Abstract
Biological visual systems have evolved to process natural scenes. A full understanding of visual cortical functions requires a comprehensive characterization of how neuronal populations in each visual area encode natural scenes. Here, we utilized widefield calcium imaging to record V4 cortical response to tens of thousands of natural images in male macaques. Using this large dataset, we developed a deep-learning digital twin of V4 that allowed us to map the natural image preferences of the neural population at 100-µm scale. This detailed map revealed a diverse set of functional domains in V4, each encoding distinct natural image features. We validated these model predictions using additional widefield imaging and single-cell resolution two-photon imaging. Feature attribution analysis revealed that these domains lie along a continuum from preferring spatially localized shape features to preferring spatially dispersed surface features. These results provide insights into the organizing principles that govern natural scene encoding in V4.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Introduction
The visual system has evolved to represent and process natural scenes efficiently1. Behaviorally relevant information in visual scenes, such as objects’ identities and locations, are not immediately accessible from the retinal inputs2,3. It is thought that disentangling scene information is achieved by transforming and re-representing the retinal image through a series of visual areas in primate visual cortex, from primary visual cortex, through visual area V4, to inferotemporal cortex (IT)2,4. Each of these visual areas provides a unique stage of representation for the visual world. Yet, understanding how natural scenes are represented by the neuronal populations of a specific visual area remains an incredibly difficult task.
One of the challenges arises from the sheer number of neurons involved in the representation. With tens of millions of neurons in each visual area4,5, it is impossible to assess the neural codes of all these neurons with the recording capabilities of current techniques. However, there are principles in cortical organization that can help. Nearby neurons in the cortex tend to have similar functional properties. It is believed that local circuits formed by these neurons could facilitate specific functional computations4,6,7. By identifying the visual features that drive the excitation of a local population of neurons, we can gain insights into what features of natural scenes are explicitly extracted and represented by the local circuits. A complete though coarse picture of how natural scenes are represented in a visual area can thus be obtained by characterizing the feature preferences of all local populations within that area. In the macaque visual cortex, the basic unit of this local population corresponds to the cortical column with a diameter of 200–400 µm8,9,10,11,12. Current imaging techniques13,14 allow us to simultaneously record neural responses at this scale over a large span of the cortical surface, giving us a relatively complete sampling of the functional units as well as their topological organization.
However, the complexity and diversity of image features in natural scenes15 pose an additional challenge for characterizing the neural codes. Earlier studies had to resort to making hypotheses on the visual features that neurons care about, and utilized simplified parametric artificial stimuli to study neural coding. Such an approach introduces a sampling bias, and may miss the visual features that a neuron is best tuned to. This problem is particularly acute for higher visual areas such as V4 and IT, where neurons have relatively large receptive fields and more complex selectivities16. Thus, to understand the neural representation of natural scenes, it is crucial to capture the rich featural variations in natural scenes by sampling an extensive set of natural images17.
In this work, we achieved long-term stable widefield calcium imaging14 in awake monkeys, which allowed us to obtain a large-scale dataset of V4. We recorded cortical responses spanning ten millimeters of the cortical surface of dorsal V4 in three monkeys, each to over 17,000 color natural images at 0.1 mm spatial resolution. This dataset enabled us to train deep-learning models18,19,20,21 that accurately predict the cortical response to arbitrary images. By identifying each cortical location’s preferred natural images using our model, and then verifying them using additional widefield and two-photon imaging, we found V4 contains more diverse functional domains than previously believed, with each domain encoding distinct natural image features. Further feature attribution analysis to preferred images shows that domains encoding shape-related attributes in V4 tend to prefer features that are spatially localized within the receptive field, while domains encoding texture or surface-related attributes prefer features dispersed in the receptive field. This separate processing of features with different dispersity provides a functional organization principle for V4 neural coding.
