing the conidial binding pattern of PTX3, directly or after complexing with SP-D, C1q or C3b. Data acquired from one experiment is presented; therefore, the control and PTX3 counts were the same in the SP-D + PTX3, C1q + PTX3 and C3b + PTX3 panels, while the binding assay was repeated with two independent batches of A. fumigatus (biological replicates), each time in duplicates (technical replicates). Source data are provided as a Source data file.
Interaction of PTX3 with A. fumigatus, directly or interplaying with other humoral PRMs, alters cytokine and chemokine production by human primary innate immune cells
We then investigated the biological implications of the direct interaction of PTX3 with germlings and of the complexes between PTX3 and other humoral PRMs on dormant conidia of A. fumigatus. To this end, we set up immunomodulation experiments where human monocyte-derived macrophages (hMDMs) and neutrophils were used, that are known to play key roles in clearing inhaled conidia of A. fumigatus. PFA-fixed conidia and serum-deficient RPMI medium were used here to avoid any off-target effects due to conidial germination and other serum factors. SP-D, C1q, C3b, PTX3 alone, unopsonized PFA-fixed conidia as well as fixed conidia that had been pre-incubated with PTX3 failed to elicit immunostimulatory responses in hMDMs and neutrophils. On the other hand, SP-D, C1q or C3b-opsonized conidia stimulated the secretion of pro-inflammatory cytokines (TNF-α, IL-6, IL-1β and IL-12p70) and IL-8 (chemokine) by hMDMs and mainly IL-8 by neutrophils (Fig. 5, and Supplementary Fig. S6A). This data suggests that individually the humoral PRMs are not immunomodulators and confirms our previous study that dormant conidial surface rodlet layer masks their recognition by host immune cells28. Whereas the opsonization with humoral PRMs (SP-D, C1q or C3b) facilitates the recognition of A. fumigatus by immune cells, resulting in a defensive host response.
PFA-fixed dormant/germinating conidia were used; hMDMs and neutrophils were interacted with these fungal samples for 24 h and 20 h, respectively, in a CO2 incubator at 37oC. hMDMs or neutrophils cultured only with medium served as the control, stimulated with lipopolysaccharide (LPS) served as a positive control. Individual humoral PRMs alone were also added to hMDMs/neutrophils that served as another set of controls. Supernatants of these cultures were collected after indicated times of interaction and analyzed for cytokines/chemokine; (A) hMDMs (number of donors, n = 8) and (B) neutrophils (n = 10). Median values are presented for each assay condition. Statistical analysis was performed by one-way ANOVA with Tukey’s multiple comparison test (ns-nonsignificant). At a time, Neutrophils or hMDMs isolated/obtained from two donors were subjected to conidial interaction assays; accordingly, 4–5 independent cultures of A. fumigatus conidia (biological replicates) were used for this entire study. Source data are provided as a Source data file.
Interestingly, addition of PTX3 to dormant conidia that had been pre-opsonized with SP-D, C1q or C3b resulted into a significant reduction in the levels of pro-inflammatory cytokines and chemokines secreted by hMDMs and neutrophils. Also under these conditions, we observed a significant increase in the concentration of the anti-inflammatory cytokine IL-10 secreted by hMDMs stimulated with germinating conidia opsonized with PTX3 compared to the unopsonized counterparts (Fig. 5). No major changes were observed in secretion of other cytokines by neutrophils stimulated with any of these fungal samples (Supplementary Fig. S6B; representative data for TNF-α is presented). Collectively, this study suggested that the direct opsonization of A. fumigatus germlings by PTX3 or indirect interaction of PTX3 with dormant conidia through forming complexes with other humoral PRMs (SP-D, C1q or C3b) contributes to modulate the inflammatory responses of the host innate immune cells that encounter the fungus. Further, we analyzed the expression of PD-L1 on neutrophils upon stimulation with PRM-opsonized conidia, as PD-L1 suppresses the protective immune responses and negatively regulates antifungal immunity37,38. However, both unopsonized and PRM-opsonized dormant conidia failed to stimulate PD-L1 expression on neutrophils (Supplementary Fig. S7). Germinating conidia (unopsonized as well as PTX3 opsonized) also failed to stimulate PD-L1 expression on neutrophils, although opsonization with PTX3 significantly increased the capacity of germinating conidia to stimulate the release of IL-10 in hMDMs compared to unopsonized germinating conidia (Fig. 5).
We then analyzed killing capacity of hMDMs towards unopsonized and humoral PRMs opsonized metabolically active conidia. After 6 h of conidia-hMDMs co-incubation, killing of unopsonized conidia and conidia incubated with PTX3 were significantly lower compared to SP-D, C1q, C3b, SP-D-PTX3, C1q-PTX3 or C3b-PTX3 opsonized conidia (Supplementary Fig. S8; p < 0.0001). This was in agreement that opsonization with PRMs (individually or through their interplay) facilitates an increased conidial phagocytosis and killing compared to PTX3 alone. Conidial opsonization by PTX3 (through surface-associated proteins), though weaker, should facilitate conidial uptake and killing. Indeed, killing of conidia incubated with PTX3 was slightly higher compared to unopsonized conidial killing (p < 0.05). This study suggests that although interplay between SP-D, C1q or C3b and PTX3 induces an anti-inflammatory response, this interplay does not affect conidial phagocytosis through PTX3. In support, PTX3 is demonstrated to be recognized by Fc-γ (immunoglobulin) receptors17.
Human neutrophils and monocyte-derived macrophages stimulated with metabolically active A. fumigatus conidia release PTX3
C1q, C3, SP-D are present in the alveolar fluid of individuals under healthy conditions39,40. C3, a central component of the complement system, has been shown to be cleaved to C3b within an hour of interaction with A. fumigatus conidia41. Also, these humoral immune components are responsible for pro-inflammatory stimulation (e.g., via complement activation) and recruitment of neutrophils. PTX3 is an acute phase protein that is locally induced at sites of infection (and inflammation)42. To effectively model the interplay of these humoral PRMs (C1q, C3b, SP-D and PTX3) in a timely manner, we assessed the release of PTX3 by neutrophils and hMDMs following conidial stimulation. This was investigated by incubating neutrophils or hMDMs with metabolically active A. fumigatus conidia and measuring the PTX3 levels at 0, 4, 6, 8 and 24 h of incubation. PTX3 release was observed at 4 to 8 h from A. fumigatus conidial challenge, with a peak at 6 h (Fig. 6). Interestingly, neutrophils released PTX3 when stimulated with conidia in a culture medium without the supplementation of NHS, while PTX3 release by hMDMs stimulated with conidia required NHS. This data suggests that under physiological conditions, the opsonization of dormant conidia by SP-D, C1q or C3b can precede their interaction with PTX3, and thereby shifting the immune response from pro-to-anti-inflammation. This is likely instrumental in mitigating an excessive or detrimental pro-inflammatory response when the immune system of a healthy host encounters dormant conidia of A. fumigatus, possibly acting as a negative feedback loop.
Neutrophils and hMDMs were incubated with metabolically active (live) A. fumigatus conidia in a medium without or supplemented with normal human serum (NHS) at 37oC in a CO2 incubator. The culture supernatants were collected at intervals of 0, 4, 6, 8, and 24 h post-stimulation. In the collected culture supernatants, PTX3 (both neutrophils and hMDMs), TNF-α (for hMDMs) and IL-10 (for hMDMs) levels were quantified by ELISA, using respective detection kits. A, B PTX3 released by neutrophils stimulated with conidia in medium without or with NHS; C, D PTX3 produced by hMDMs stimulated with conidia in medium without or with NHS; E, F TNF-α and IL-10 secreted by hMDMs stimulated with conidia in medium supplemented with NHS; (number of donors = 4). Data are presented as mean values ± SEM. Source data are provided as a Source data file.
Neutrophils have been shown to release PTX3 stored in granules in response to the recognition of microbes or inflammatory stimulation15. On the other hand, phagocytes synthesize de novo and secrete PTX3 at the site of infection (and inflammation)16. This neosynthesis has been demonstrated to be strongly induced by TNF-α, IL-1β, and by TLR agonists43. To analyze this in our study setting, we quantified TNF-α level in the culture supernatants of hMDMs stimulated with metabolically active conidia in presence of NHS, at 0, 4, 6, 8 and 24 h of intervals. IL-10 was also quantified, in parallel, in these culture supernatants (Fig. 6). TNF-α level followed the secretion pattern of PTX3 by hMDMs, whereas IL-10 secretion was increased during the course of their interaction with conidia.
PTX3 is increased in the serum of patients with invasive pulmonary aspergillosis (IPA) and COVID-19-associated pulmonary aspergillosis (CAPA)
In a recent comparative proteomic analysis, we observed that PTX3 was present in higher amount in the pooled bronchoalveolar lavage fluids of patients colonized (n = 4) and infected with A. fumigatus [3 patients each with IPA and chronic pulmonary aspergillosis (CPA)]44. Studies have been made to exploit serum/plasma or bronchoalveolar lavage fluid PTX3 level as the diagnostic marker of IPA or to distinguish IPA from CPA26,45,46,47. We compared PTX3 levels in the sera of patients with COVID-19-associated pulmonary aspergillosis (CAPA), candidemia, mucormycosis along with IPA and CPA. The median (interquartile range; IQR) serum PTX3 levels in invasive pulmonary aspergillosis (IPA) [1.93 (0.86 − 3.36) ng/mL] and CAPA [0.83 (0.58 − 2.01) ng/mL] were significantly higher than in the controls [0.18 (0.09 − 0.59) ng/mL] (p < 0.001; p < 0.01) (Fig. 7A). The median (IQR) serum PTX3 was similar in CPA (0.12 (0.08 − 0.17) ng/mL) and the control group (p = 0.20). In candidemia, the median (IQR) PTX3 level was significantly higher [3.07 (0.67 − 11.3) ng/mL] than in healthy controls (p < 0.01) and not statistically different from patients with IPA (p = 0.63). In mucormycosis, the median (IQR) serum PTX3 level [0.29 ( < 0.17 − 12.2) ng/mL] was not significantly different from controls. Of note, for IPA, 3 patients received antifungal treatment 24–48 h before serum was collected (that was used to measure PTX3 levels); PTX3 levels were not statistically different between treated and untreated groups [1.79 (1.49 − 1.95) ng/mL and 1.99 (0.72 − 3.28) ng/mL respectively; p = 0.55]. For CPA, 3 out of 12 patients were prescribed antifungal treatment respectively 4, 7 and 211 days before collection of their sera. In these treated and untreated CPA groups also the PTX3 levels were not statistically different [0.14 (0.11 − 0.18) ng/mL and 0.11 (0.059 − 0.17) ng/mL respectively; p = 0.59]. For candidemia, 2 out of 5 patients were prescribed antifungal treatment 2-days before collection of their sera; nonetheless, PTX3 levels were not statistically different between treated and untreated groups [2.07 (1.08 − 3.07) ng/mL and 4.87 (0.26 − 17.75) ng/mL, respectively; p = 0.80]. All CAPA (n = 6) and mucormycosis (n = 6) patients were treated after the date of the sampling.
A PTX3 and B IL-10 levels were higher in the serum from patients with invasive pulmonary aspergillosis (IPA) and COVID-19-associated pulmonary aspergillosis (CAPA; only PTX3 level). Serum samples were retrospectively collected from the patients with IPA (n = 19), chronic pulmonary aspergillosis (CPA; n = 12), CAPA (n = 6), candidemia (n = 5), mucormycosis (n = 6) and healthy controls (n = 11). Median values are presented. Statistical analysis was performed by two-tailed Mann Whitney test for comparison of control with infected patient groups and by unpaired t-test between two infected patient groups. Source data are provided as a Source data file.
We determined serum IL-10 levels in these groups of control and patients. IPA and CAPA groups showed significantly higher IL-10 levels compared to control group, but not CPA group (Fig. 7B) [median (IQR) levels: control, 5.82 (5.31 − 6.47) pg/mL; IPA, 12.52 (11.14 − 18.11) pg/mL; CAPA, 12.29 (10.88 − 15.24) pg/mL; CPA, 6.97 (5.69 − 9.0) pg/mL]. Patients with candidemia and mucormycosis also showed higher serum IL-10 levels [respectively, 21.14 (18.63 − 30.38) pg/mL and 11.30 (10.03 − 16.24) pg/mL]. In the candidemia group, serum IL-10 level was significantly higher compared to IPA patients; but elevated IL-10 levels were not statistically different between IPA and mucormycosis groups.
Discussion
In this study, we demonstrate that PTX3 interacts with swollen conidia and germlings of A. fumigatus by recognizing its cell wall polysaccharide galactosaminogalactan (GAG) as the ligand. On the other hand, the binding of PTX3 to the dormant morphotype of A. fumigatus is mediated by conidial surface-associated proteins instead and occurs through low affinity interaction. While formation of complexes with other humoral pattern recognition molecules (PRMs; mainly SP-D, C1q or C3b) enhanced the binding of PTX3 to dormant conidia. Importantly, morphotype-specific opsonization of A. fumigatus by PTX3, either directly or through PRM complexes, results in the modulation of immune responses to this fungal pathogen. In fact, recruitment of PTX3 onto the conidial surface led to decreased synthesis and release of pro-inflammatory cytokines and chemokines, and increased production of anti-inflammatory cytokines by hMDMs and neutrophils challenged with A. fumigatus. Moreover, we observed that the concentration of PTX3 in the serum of patients with IPA and CAPA is significantly higher than that in the serum of healthy controls or patients with CPA or mucormycosis.
Initial works showed that PTX3 could interact with viable as well as heat-inactivated dormant conidia of A. fumigatus, and this interaction was abolished in the presence of galactomannan4, suggesting that this polysaccharide, a constituent of the A. fumigatus cell wall, is the ligand of PTX3 in dormant conidia. In our study, however, by immunolabelling and microscopic analysis we did not observe any interaction between PTX3 and dormant conidia, at close-to physiological concentrations of PTX3. However, flow cytometry indicated a weak binding of PTX3 to this morphotype, and direct ELISA did not show any interaction between galactomannan purified from the A. fumigatus cell wall and PTX3. This discrepancy could be due to: (a) the PTX3 concentration applied in previous binding studies was up to 200 μg/mL, which is about 20,000-fold higher than the physiological concentration of PTX3 in the serum and bronchoalveolar lavage fluid26,47; here, we used near physiological or pathological concentration of PTX3, and (b) in previous studies, for the inhibition assay a commercially available galactomannan from a plant source was used, which has a very different structure compared to galactomannan present in the A. fumigatus cell wall. Indeed, A. fumigatus galactomannan has a linear backbone made of α-mannan with a -6Manα1-2Manα1-2Manα1-2Manα1- repeating unit and sidechains composed of β-1,5-linked galactofuranose residues48. While galactomannan from plant sources has a mannan backbone in which mannose residues are connected by β-1,4-glycosidic bonds and galactose sidechains are linked to this backbone by α-1,6-linkages49. Also, in a pioneer study on A. fumigatus and PTX34, no interaction between the hyphal morphotype of A. fumigatus and this protein was observed. This is likely due to that flow cytometry was used in this study to assess the binding, which is not a suitable technique for the hyphal morphotype of A. fumigatus.
In our study, we demonstrate that the prior opsonization with other humoral PRMs enhances the interaction of PTX3 with dormant conidia of A. fumigatus. PTX3 has been shown to interact with a number of complement proteins, including C1q, mannose-binding lectin (MBL) and ficolin, thereby modulating activation and effector functions of this system33. We are also reporting a direct interaction of PTX3 with SP-D, a major component of the lung surfactant that exerts key roles in pulmonary homeostasis and antimicrobial immunity50. Here we observe that pre-opsonization of A. fumigatus dormant conidia with SP-D (which we demonstrated earlier to recognize dormant conidial surface exposed melanin pigments51) promotes recruitment of PTX3, and this results into control of the inflammatory reaction induced by A. fumigatus in human hMDMs and neutrophils. To our knowledge, this is the first evidence of a direct interaction between PTX3 and SP-D with functional implications in antifungal immunity. PTX3 has been shown to promote the deposition of ficolin-2 and MBL on the pathogen surface; here we show the inverse pattern of enhanced recruitment of PTX3 through SP-D, C1q or C3b. A previous study has demonstrated that ficolin-2 interacts with PTX3 and the binding of ficolin-2 to A. fumigatus conidia is enhanced by PTX3, and vice versa22. Nonetheless, in our experimental setting, we only observed a weak binding between ficolin-2/MBL and PTX3. This could be explained by the fact that we did not supplement ficolin-2/MBL and PTX3 interacting medium with Ca+2 as this divalent ion has been demonstrated to be instrumental in their interaction22,23. Moreover, in the previous study, for synergistic interaction between ficolin-2 and PTX3 with A. fumigatus conidia, the binding medium was supplemented with heat-inactivated fetal calf serum that does not rule out the involvement of other humoral PRMs during this interaction. We have reported that PTX3 recognizes C3b and acts as a molecular scaffold for assembly of a ternary complex with factor H (major soluble inhibitor of the alternative complement pathway) that tames complement activation52. Here, we observe that conidia bound C3b enhances recruitment of PTX3 to the dormant conidia of A. fumigatus even in the absence of additional serum (and complement) factors, including factor H, and this results into immunomodulatory outcomes. This finding is of major relevance, given that the alternative pathway of complement activation has been reported to be necessary to support the opsonophagocytic properties of PTX3 towards A. fumigatus17.