Results
A large dataset of macaque V4 cortical responses to natural images
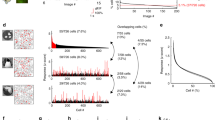
We performed widefield calcium imaging to record V4 cortical responses to a large set of natural stimuli (Fig. 1a). AAVs expressing the calcium indicator GCaMP5G22 were injected into macaque visual cortical area V4. A 10mm-diameter optical window was implanted for imaging (Fig. 1b). During calcium imaging, a 470 nm blue light was used to illuminate the cortex through the optical window, and the green fluorescence excitation signals were recorded using a CCD camera (Camera A in Fig. 1a).
a Schematic of the widefield calcium imaging setup. A shutter was used to control intermittent illumination. On the detection side, a 525 nm dichroic mirror splits the reflectance light into green and blue, projecting them onto Camera A and Camera B, respectively. b Example blue reflectance image recorded by Camera B in V4. A, anterior; M, medial; io, inferior occipital sulcus; lu, lunate sulcus. c Top: Average time course of V4 responses to a stimulus, averaged over the responses to 100 natural images. Blue areas indicate “on” illumination. Image frames used for computing cortical responses are extracted from the periods indicated by the black labels on the response curve within the shutter “on” periods. Bottom: Control signal for shutter on-off; black bar denotes the stimulus presentation period. Source data are provided as a Source Data file. d Example cortical responses to natural images across trials and days. The last two columns show the average responses of 5 repeats on Day 4 and Day 5. e The dataset, used for training the deep neural network, contains cortical responses to 19,900 natural images from monkey C.
To obtain neural responses to tens of thousands of stimuli we needed to integrate recordings across multiple days. However, we found that continuous blue light exposure causes severe photobleaching that results in a gradual attenuation of the fluorescence signal (Supplementary Fig. 1). To solve this problem, we developed an intermittent illumination paradigm. A shutter synchronized to the stimulus presentation was inserted into the blue light pathway to control the illumination. Each trial consisted of a 900 ms blank pre-stimulus period followed by 500 ms of stimulus presentation while the subject maintained fixation. Optical illumination lasting 250 ms occurred twice: one epoch 150 ms before, and one epoch 350 ms after stimulus onset. These corresponded to the baseline period before response initiation, and the peak response period, respectively (Fig. 1c). The fluorescence images recorded in these two periods were used to calculate cortical responses (ΔF/F0, see Methods). This intermittent illumination method significantly improved the long-term stability of signals (Supplementary Fig. 1), enabling the extensive sampling of the stimulus variations. To facilitate multi-day imaging registration, we recorded the blue reflectance images with another camera (Camera B in Fig. 1a) and used the cortical capillaries (Fig. 1b) as the reference for image registration. We found that the calcium signal was robust between different trials and across days (Fig. 1d), allowing us to more confidently integrated data across multiple days into a large dataset.
We obtained the widefield calcium imaging dataset from the dorsal V4 of three monkeys. Each monkey’s dataset includes a training set for fitting neural network models and a validation set for evaluating the model’s prediction performance to novel stimuli. The stimulus presentation area was selected such that the receptive field of the imaged cortex, hand-mapped using small grating patches, was positioned at the center of the stimulus (Supplementary Fig. 2b). The training set consists of single-trial cortical responses to 17,000–20,000 distinct color natural stimuli drawn from ImageNet23 (see Fig. 1e for examples). 500 natural images, distinct from the training set, were used as validation stimuli. We also added a set of 56 conventional artificial stimuli (Supplementary Fig. 2c), including gratings of 8 orientations, 2 phases, and 3 spatial frequencies, as well as 8 uniform color patches, to the validation set. Each validation stimulus was repeated ten times in a randomly interleaved fashion. Data collection for each monkey spanned six consecutive days. To monitor the stability of cortical responses across these days, we tested 100 natural images selected from the validation set as fingerprint images in each recording day. The high degree of correlation observed in the neural responses to fingerprint images across days provides further confirmation of the long-term stability of the measurements, supporting data integration (Supplementary Fig. 2d, e).
DNN modelling of the cortical response dataset
We found nature images elicit much stronger and more diverse responses in V4 than conventional artificial stimuli (Supplementary Fig. 3). For regions that showed significant selectivity to the validation stimuli (Supplementary Fig. 2a, See Methods), we trained a deep learning model to capture the encoding relationship between the visual stimuli and the responses of the cortical pixels.