Flow cytometry-based investigation indicated that the complexing of PTX3 with SP-D and C3b results in conidia showing two peaks, PTX3-high and PTX3-low conidia, while complexing with C1q showed one peak. Conidial cell wall is composed of different polysaccharides [β-1,3-glucan, chitin, galactomannan (GM) and galactosaminogalactan (GAG, which is synthesized during germination)]. In dormant conidia, these polysaccharides are concealed by hydrophobic RodA protein that forms rodlet layer and melanin pigment layer. However, we showed that GM is weakly exposed on dormant conidial surface53. On the other hand, we demonstrated that (i) SP-D recognizes melanin pigments, GM, GAG51, (ii) C3b can bind to RodA, β-1, 3-glucan, GM54, and (iii) we identified that C1q interact with RodA, β-1, 3-glucan, and GAG with different affinities. This suggests that these humoral PRMs can recognize multiple ligands of A. fumigatus. Together, on dormant conidial morphotype, (a) SP-D can interact with melanin pigments and GM, (b) C3b can bind to RodAp and GM, and (c) C1q can recognize only RodA protein. Therefore, the capacity of SP-D and C3b to be able to interact with two fungal ligands with different intensities explains the two peaks (PTX3-high/PTX3-low) observed for SP-D-PTX3 and C3b-PTX3 deposition on dormant conidia by flow cytometry.
Indirect binding of PTX3 to conidia through SP-D, C1q or C3b and its direct binding to germinating conidia both reduced the secretion of pro-inflammatory cytokines and chemokine but increased the release of anti-inflammatory cytokine (for germinating morphotype) by hMDMs/neutrophils in vitro. PTX3 may have an opposite impact on modulating the immune response. It was shown to amplify a pro-inflammatory effect of recognized microbial moieties through activation of the complement cascade55. On the other hand, PTX3 can also interact with factor H and C4-binding protein, a regulator of the complement system, inhibiting the alternative pathway of complement activation56. However, our experimental setting was devoid of serum and thereby complement factors, and thus the complement cascade could neither be activated nor be inhibited. PTX3 has also been observed to have a regulatory role on inflammation by acting as a feedback mechanism of inhibition of leukocyte recruitment57. It is thus conceivable that binding of PTX3 to serum factors could decrease recognition by macrophages or partially mask microbe-associated molecular patterns (MAMPs) on hyphae, thereby resulting in a decreased pro-inflammatory immune response as observed.
Given the timely opsonization contingent on the presence of C1q, SP-D, and the rapid conversion of C3 to C3b at the A. fumigatus surface coupled with the subsequent release of PTX3 by recruited neutrophils, it is plausible to hypothesize that PTX3 plays a pivotal role in a feedback mechanism crucial for attenuating the detrimental excess of inflammation, which is responsible for tissue damage. RNA-sequencing analysis of a murine model of systemic aspergillosis revealed that the expression of PTX3 and complement (C1q and C3) is high between 5- and 8-days post-infection reinforcing the hypothesis of a retro-control mechanism58 via the reduction of IL-1β, IL-6, TNF-α pro-inflammatory cytokine production and the secretion of the anti-inflammatory cytokine IL-10. Of note, the excess of IL-10 has been reported to impair the anti-fungal immunity59. In this regard, it has been reported that, upon binding to MD-2, PTX3 activates the TLR4/Toll/IL-1R domain-containing adapter inducing IFN-β (TRIF)–dependent signaling pathway that leads to induction of indolamine-2,3-dioxygenase (IDO) and IL-10, thus contributing to limit immunopathology in A. fumigatus infections60. Alternatively, PTX3 could trigger intracellular cascades leading to transcriptional changes responsible for immune modulation in macrophages. To support this hypothesis, it was shown that PTX3 acts as a physiological break of granuloma formation by restraining the pathogenic activation of complement and the downstream metabolic reprogramming of macrophages to sustain their functional activity as well as proliferation61. Chronic granulomatous disease (CGD) is an immunodeficiency caused by the lack of the superoxide-producing phagocyte NADPH oxidase with increased susceptibility to IPA. In a mice model of CGD, PTX3 limited Th17-driven inflammation, enhanced antifungal protective Th1 response and anti-inflammatory Tregs62.
The collection of bronchoalveolar lavage fluid is an invasive protocol. Therefore, the preferred body fluid to detect biomarkers of invasive fungal infection is serum. Of note, in a recent comparative proteomic analysis of the bronchoalveolar lavages from control and A. fumigatus colonized/infected patients, we observed that PTX3 was significantly higher in the bronchoalveolar lavage fluids of A. fumigatus infected patients compared to that in controls44. Therefore, we opted to quantify PTX3 in the sera, and found that PTX3 levels were increased compared to healthy controls in IPA, CAPA but not in CPA patients. Few studies have measured the concentration of PTX3 in bronchoalveolar lavage fluid and serum of patients with aspergillosis or other fungal infections26,45,46,47. In healthy human serum, PTX3 concentration was reported to be <2 ng/mL. A significant increase in the serum PTX3 was reported in children with hematological malignancies and IPA compared to a control pediatric cohort; however, as the ranges of concentrations overlapped, no potential cut-off was proposed45. In another study, although median PTX3 concentration was significantly much lower in CPA (n = 42; 2.84 ng/mL) than IPA (12.11 ng/mL), CPA patients had significantly higher serum PTX3 concentration than healthy controls (1.10 ng/mL), and the authors proposed a cut-off value of 2.3 ng/mL in serum26. Using this cut-off, we found a sensitivity and specificity (when compared only to the healthy controls) in our IPA cohort of 42.1% [confidence interval (CI)95% 23.1–63.7%] and 100% (CI95% 74.1–100%), respectively.
When studying other invasive fungal infections to appraise the specificity of increased PTX3 levels for aspergillosis, Li et al. included patient with pulmonary cryptococcosis (n = 26), which did not have an increased PTX3 level (0.64 ng/mL) in serum nor in bronchoalveolar lavage fluids (BALF)26. However, we found that PTX3 level is increased in patients with candidemia stressing the lack of specificity of PTX3 as a potential immune biomarker. Different species of Mucorales are responsible for mucormycosis. A previous study reported a median PTX3 concentration of 4326 pg/mL in the BALF sample46 where all four samples were collected from patients infected with Rhizopus. Although not statistically significant, in our study, two patients with mucormycosis had higher levels of PTX3 in their sera compared to the remaining four; these patients were infected with different Mucorales species (see “Methods” section). This observation leads to hypothesize that the species responsible for mucormycosis may have an impact on PTX3 level in the body fluids. However, the scarcity of clinical data on this small retrospective cohort does not allow any further conclusion. It was shown that PTX3 levels can also be increased in lung cancer, tuberculosis, small-vessel vasculitis, and septic shock26. Although PTX3 levels were increased in CAPA patients, we lack a control group of COVID-19 patients. However, a study suggested the prognostic value of PTX3 level in severe COVID-19 conditions, with higher PTX3 level associated to a poorer outcome of the patients63. Overall, the diagnosis of IPA remains a challenge, while PTX3 is a promising additional biomarker, but to be used in combination with existing biomarkers. Also, evaluation is required to identify adequate cut-offs and to calculate useful negative predictive value. Nonetheless, quantification of PTX3 could be of particular interest to monitor a response to antifungal treatment45.