Earlier studies24,25,26,27 suggest that deep neural networks (DNNs) optimized for object recognition provide a state-of-the-art model of the primate ventral visual stream. These task-driven DNN’s internal representations can be used to fit neural responses to image stimuli via transfer learning. Typically, this is done by fitting the neural responses with a linear transform of feature activations of a specific DNN layer in response to the input image. This feature transfer approach has been shown to produce acceptable response prediction performance, even with relatively small training dataset20,25,28. One drawback of this approach however is that data fitting is restricted to the feature space of the pre-trained DNN, and can potentially fail to capture some of the characteristic of the feature space of the brain. One solution is to use the neural data to fine-tune the feature detectors upstream of the selected DNN layer via backpropagation. However, given the large number of trainable parameters in the DNN for recognition tasks, coupled with the modest size of the neural data, such models tend to overfit, which could account for the decrease in their generalization performance (Supplementary Table 1). To address this issue, we developed a neural modeling strategy that leveraged knowledge distillation for transfer learning (Fig. 2a). Specifically, we used the feature transfer model mentioned above as the teacher to train a student network with significantly fewer parameters, known as knowledge distillation29,30. Subsequently, the student network was fine-tuned on the targeted neural data again to obtain a final model that exhibit improved generalization performance.
a Schematic of transfer learning with knowledge distillation. We first used the neural data to train a feature transfer model which uses a two-layer perceptron to map the responses of the Add-6 layer of the Xception to input images to their evoked cortical responses. We then performed knowledge distillation to condense this feature-transfer model to a smaller DNN. This step was completed by training the small DNN using the responses of feature transfer model to 100 K natural images. We finally fine-tuned the small DNN on the recorded neural dataset. Parameters of the network layers in blue are optimized on neural data. b Neural response prediction performance of the feature transfer model (green), the data-driven model (Direct, light blue), and the knowledge distillation transfer model (KD Transfer, dark blue) on the data of 3 monkeys, across all imaged cortical pixels. For each cortical pixel, the model’s performance is quantified by computing the Pearson correlation between the predicted responses and recorded responses on validation images and then normalizing it with the noise ceiling of the pixel (see Methods, Supplementary Fig. 2f). The KD Transfer model performed significantly better than the data-driven model and the feature transfer model (P = 0.0 in all cases; one-sided Wilcoxon signed-rank test) over n = 7750 cortical pixels pooled across 3 monkeys. The data distribution is shown in the boxplot. Boxplot center is median, box extends 25th and 75th percentiles, whiskers extend to the most extreme data that are not considered outliers, dots denote outliers. Source data are provided as a Source Data file. c Performance measured in raw Pearson correlation as a function of training sample size relative to the noise ceiling is shown. The plotted lines and error bars indicate the mean and standard deviation of the performance of 27 sets of models. Each set includes one model from each of the 3 monkeys, and each model is trained on a random subsample of the data of a particular size. The performance of a set of models is the averaged correlation over the total 7750 cortical pixels. Source data are provided as a Source Data file.
We found this modelling strategy to be very effective in producing a model with superior neural response prediction performance on our measured data (Fig. 2). We trained the feature transfer model with the Add-6 layer of ImageNet pre-trained Xception31 (Supplementary Fig. 4a, b). Knowledge distillation was then performed by training a small Xception-like DNN (Supplementary Fig. 4c, See Methods) on 100 K image-response pairs predicted by the feature transfer model. The prediction performance of the transfer learning model obtained with knowledge distillation (KD Transfer, 73.1% of the achievable performance Fig. 2b) is significantly better than the original feature transfer model (Feature Transfer, 68.2%; P = 0.0, one-sided Wilcoxon signed-rank test) and the small Xception-like DNN directly training with neural data (Direct, 68.8%; P = 0.0, one-sided Wilcoxon signed-rank test). We also evaluated the dependence of the performance of these three models on the size of the training data set (Fig. 2c). As the data size increased, the direct data-driven model exhibited the most significant improvement in performance. Its performance was much weaker than the feature transfer model with limited data, but the two became on par when the data size reached 17 K. This suggests that our dataset is large enough for the data-driven model to learn a feature space that is as effective as the feature space of the pre-trained networks. Our KD Transfer model is consistently better than the other two models regardless of the size of the training dataset, suggesting it is a superior approach for modeling V4 neural tuning.