Parallel to serum PTX3 level, we also quantified serum IL-10 levels in the control and patient groups, which were significantly higher in patients with IPA, CAPA, candidemia and mucormycosis, but not in CPA patients, compared to control group. These quantifications were in agreement with the earlier studies comparing control group with IPA, CAPA, CPA or candidemia59,64,65,66. Interestingly, serum PTX3 levels were significantly lower in mucormycosis patients compared to IPA group, whereas there was no difference between these two patient groups for IL-10. Both being filamentous fungal pathogens, possibly serum PTX3 and IL-10 could discriminate mucormycosis from aspergillosis, but this needs to be validated with additional samples from these two groups of patients.
Single nucleotide polymorphisms (SNPs) in the PTX3 have been described to contribute to the susceptibility to IPA9,10,67. A genetic variant of PTX3 (GG genotype at rs2305619) has been described to impair its expression and influence the levels of IL-6 and IL-8 in the lungs of the patients with IPA68. Restoring PTX3 levels has been shown to revert functional defects of neutrophils in vitro and increase antifungal drug efficacy in animal models of IPA9,69,70, suggesting that PTX3-based immunotherapy could be sought to treat/prevent IPA in at-risk patients. IPA patients harboring a SNP in PTX3 (rs1840680 AA genotype) showed decreased plasma PTX3 levels, but those IPA patients without this SNP showed increased plasma PTX3 level12. Moreover, patients with chronic obstructive pulmonary disease (COPD) harboring this PTX3-SNP (rs1840680 AA genotype) showed high susceptibility to IPA12. On the other hand, SNPs in SFPTD (SP-D gene) affecting the systemic level of SP-D is a risk factor for COPD patients, while IPA is common in these patients71, suggesting a possible association between PTX3, SP-D (that interplay for A. fumigatus binding) and COPD.
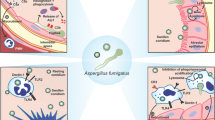
Our data allowed us to propose a scheme for the A. fumigatus interaction dependent release of, and the immunomodulatory function executed by PTX3 (Fig. 8). We observed an anti-inflammatory effect of PTX3 against A. fumigatus, suggesting that in healthy condition this humoral PRM maintains immune homeostasis. Increased susceptibility of PTX3-/- mice to IPA could be due to lack of immune homeostasis resulting in hyperinflammation leading to tissue damage. Indeed, excess inflammation is the hallmark of allergic broncho-pulmonary aspergillosis (ABPA), which leads to a progressive and irreversible destruction of lung tissues. In CGD patients, excessive inflammation is the result of unresolved infection and intrinsic defect in the control of inflammation. Nonetheless, in both of these patient groups, the levels of PTX3 in body fluids has never been quantified. If found to decrease, PTX3 could be a promising candidate for adjuvant immunotherapy in these disease conditions.
Inhaled conidia opsonized with SP-D, C1q or C3b will be phagocytosed by alveolar macrophages, which stimulate macrophages to release pro-inflammatory cytokines TNF-α, IL-1β, chemokine IL-8 as well as other cytokines. IL-8 recruits neutrophils, TNF-α, IL-1β stimulate macrophages to synthesize-secrete PTX3 and neutrophils to release PTX3. As conidial inhalation is a constant process, additional conidia entering lung alveoli encounter immune environment with existing pro-inflammatory cytokines and chemokine. While these conidia will be phagocytosed through PTX3 that complexes with SP-D, C1q or C3b but without stimulating additional cytokine/chemokine secretions. Conidia that occasionally escape from phagocytes and germinating in the alveoli will be opsonized by PTX3 and killed by phagocytes. PTX3 mediated phagocytosis prevents additional/undue immune activation, maintaining immune homeostasis [Image created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs (CC-BY-NC-ND) 4.0 International license].
On the other hand, in patients with IPA, we and others observed a significant increase of PTX3 levels in the bronchoalveolar lavage fluid, and we observed a concomitant decrease in the levels of SP-D and C1q44. Possible explanations for increased PTX3 levels and consequences in IPA patients are: (a) inhaled conidia fail to mount a protective immune response due to a reduction in the SP-D/C1q dependent conidial recognition and phagocytosis; (b) conidia escaping from immune defenses invade alveolar epithelial cells; (c) mechanical stretch induces the production of PTX3 by alveolar epithelial cells72; fungal invasion should induce mechanical stretch, increasing PTX3 production by epithelial cells; (d) elevated PTX3 levels is associated with endothelial dysfunction in humans73,74,75; this might explain the invasion of endothelium by A. fumigatus during IPA. Based on these, our study suggests that supplementing SP-D and/or C1q could be a therapeutic strategy for IPA. We showed that SP-D directly inhibits the growth of A. fumigatus76; thus, supplementing SP-D might reduce the mechanical stretching of alveolar epithelial cells to overproduce PTX3, thereby preventing its undue activity.
In essence, antifungal immunotherapy continues to be an attractive adjunct to the existing antifungal drug treatment, as, albeit effective, these drugs have limitations and despite treatment, the morbidity and mortality associated with invasive fungal diseases remain high. Here, we provide a mechanistic insight into the antifungal activities of humoral PRMs, which suggests that they have the potential as selective immunotherapeutic molecules against specific infection caused by A. fumigatus.
Methods
Ethics statement
Blood samples from healthy human donors were obtained from the Etablissement Français du Sang Trinité (Paris, France) with written informed consent as per Institutional (Institut Pasteur) Ethics Committee guidelines (convention 12/EFS/023). Serum samples used in our study, which were collected from the groups of controls and infected patients. This study complies with the ethical and legal requirements of the French law (15th April 2019) and the Declaration of Helsinki. This is a non-interventional retrospective study without any change in the usual diagnostic and care procedures. The analysis relied on existing leftover samples acquired through routine clinical work according to physicians’ prescriptions. Identifiable information of the patients had been anonymized prior to the analysis. According to the French Health Public Law (CSP Art L1121-1.1), such protocols do not require approval by an ethics committee and patient’s consent (CSP Art L1122-1.1).
Fungal strains and culture conditions
A. fumigatus clinical isolate CBS144-89 was used as the wild-type strain77 and ∆gt4bc was used as the surrogate strain lacking galactosaminogalactan78. The reference strain Af293, the parental strain Δku80 pyrG+ and the ∆rodA as well as ∆pksP generated in this parental strain are the other mutants used in this study17,32,79. All the strains were cultured on malt agar slants (2%) at ambient temperature. Conidia were harvested from the slants of 12–15-days old cultures using 0.05% aqueous Tween-80, and then filtered through 40 µm FalconTM cell strainer (ThermoFisher Scientific) to remove any mycelia, washed, resuspended in 0.05% aqueous Tween-80. Unless stated otherwise, the clinical isolate CBS144-89 is the major strain used in this study. Swollen and germinating conidial morphotypes were prepared by culturing dormant conidia in Sabouraud liquid medium at 37 °C for 5 h and 7 h respectively. These conidial morphotypes (dormant, swollen and germinating) were then fixed with p-formaldehyde [PFA; 2.5% in phosphate buffered saline (PBS; pH 7.4; Gibco), for overnight at 4 °C], quenched with 0.1 M NH4Cl (3 times), washed, and resuspended in PBS as described earlier77 for further applications.
Proteins
Human PTX3 was expressed and purified as described previously80. Recombinant PTX3 (ab85335) and SP-D (ab152069) were also purchased from Abcam; another recombinant SP-D was also purchased from R&D Systems (1920-SP). C1q (204876), C3 (204885), C3b (204860) and C4b (204897) were purchased from Calbiochem. Ficolin-1 (4209-FC; FCN1), Ficolin-2 (2428-FC; FCN2), Ficolin-3 (2367-FC; FCN3) and mannose binding lectin (9086-CF; MBL) were purchased from R&D Systems. Bovine serum albumin (BSA; A3059) and other reagents were from Sigma-Aldrich. Anti-human PTX3 antibodies were either rabbit polyclonal or mouse monoclonal (Clone 5B7) purchased from Abnova. A. fumigatus recombinant proteins Crf1, catalase B (CatB), alkaline protease 1 (Alp1), aspergillopepsin, exo-/endo-1,3-β-glucanase were expressed and purified as described elsewhere81.
Immunolabelling
(A) Imaging: PFA-fixed wild-type conidial morphotypes or swollen/germinating morphotypes of ∆gt4bc (2 × 106) were incubated with recombinant PTX3 (1 µg/mL) or normal human serum (NHS; Zen-Bio) for 1 h at 37 °C in HEPES buffer (20 mM) supplemented with 5 mM CaCl2 and MgCl2. Thereafter, they were incubated with anti-human PTX3 antibodies (rabbit polyclonal; 1:500 dilution) and secondary anti-rabbit IgG conjugated with Alexa Fluor®647 (Abcam, ab150079; dilution 1:100) or FITC (Sigma-Aldrich; F0382; dilution 1:100). To study the interplay between SP-D, C1q or C3b and PTX3, conidia were sequentially incubated with these proteins at 1 µg/mL for 1 h at 37 °C. Further, SP-D was labeled by purified mouse monoclonal IgG2B clone #292201 raised against human SP-D (R&D System; dilution 1:200) and secondary anti-mouse IgG-FITC (Sigma-Aldrich; F0257; 1:200 dilution). C1q was labeled with FITC conjugated anti-C1q antibodies (Abcam; ab4223, dilution 1:200). C3b was labeled with FITC conjugated anti-C3 antibodies (clone 2D8, mouse monoclonal, InvitrogenTM ThermoFisher Scientific; HYB 005-01-02; dilution 1:200). PTX3 was labeled with primary anti-human PTX3 antibodies (rabbit polyclonal) and secondary anti-rabbit IgG-FITC (Sigma-Aldrich; F0382; dilution 1:100). Three washes with PBS were performed between each step. Immunolabelled conidia were observed under an inverted Zeiss LSM 700 Laser Scanning confocal microscope; images were processed with Zeiss software. To visualize the colocalization of PTX3 and galactosaminogalactan (GAG; the germination specific cell wall polysaccharides), (i) GAG was immunolabelled with mouse monoclonal anti-GAG antibodies (5 μg/mL)27 and mouse IgG conjugated with TRITC (Sigma-Aldrich; T5393; dilution 1:200), and (ii) PTX3 was labeled with rat monoclonal anti-PTX3 antibodies (MNB4; Hycult Biotech; HM2242; 5 μg/mL) and anti-rat IgG conjugated with Alexa Fluor®405 (CliniSciences; NB-22-39129-1; 1:100 dilution), followed by confocal microscopy. (B) Flow cytometry: Fixed conidia (2 × 106) were incubated with PBS supplemented with 1% BSA for 30 minutes. Then conidia were opsonized with 50 µg/mL of SP-D, C3, C1q in PBS or buffer (PBS) only for 1 h at 37 °C followed by 50 µg/mL PTX3 in 100 µL (25, 50 or 100 µg/mL for concentration-dependent direct binding study) and anti-human PTX3 monoclonal antibody-FITC (Hycult Biotech; HM2242F; 1:100 dilution) for 1 h at 37 °C. Conidia were washed thrice with PBS between each step. Samples were then acquired by using LSR II (BD Biosciences) and the data was analyzed by FlowJo. The data presented as percentage of conidia positive for indicated markers or median fluorescence intensities of their binding.
Extraction of fungal cell wall components
Different cell wall components of A. fumigatus were extracted or purified as described earlier27,28,38,82,83. De-N-acetylation and acetylation of GAG were performed as described earlier84. Briefly, for de-N-acetylation, GAG was sonicated in 10 mM HCl, 18.8 M NaOH was added at a 1:1 ratio and incubated at 100 °C for 5 h. The reaction was stopped on ice and neutralized with 12 M HCl. De-N-acetylated GAG (dGAG) was dialyzed against water. Acetylated GAG (aGAG) was obtained by solving dry dGAG in 400 mM acetic acid and CH3OH at a 1:4 ratio. Acetylation was initiated by adding acetic anhydride for 1 h. Solvents were evaporated and samples were desalted by repeated dissolution in water. Oligosaccharides of GAG were produced as described earlier84. Concisely, dGAG was hydrolyzed by 2 M HCl at 100 °C for 1.5, 3 and 4.5 h. HCl was evaporated under vacuum and soluble oligosaccharides were dissolved in 150 mM ammonium acetate and subjected to size exclusion chromatography. Nine fractions were obtained with various oligosaccharide chain lengths labeled I to IX. To extract proteins from the conidial surface, conidia were incubated for 2 h in 0.2 M NaCl solution at room temperature at a ratio of 1010 conidia/mL. NaCl supernatant was recovered after centrifugation and concentrated using Speed-Vac Concentrator (Savant SPD1010, ThermoFisher Scientific).
Proteomics of conidial surface proteins
Surface proteins were extracted by incubating conidia (2 × 108) in 0.2 M NaCl (200 µL) at ambient temperature for 2 h in a rotospin (tube rotor). Conidia were separated by centrifugation and the supernatant was subjected to proteomic analysis. In solution digestion: Proteins in the supernatant were precipitated with 20% trichloroacetic acid for 30 min on ice. After centrifugation (20 min, 20000 g, 1 °C) the precipitate was washed with 90% chilled (−20 °C) acetone and centrifuged again. The precipitate was resolubilized in 50 µl of 50 mM triethylammonium bicarbonate (TEAB) in 50/50 2,2,2-trifluoroethanol (TFE)/H2O. Cysteine thiols were reduced and alkylated in a thermomixer (500 rpm) with 10 mM TCEP (tris(2-carboxyethyl)-phosphine) and 12.5 mM 2-chloroacetamide at 70 °C for 30 min. The proteins were evaporated to dryness in a vacuum concentrator and subsequently resolubilized in 50 µl 100 mM TEAB in 5% TFE. Proteolytic digestion was performed with Trypsin/LysC protease mix (Promega) at a protease:protein ratio of 1:25 for 18 h at 37 °C. Tryptic peptides were evaporated with a vacuum concentrator to dryness, resolubilized in 30 µl aqueous 0.05% TFA and 2% ACN and filtered through 10 kDa cut-off spin filters (15 min, 16000 g, and at 8 °C). The filtrate was transferred to HPLC vials and injected into the LC-MS/MS instrument. LC-MS/MS analysis: Each sample was measured in triplicate (3 analytical replicates). LC-MS/MS analysis was performed on an Ultimate 3000 nano RSLC system connected to an Orbitrap Exploris 480 mass spectrometer (both ThermoFisher Scientific, Waltham, MA, USA) with FAIMS. Peptide trapping for 5 min on an Acclaim Pep Map 100 column (2 cm × 75 µm, 3 µm) at 5 µL/min was followed by separation on an analytical Acclaim Pep Map RSLC nano column (50 cm × 75 µm, 2 µm). Gradient elution (eluent A: 0.1% formic acid in water; eluent B: 0.1% formic acid in 90/10 acetonitrile/water) was performed: 0 min at 4% B, 20 min at 6% B, 45 min at 10% B, 75 min at 16% B, 105 min at 25% B, 135 min at 45% B, 150 min at 65%, B 160–165 min at 96% B, 165.1–180 min at 4% B. Positively charged ions were generated at spray voltage of 2.2 kV using a stainless steel emitter attached to the Nanospray Flex Ion Source (ThermoFisher Scientific). The quadrupole/orbitrap instrument was operated in Full MS/data dependent MS2 mode. Precursor ions were monitored at m/z 300–1200 at a resolution of 120,000 FWHM (full width at half maximum) using a maximum injection time (ITmax) of 100 ms and 300% normalized AGC (automatic gain control) target. Precursor ions with a charge state of z = 2–5 were filtered at an isolation width of m/z 4.0 amu for further fragmentation at 28% HCD collision energy. MS2 ions were scanned at 15,000 FWHM (ITmax=100 ms, AGC = 200%). Three compensation voltages were applied (-48 V, -63 V, -78 V). Protein database search: Tandem mass spectra were searched against the UniProt database of the Neosartorya fumigata pan proteome (https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/pan_proteomes/UP000002530.fasta.gz) using Proteome Discoverer (PD) 3.0 (Thermo) and the database search algorithms (threshold search enginge scores in parenthesis) Chimerys (> 2), Mascot 2.8 ( > 30), Comet (> 3), MS Amanda 2.0 ( > 300), Sequest HT ( > 3) with and without INFERYS Rescoring. Two missed cleavages were allowed for the tryptic digestion. The precursor mass tolerance and the fragment mass tolerance were set to 10 ppm and 0.02 Da, respectively. Modifications were defined as dynamic Met oxidation, phosphorylation of Ser, Thr, and Tyr, protein N-term acetylation with or without Met-loss and static Cys carbamidomethylation. A strict false discovery rate <1% (peptide and protein level) was required for positive protein hits. Percolator node of PD3.0 and a reverse decoy database was used for q-value validation of spectral matches. Only rank-1 peptides of the top scored proteins were counted. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE85 partner repository (dataset identifier PXD043546).