Natural image preference maps in V4
Our neural network model that predicts V4 cortical responses with high accuracy provides us with a digital twin of V4. This allows us to perform extensive tests in silico to dissect and characterize the neural coding in V418,19,20,32,33. We first employed the digital twin to identify the natural stimuli that elicit the strongest responses in each cortical pixel, which reflects the preference of a local population of neurons. Specifically, we used the KD Transfer model to search for the preferred stimuli for all the cortical pixels across a set of 50,000 natural images. The model’s top nine preferred images for each location were then showed in a 3 × 3 array over that location of the cortical surface (Fig. 3a). The resulting map indicates the stimulus preference for each visually responsive pixel across the imaged area (Fig. 3b, Supplementary Data 1–3). What appeared to be distinct clusters preferring different kinds of natural images were observed. To characterize the organizational structure of the map, we performed hierarchical clustering of cortical pixels based on the similarity of their top nine preferred images. The similarity in image preference between two cortical pixels was computed based on the Pearson correlation between the averaged cortical response patterns to their respective preferred images (Supplementary Fig. 5, See Methods). Hierarchical clustering was then employed to group the cortical pixels into multiple domains. Each of these domains shows a preference for some specific features. Some of these preferred features emphasize specific colors or specific textures. Some prefer image patterns that are marked by transitions in color or luminance, while others prefer specific objects, such as round objects or even faces (Fig. 3c, d). These findings suggest V4 has a diverse and rich set of functional domains, encoding a variety of distinct natural image features.
a The KD Transfer model was used to predict cortical responses to a 50,000-image set. For each cortical pixel (90 × 90 μm physical size; the single pixel example is marked with an orange cross), the top 9 ranked images are shown as a 3 × 3 grid (only the center 4×4 degree of each image is visualized). b Overall preference map obtained from monkey C. Domains preferring different colors can easily be observed. Zooming into the two regions marked by white rectangles, we observe cortical pixels preferring distinct shapes and texture attributes. c The cortical pixels in monkey C were hierarchically clustered based on the similarity of the cortical responses to their top 9 preferred images. Clusters that contained connected region with more than 40 pixels were identified as functional domains, marked by distinct color. Clusters that did not meet the above criteria were marked in gray. Left: The dendrogram of the hierarchical clustering. Right: Cortical map with functional domains colored. Bottom: the images predicted to evoke strong responses for the identified domains. The same color scheme, indicating the functional domain categories, is used for the dendrogram, the cortical map and the image frames. d Same as in (c), for monkey B.
To evaluate the validity of these model-predicted domains empirically, we performed additional widefield calcium imaging on monkey B and monkey C. For each domain, 16 preferred images predicted by the model were selected as test stimuli (Fig. 4a and Supplementary Fig. 6a). We found that stimuli selected for different domains elicited distinct cortical responses, and the measured activation patterns were consistent with model predictions (Fig. 4a, b). Figure 4c shows the responses of each cortical pixel to each group of preferred images associated with the different functional domains (labeled in different colors). We found each group of preferred images could locate a cortical region with the strongest response to them. These regions were largely consistent with the model-identified functional domains (monkey C consistency = 77.9%, monkey B consistency = 87.3%, Fig. 4d). These results indicated that the model-predicted natural image preference map is a good reflection of the functional organization of V4, comprising a wide variety of domains preferring different visual features. Note that this finding critically relies on the use of a large and diverse set of natural stimuli. When testing V4 with conventional artificial stimuli, we found that we missed the true preference of many regions, overlooking the underlying functional differences among them (Supplementary Fig. 7).