Binding assays
A. fumigatus cell wall components dissolved or suspended in sodium carbonate buffer (0.1 M, pH 9.6; 5-10 µg/mL for polysaccharides, 10 µg/mL for melanin pigment, and 250 ng/well for protein; 100 µL/well) were coated on 96-well Microtiter plates, overnight. Wells were blocked with PBS containing 1% BSA for 1 h and sequentially interacted with PTX3 in PBS-BSA (1 µg/mL; 100 µL/well) for 2 h, anti-human PTX3 (Abnova 5806; mouse monoclonal IgG; dilution 1:100) for 1 h, and peroxidase conjugated anti-mouse IgG (Sigma-Aldrich; A4416; dilution 1:1000) for 1 h. Between each step, wells were washed four times with PBS-Tween (0.05%). To quantify PTX3 binding to different cell wall components, peroxidase substrate 3,3’,5,5’-tetramethylbenzidine (TMB; Tebu-bio laboratories) was added, the reaction was arrested with 4% H2SO4 and the optical density (OD) was measured at 450 nm.
Immunomodulatory assays
Monocyte isolation and differentiation into macrophages: Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples by a density-gradient separation method using ficoll (Eurobio, France). Isolated PBMCs were suspended in RPMI 1640 medium + GlutaMAXTM (Gibco, The Netherlands) at a cell count of 8 × 106 PBMCs/mL, from which 500 µL/well were distributed in a 48-well cell culture plates (TPP). Following overnight incubation, non-adhering cells were washed twice with PBS and adhering monocytes were differentiated into macrophages by adding RPMI supplemented with 10% pooled normal human serum (Zen-Bio) and granulocyte macrophage colony stimulating factor (GM-CSF; 10 ng/mL; R&D systems). After 6 days (with medium change once after 3 days), the medium was discarded and differentiated monocyte-derived macrophages (hMDM) were washed with PBS. Neutrophils: Isolated using EasySepTM Direct Human Neutrophil isolation kit (STEMCELL Technologies) according to the manufacturer protocol; per well, 1 × 105 neutrophils suspended in 100 µL RPMI were distributed into 48-well cell culture plates. Conidia opsonization: PFA-fixed dormant conidia were incubated with SP-D, C1q, C3b or PTX3 (1 µg/107 conidia in 50 µL) for 1 h at 37 °C; for double labeling, SP-D, C1q or C3b opsonized conidia were added with PTX3 in PBS (1 µg/107 conidia in 100 µL) and incubated for 1 h at 37 °C. After opsonization with each humoral factor, conidia were washed twice with PBS and finally resuspended in RPMI medium without serum. HMDM stimulation: 2.5 × 106 conidia per well in a volume of 500 µL RPMI were added to hMDM and incubated for 24 h at 37 °C in a CO2 incubator. Neutrophils stimulation: 2.5 × 106 conidia suspended in 300 µL of RPMI were added per well containing 1 × 105 neutrophils (in 100 µL of RPMI) and incubated for 20 h at 37 °C in a CO2 incubator. Unopsonized conidia and different humoral PRMs opsonized conidia were used for the stimulation of hMDMs/neutrophils. In stimulation studies, lipopolysaccharide was used as a positive control (10 ng/well) and medium alone was the negative control. Cells (hMDMs and neutrophils) were also stimulated with individual humoral PRMs (0.25 µg/well). Supernatants were collected and stored at −20 °C until further analysis. Cytokines in the supernatants were quantified by DuoSet ELISA kits (R&D Systems).
Programmed death ligand 1 (PD-L1) expression on neutrophils
Neutrophils isolated from whole blood samples of healthy donors were stimulated with PFA-fixed unopsonized conidia and different humoral PRMs opsonized conidia (as detailed above) for 20 h at 37 °C in a CO2 incubator. Then the neutrophils were recovered by repeated washing of the cell culture plate wells with incomplete RPMI. Collected neutrophils were labeled for CD11b (PE rat anti-CD11b antibodies; clone M1/70, Cat No. 557397, BD Biosciences) and PD-L1 (FITC-conjugated mouse anti-human CD274; clone MIH1, Cat No. 558065, BD Biosciences) for 30 minutes in the dark at 4oC. An FITC-conjugated mouse IgG1 isotype control (Cat No. 555748, BD Biosciences) was used as a control. Dead cells were excluded with the fixable viability dye eFluor 506 staining-kit (Life Technologies). The samples were acquired using LSR II (BD Biosciences), and the data were analyzed by BD FACS DIVA (BD Biosciences) and FlowJo. The performance anti-PD-L1 antibodies was checked on human PBMCs stimulated with lipopolysaccharide (100 ng/mL). Data presented as neutrophils (percent) positive for indicated markers or median fluorescence intensities (MFI) of their binding.
Conidial killing by macrophages
Metabolically active unopsonized or humoral PRMs opsonized conidia (1 × 106/well in 100 μL RPMI) was added to hMDMs (1 × 106/well in 100 μL RPMI; n = 4) in 96 well cell-culture plate and incubated at 37 °C in a CO2 incubator for 6 h. Following, the culture medium was removed, hMDMs were added with 200 μL of cold water and incubated at 4oC for 15 min to lyse hMDMs; lysis was ensured by observing under microscope. Lysate was pooled with collected culture medium. Wells were washed thrice with water (each time with 200 μL) and the washings were also pooled with culture medium and lysate mixture. After appropriate dilution, aliquot of pooled sample (volume corresponding to 250 conidia) was spread on Sabouraud agar plate and incubated at 37oC for 24 h. Following, fungal colony forming units (CFUs) formed were counted. Positive control was conidia (1 × 106/well) added to the well without any hMDMs and processed like unopsonized or PRMs opsonized conidia interacting with hMDMs. The difference between the CFUs for control versus hMDM-treated conidia was then presented as the percentage of killed conidia.
Stimulation and quantification of PTX3 released by neutrophils and macrophages
To quantify PTX3 released/produced, respectively, neutrophils (1 × 105) or hMDMs (5 × 105) were stimulated with metabolically active conidia [multiplicity of infection, 1:1] in RPMI medium with/without serum, the culture supernatants were collected at 0, 4, 6, 8 and 24 h intervals. PTX3 in the culture supernatants was quantified using LEGEND MAXTM Human PTX3 ELISA kit (BioLegend, USA).
Quantification of PTX3 in the serum of patients
Serum samples were collected from patients with probable invasive (n = 19), chronic (n = 12) and probable coronavirus disease-2019 (COVID-19)-associated (n = 6) pulmonary aspergillosis diagnosed according to EORTC/MSGERC guidelines, ERS guidelines and ISHAM definitions, respectively86,87,88. Control populations were the healthy individuals who consulted for serology test for suspected sexually transmitted diseases or before pregnancy (n = 11) but with a negative viral serology. Serum samples from patients diagnosed with candidemia (n = 6) or mucormycosis (n = 6; one each with Rhizopus, Lichtheimia, Rhizomucor, and three with un-distinguishable Mucor or Rhizopus infections) were collected as other sets of controls, to compare with that of the patients with A. fumigatus infection. Serum samples were collected within the first 7-days of diagnosis. Male/female ratio was 1.6 with the median age of 63-years (interquartile range: 47–72). The quantification of PTX3 in these serum samples was performed using LEGEND MAXTM Human PTX3 ELISA kit (BioLegend, USA; sensitivity 17.1 pg/mL).
Statistical analysis
Performed using GraphPad Prism software version 10.3.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE85 partner repository (dataset identifier PXD043546; https://doi.org/10.1038/S41467-024-51047-9). All data needed to evaluate the conclusions of this study are presented in the manuscript and/or in the Supplementary Information. Source data are provided with this paper. Recombinant Pentraxin-3 (PTX3) is available with Antonio Inforzato (Antonio.inforzato@hunimed.eu). Galactosaminogalactan (GAG) and its oligosaccharidic fractions are available with Thierry Fontaine (Thierry.fontaine@pasteur.fr). Source data are provided with this paper.
References
Delliere, S., Sze Wah Wong, S. & Aimanianda, V. Soluble mediators in anti-fungal immunity. Curr. Opin. Microbiol 58, 24–31 (2020).
Tsoni, S. V. et al. Complement C3 plays an essential role in the control of opportunistic fungal infections. Infect. Immun. 77, 3679–3685 (2009).
Shende, R. et al. Protective role of host complement system in Aspergillus fumigatus infection. Front Immunol. 13, 978152 (2022).
Garlanda, C. et al. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature 420, 182–186 (2002).
Rhodes, J. C. Contribution of complement component C5 to the pathogenesis of experimental murine cryptococcosis. Sabouraudia 23, 225–234 (1985).
Naik, B., Ahmed, S. M. Q., Laha, S. & Das, S. P. Genetic susceptibility to fungal infections and links to human ancestry. Front Genet 12, 709315 (2021).
Doni, A. et al. Serum amyloid P component is an essential element of resistance against Aspergillus fumigatus. Nat. Commun. 12, 3739 (2021).
Campos, C. F. et al. PTX3 polymorphisms influence cytomegalovirus reactivation after stem-cell transplantation. Front Immunol. 10, 88 (2019).
Cunha, C. et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N. Engl. J. Med 370, 421–432 (2014).
Cunha, C. et al. PTX3-based genetic testing for risk of aspergillosis after lung transplant. Clin. Infect. Dis. 61, 1893–1894 (2015).
Fisher, C. E. et al. Validation of single nucleotide polymorphisms in invasive aspergillosis following hematopoietic cell transplantation. Blood 129, 2693–2701 (2017).
He, Q. et al. Pentraxin 3 gene polymorphisms and pulmonary aspergillosis in chronic obstructive pulmonary disease patients. Clin. Infect. Dis. 66, 261–267 (2018).
Herrero-Sanchez, M. C. et al. Polymorphisms in receptors involved in opsonic and nonopsonic phagocytosis, and correlation with risk of infection in oncohematology patients. Infect. Immun. 86, e00709-18 (2018).
Wojtowicz, A. et al. PTX3 Polymorphisms and invasive mold infections after solid organ transplant. Clin. Infect. Dis. 61, 619–622 (2015).
Jaillon, S. et al. The humoral pattern recognition receptor PTX3 is stored in neutrophil granules and localizes in extracellular traps. J. Exp. Med 204, 793–804 (2007).
Cieslik, P. & Hrycek, A. Long pentraxin 3 (PTX3) in the light of its structure, mechanism of action and clinical implications. Autoimmunity 45, 119–128 (2012).
Moalli, F. et al. Role of complement and Fcgamma receptors in the protective activity of the long pentraxin PTX3 against Aspergillus fumigatus. Blood 116, 5170–5180 (2010).
Latge, J. P. 30 years of battling the cell wall. Med Mycol. 55, 4–9 (2017).
Ma, Y. J. et al. Ficolin-1-PTX3 complex formation promotes clearance of altered self-cells and modulates IL-8 production. J. Immunol. 191, 1324–1333 (2013).
Rosbjerg, A. et al. Complementary roles of the classical and lectin complement pathways in the defense against aspergillus fumigatus. Front Immunol. 7, 473 (2016).
Hummelshoj, T. et al. The interaction pattern of murine serum ficolin-A with microorganisms. PLoS One 7, e38196 (2012).
Ma, Y. J. et al. Synergy between ficolin-2 and pentraxin 3 boosts innate immune recognition and complement deposition. J. Biol. Chem. 284, 28263–28275 (2009).
Ma, Y. J. et al. Heterocomplexes of mannose-binding lectin and the pentraxins PTX3 or serum amyloid P component trigger cross-activation of the complement system. J. Biol. Chem. 286, 3405–3417 (2011).
Monga, D. P. Studies on experimental aspergillosis in immunodeficient mice. Zentralbl Bakteriol. Mikrobiol. Hyg. A Med Mikrobiol. Infekt. Parasitol. 254, 552–560 (1983).
Cunha, C., Kurzai, O. & Carvalho, A. PTX3 deficiency and aspergillosis. N. Engl. J. Med 370, 1666–1667 (2014).
Li, H. et al. Pentraxin 3 in bronchoalveolar lavage fluid and plasma in non-neutropenic patients with pulmonary aspergillosis. Clin. Microbiol Infect. 25, 504–510 (2019).
Fontaine, T. et al. Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus. PLoS Pathog. 7, e1002372 (2011).
Aimanianda, V. et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460, 1117–1121 (2009).
Reading, P. C. et al. Antiviral activity of the long chain pentraxin PTX3 against influenza viruses. J. Immunol. 180, 3391–3398 (2008).
Jeannin, P. et al. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity 22, 551–560 (2005).
Bottazzi, B. et al. Recognition of neisseria meningitidis by the long pentraxin PTX3 and its role as an endogenous adjuvant. PLoS One 10, e0120807 (2015).
Valsecchi, I. et al. The puzzling construction of the conidial outer layer of Aspergillus fumigatus. Cell Microbiol 21, e12994 (2019).
Doni, A. et al. The long pentraxin PTX3 as a link between innate immunity, tissue remodeling, and cancer. Front Immunol. 10, 712 (2019).
Parente, R., Doni, A., Bottazzi, B., Garlanda, C. & Inforzato, A. The complement system in Aspergillus fumigatus infections and its crosstalk with pentraxins. FEBS Lett. 594, 2480–2501 (2020).
Ma, Y. J., Lee, B. L. & Garred, P. An overview of the synergy and crosstalk between pentraxins and collectins/ficolins: their functional relevance in complement activation. Exp. Mol. Med 49, e320 (2017).
Foo, S. S., Reading, P. C., Jaillon, S., Mantovani, A. & Mahalingam, S. Pentraxins and collectins: friend or foe during pathogen invasion? Trends Microbiol 23, 799–811 (2015).
Yu, Y. et al. PD-L1 negatively regulates antifungal immunity by inhibiting neutrophil release from bone marrow. Nat. Commun. 13, 6857 (2022).
Stephen-Victor, E. et al. Aspergillus fumigatus cell wall alpha-(1,3)-glucan stimulates regulatory T-Cell polarization by inducing PD-L1 expression on human dendritic cells. J. Infect. Dis. 216, 1281–1294 (2017).
Watford, W. T., Ghio, A. J. & Wright, J. R. Complement-mediated host defense in the lung. Am. J. Physiol. Lung Cell Mol. Physiol. 279, L790–L798 (2000).
Chen, J., Ryu, S., Gharib, S. A., Goodlett, D. R. & Schnapp, L. M. Exploration of the normal human bronchoalveolar lavage fluid proteome. Proteom. Clin. Appl 2, 585–595 (2008).
Kozel, T. R., Wilson, M. A., Farrell, T. P. & Levitz, S. M. Activation of C3 and binding to Aspergillus fumigatus conidia and hyphae. Infect. Immun. 57, 3412–3417 (1989).
Mantovani, A., Garlanda, C., Doni, A. & Bottazzi, B. Pentraxins in innate immunity: from C-reactive protein to the long pentraxin PTX3. J. Clin. Immunol. 28, 1–13 (2008).
Alles, V. V. et al. Inducible expression of PTX3, a new member of the pentraxin family, in human mononuclear phagocytes. Blood 84, 3483–3493 (1994).
Delliere, S. et al. Proteomic analysis of humoral immune components in bronchoalveolar lavage of patients infected or colonized by aspergillus fumigatus. Front Immunol. 12, 677798 (2021).
Biagi, E. et al. PTX3 as a potential novel tool for the diagnosis and monitoring of pulmonary fungal infections in immuno-compromised pediatric patients. J. Pediatr. Hematol. Oncol. 30, 881–885 (2008).
Dobias, R. et al. Distinguishing invasive from chronic pulmonary infections: host pentraxin 3 and fungal siderophores in bronchoalveolar lavage fluids. J. Fungi (Basel) 8, 1194 (2022).
Kabbani, D. et al. Pentraxin 3 levels in bronchoalveolar lavage fluid of lung transplant recipients with invasive aspergillosis. J. Heart Lung Transpl. 36, 973–979 (2017).
Latge, J. P. et al. Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect. Immun. 62, 5424–5433 (1994).
Prajapati, V. D. et al. Galactomannan: a versatile biodegradable seed polysaccharide. Int J. Biol. Macromol. 60, 83–92 (2013).
Sorensen, G. L. Surfactant protein D in respiratory and non-respiratory diseases. Front Med (Lausanne) 5, 18 (2018).
Wong, S. S. W. et al. Fungal melanin stimulates surfactant protein D-mediated opsonization of and host immune response to Aspergillus fumigatus spores. J. Biol. Chem. 293, 4901–4912 (2018).
Stravalaci, M. et al. Control of Complement activation by the long pentraxin ptx3: implications in age-related macular degeneration. Front Pharm. 11, 591908 (2020).