a Test stimuli for the four example domains and their corresponding average activation patterns. The first column shows the central 4×4 degrees of each stimulus. The second and third columns show the measured and model-predicted average activation patterns, respectively. R denotes the correlation coefficient between the measured and the predicted pattern. The scale bar denotes 2 mm. b The blue bars in the histogram show the distribution of correlation coefficients between the measured and the model-predicted activation patterns. As a control, the red bars in the histogram display the correlation coefficients between measured activation patterns of all pairs of different stimulus categories. Arrows indicate means of the respective distributions. Source data are provided as a Source Data file. c Population response matrices (z-scored, color scale lower left) to the test stimulus set for all classified cortical pixels (Fig. 3c). Cortical pixels were sorted from top to bottom based on their responses to the test stimuli for their respective category. d Measured stimulus preferences across the cortical surface. For each cortical pixel, we averaged its response to the test stimuli for each domain and identified the one with the highest response as its preferred category. The color of the cortical pixel represents its preferred category, the black contour outlines the model-predicted domains, and the hatched area represents the unclustered regions (grey in Fig. 3c, d).
Testing single-neuron selectivity on the preference map
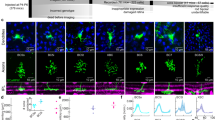
Having demonstrated a correspondence between the neural response at the columnar scale and the model-predicted preference map, we next tested the relationship between columnar-scale preference and single-neuron selectivity. We performed a series of two-photon calcium imaging recordings34 on ten selected fields of view (FOVs) of monkey C and monkey B respectively (Fig. 5a, b and Supplementary Fig. 8a). To ensure that the test stimuli could effectively activated the neurons, we used the model to select a set of images preferred by dozens of representative cortical pixels to compose the test stimulus set (see Methods). The stimulus sets for monkey C and monkey B include 905 and 537 natural stimuli, respectively. We identified soma and dendrites that responded robustly to test stimuli as ROIs for the two-photon imaging analysis (see Methods, Supplementary Fig. 8b). We found that the cortical preferences obtained by widefield imaging are roughly consistent with the stimulus preferences of single neurons in the corresponding region (Fig. 5c, d, f and Supplementary Fig. 8c). As shown in Fig. 5c, d, the neurons at the face and dot domains also preferred face or dot stimuli. The average tuning of single-cell responses within each FOV is in good agreement with that measured by widefield imaging (Fig. 5e, f). However, single cells (ROIs) exhibited a much greater degree of diversity and sparsity of tuning compared to the FOV responses, indicating that single neurons can encode and discriminate subtler variations of the image features (Fig. 5c, d, f).
a Two-photon imaging recording sites in monkey C. The color hue represents the measured cortical preference, following the same color scheme as Fig. 4d. b The two-photon fluorescence image of an example field of view (FOV) averaged over all stimuli, located at the junction of the face and dot domains is shown. c Responses of two example cell ROIs marked in (b) are shown. The error bar represents the SEM across n = 8 repetitions. The red line denotes the cortical responses of this FOV measured with widefield imaging. The stimuli are ranked according to the FOV’s cortical responses. There is a significant correlation between the response of ROI and FOV, as shown. The central 4×4 degrees of stimuli with the top 10 neuronal responses are shown on the right side. For each stimulus, the number on the top indicates its neuronal response, and the number on the bottom indicates its ranked stimulus index based on widefield imaging. Source data are provided as a Source Data file. d Population response matrix of cell ROIs across the example FOV region shown in (b). e Average responses of all ROIs (n = 298) in the example FOV region (error bar shows the SEM over the ROIs). As in (c), the red line denotes the FOV’s response measured by widefield imaging, rescaled to minimize mean square error with the averaged ROI responses. The correlation between the two is 0.83. Source data are provided as a Source Data file. f The blue histogram shows the distribution of correlation between the cortical response of each FOV measured by widefield imaging and the average responses of the ROIs within the FOV measured by two-photon imaging (FOV counts: n = 10 for both monkey). The red histogram shows the distribution of correlation between the cortical response of each FOV and the single ROIs responses within the FOV (ROI counts: n = 1650 for monkey B, n = 3439 for monkey C). Arrows indicate means. Source data are provided as a Source Data file.