Alsteens, D. et al. Unraveling the nanoscale surface properties of chitin synthase mutants of Aspergillus fumigatus and their biological implications. Biophys. J. 105, 320–327 (2013).
Wong, S. S. W. et al. Differential interactions of serum and bronchoalveolar lavage fluid complement proteins with conidia of airborne fungal pathogen aspergillus fumigatus. Infect Immun 88, e00212-20 (2020).
Cotena, A. et al. Complement dependent amplification of the innate response to a cognate microbial ligand by the long pentraxin PTX3. J. Immunol. 179, 6311–6317 (2007).
Inforzato, A. et al. PTX3 as a paradigm for the interaction of pentraxins with the complement system. Semin Immunol. 25, 79–85 (2013).
Deban, L. et al. Regulation of leukocyte recruitment by the long pentraxin PTX3. Nat. Immunol. 11, 328–334 (2010).
Shankar, J., Cerqueira, G. C., Wortman, J. R., Clemons, K. V. & Stevens, D. A. RNA-Seq profile reveals Th-1 and Th-17-type of immune responses in mice infected systemically with aspergillus fumigatus. Mycopathologia 183, 645–658 (2018).
Cunha, C. et al. IL-10 overexpression predisposes to invasive aspergillosis by suppressing antifungal immunity. J. Allergy Clin. Immunol. 140, 867–870 e869 (2017).
Bozza, S. et al. PTX3 binds MD-2 and promotes TRIF-dependent immune protection in aspergillosis. J. Immunol. 193, 2340–2348 (2014).
Goncales, R. A. et al. Pentraxin 3 inhibits complement-driven macrophage activation to restrain granuloma formation in sarcoidosis. Am. J. Respir. Crit. Care Med 206, 1140–1152 (2022).
D’Angelo, C. et al. Exogenous pentraxin 3 restores antifungal resistance and restrains inflammation in murine chronic granulomatous disease. J. Immunol. 183, 4609–4618 (2009).
Capra, A. P. et al. The prognostic value of pentraxin-3 in COVID-19 patients: a systematic review and meta-analysis of mortality incidence. Int. J. Mol. Sci. 24, 3537 (2023).
Ito, Y. et al. Serum cytokines usefulness for understanding the pathology in allergic bronchopulmonary aspergillosis and chronic pulmonary aspergillosis. J. Fungi (Basel) 8, 436 (2022).
Russo, A., Morrone, H. L., Rotundo, S., Trecarichi, E. M. & Torti, C. Cytokine profile of invasive pulmonary aspergillosis in severe COVID-19 and possible therapeutic targets. Diagnostics (Basel) 12, 1364 (2022).
Taj-Aldeen, S. J., Mir, F. A., Sivaraman, S. K. & AbdulWahab, A. Serum cytokine profile in patients with candidemia versus bacteremia. Pathogens 10, 1349 (2021).
Kang, Y., Yu, Y. & Lu, L. The role of pentraxin 3 in aspergillosis: reality and prospects. Mycobiology 48, 1–8 (2020).
Goncalves, S. M. et al. Evaluation of bronchoalveolar lavage fluid cytokines as biomarkers for invasive pulmonary aspergillosis in at-risk patients. Front Microbiol 8, 2362 (2017).
Marra, E. et al. Efficacy of PTX3 and posaconazole combination in a rat model of invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 58, 6284–6286 (2014).
Lo Giudice, P., Campo, S., De Santis, R. & Salvatori, G. Effect of PTX3 and voriconazole combination in a rat model of invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 56, 6400–6402 (2012).
Obeidat, M. et al. Surfactant protein D is a causal risk factor for COPD: results of Mendelian randomisation. Eur Respir J 50, 1700657 (2017).
Wu, Q., Shu, H., Yao, S. & Xiang, H. Mechanical stretch induces pentraxin 3 release by alveolar epithelial cells in vitro. Med Sci. Monit. 15, BR135–BR140 (2009).
Carrizzo, A. et al. Pentraxin 3 induces vascular endothelial dysfunction through a P-selectin/matrix metalloproteinase-1 pathway. Circulation 131, 1495–1505 (2015).
Suliman, M. E. et al. Novel links between the long pentraxin 3, endothelial dysfunction, and albuminuria in early and advanced chronic kidney disease. Clin. J. Am. Soc. Nephrol. 3, 976–985 (2008).
Witasp, A. et al. Elevated circulating levels and tissue expression of pentraxin 3 in uremia: a reflection of endothelial dysfunction. PLoS One 8, e63493 (2013).
Wong, S. S. W. et al. Surfactant protein D inhibits growth, alters cell surface polysaccharide exposure and immune activation potential of Aspergillus fumigatus. Cell Surf. 8, 100072 (2022).
Thau, N. et al. Rodletless mutants of aspergillus fumigatus. Infect. Immun. 62, 4380–4388 (1994).
Briard, B. et al. Publisher Correction: galactosaminogalactan activates the inflammasome to provide host protection. Nature 589, E3 (2021).
da Silva Ferreira, M. E. et al. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5, 207–211 (2006).
Inforzato, A. et al. Structure and function of the long pentraxin PTX3 glycosidic moiety: fine-tuning of the interaction with C1q and complement activation. Biochemistry 45, 11540–11551 (2006).
Sarfati, J. et al. Recombinant antigens as diagnostic markers for aspergillosis. Diagn. Micr Infec Dis. 55, 279–291 (2006).
Bozza, S. et al. Immune sensing of aspergillus fumigatus proteins, glycolipids, and polysaccharides and the impact on Th immunity and vaccination. J. Immunol. 183, 2407–2414 (2009).
Becker, K. L. et al. Aspergillus cell wall chitin induces anti- and proinflammatory cytokines in human PBMCs via the Fc-gamma receptor/Syk/PI3K Pathway. mBio. 7, e01823-15 (2016).
Gressler, M. et al. Definition of the anti-inflammatory oligosaccharides derived from the galactosaminogalactan (GAG) from aspergillus fumigatus. Front Cell Infect. Microbiol 9, 365 (2019).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50, D543–D552 (2022).
Donnelly, J. P. et al. Revision and update of the consensus definitions of invasive fungal disease from the european organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin. Infect. Dis. 71, 1367–1376 (2020).
Denning, D. W. et al. Chronic pulmonary aspergillosis: rationale and clinical guidelines for diagnosis and management. Eur. Respir. J. 47, 45–68 (2016).
Koehler, P. et al. Defining and managing COVID-19-associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect. Dis. 21, e149–e162 (2021).
Acknowledgements
This study was supported by the Agence Nationale de la Recherche grant (ANR-21-CE17-0032-01 grant, FUNPOLYVAC) to VA. SD was supported by Assistance Publique-Hôpitaux de Paris-Institut Pasteur Poste d’Accueil funding. SSWW was supported by Pasteur-Roux-Cantarine postdoctoral fellowship. JB was supported by the Agence Nationale de la Recherche grant (ANR-19-CE17-0021; BASIN). MG was supported by la Fondation pour la Recherche Médicale (DEQ20150331722 LATGE Equipe FRM 2015). OK and AAB were supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) Collaborative Research Center/Transregio FungiNet 124 ‘Pathogentic fungi and their human host: Networks of Interaction’ (Project No. 210879364; projects A1 and Z2). AC was supported by the Fundação para a Ciência e a Tecnologia (FCT) (PTDC/MED-OUT/1112/2021, UIDB/50026/2020, UIDP/50026/2020). VP and AI were supported by Fondazione Beppe e Nucci Angiolini. Recombinant human PTX3 was made within the frame of an Investigator Grant from the Italian Ministry of Health to AI (GR-2011-02349539).
Author information
Authors and Affiliations
Contributions
V.A., J.P.L., and A.I. designed the study; S.D., C.C., S.S.W.W., M.G., V.P., R.P. and T.K. performed the experiments; T.F. and A.I. provided the materials; O.K., J.B., A.C., A.A.B., A.I., and V.A. analyzed the data; S.D. and V.A. wrote the draft manuscript; all the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
All the authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Peter Cook, Teresa Zelante and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dellière, S., Chauvin, C., Wong, S.S.W. et al. Interplay between host humoral pattern recognition molecules controls undue immune responses against Aspergillus fumigatus. Nat Commun 15, 6966 (2024). https://doi.org/10.1038/s41467-024-51047-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51047-9
- Springer Nature Limited