Characterizing feature tuning using feature attribution analysis
Above, we characterized neural coding in terms of natural image preference. Natural images typically encompass a mixture of visual features. To gain a deeper understanding of the neural codes for visual features, it is necessary to decompose the specific features in the preferred images that drive the neural responses. Taking advantage of our V4 digital twin, we performed feature attribution analysis on our model using the SmoothGrad-Square35,36 method. For a given input image, this gradient-based algorithm generates a heatmap that reflects the contribution of each pixel in the input image to the response of the target cortical region. The heatmap thus highlights the critical features in the image that drive the neural responses. Figure 6a shows several heatmap examples. It is evident that heatmaps provide a reasonable estimation of the critical features in domains that prefer shape attributes, such as dots, edges, and curvature, as well as domains that prefer texture or surface attributes. Notably, for identified face domains, the heatmap reveals face components such as the nose and mouth are responsible for driving the neural responses.
a SmoothGrad-Square heatmaps of the preferred images highlight the critical features responsible for activating the domains. The first row shows the preferred image for example domains in monkey C; the second row shows the corresponding heatmap of each preferred image; the third row represents the image features emphasized by the heatmap. For images containing faces, only the central 4×4 degrees are shown. The box colors denote the domain categories. b For the face domain in monkey C, we experimentally tested three types of stimuli — the original preferred images of the domain, images with only critical features visible, and images with critical features masked out. Cortical responses to a set of images are shown. The scale bar denotes 2 mm. c Face domain’s responses to the example stimuli shown in (b). The error bar represents the SEM across n = 8 repetitions, and dots denote single-trial responses. Source data are provided as a Source Data file. d Face domain’s responses to the 12 sets of stimuli tested, showing a high correlation between the measured and the model-predicted responses to the three types of stimuli across the different images. Source data are provided as a Source Data file. e Comparison of face domain’s responses to the three types of stimuli. P-values for one-sided paired t-tests are: original-image vs. feature-only, P = 1.35 × 10−5; original-image vs. feature-masked, P = 5.85 × 10−8; feature-only vs. feature-masked, P = 3.38 × 10−4. P-values were corrected for multiple comparisons with Bonferroni correction. Source data are provided as a Source Data file.
We perform an in vivo widefield imaging experiment to check whether the ablation of the critical features would indeed cause a significant drop in neural response. We targeted the face domain in monkey C and tested 12 sets of images, each derived from an image that was preferred by the face domain. Each set consisted of three test images: the original preferred image, the preferred image with the critical feature masked out, the preferred image with only the critical feature remaining (Supplementary Fig. 6b). Figure 6b shows the cortical responses to an example set of test images. We obtained the face domain’s response by averaging the cortical pixels’ responses within the face domain (the salmon-colored domain in Fig. 3c). The measured responses of the face domain to the 12 sets of images are highly consistent with the model-predicted responses (Fig. 6d, Pearson correlation = 0.84). We found that, although critical features constituted only a small part of the whole image, their occlusion resulted in a greater decrease in face domain’s response compared to occlusion of all other parts of the image (Fig. 6e). This evidence suggests that the critical feature revealed by the heatmap is indeed the part of the image critical for driving the response of the target domain.
For each cortical pixel, we next averaged the heatmaps of its top 1000 images from the 50,000-image set. The resulting aggregated heatmap provides a reasonable estimate of the receptive field (RF) for the neuronal population in that pixel. We determined the location and size of the RF by fitting the aggregate-heatmap with elliptical Gaussian (Supplementary Fig. 9). Compared to traditional RF mapping approaches, we found that this method was more general, effectively estimating the RF of cortical regions that respond poorly to traditional stimuli (Supplementary Fig. 10).
Combining the receptive field and the heatmap of preferred images, we found that the strong responses of some cortical regions were driven by spatially localized features, while the strong responses of other regions depended on features that were more dispersed inside the RF (Fig. 7a). This difference may reflect the mechanisms for two distinct classes of feature computation. To quantify how dispersed the features are in the receptive field, we designed a content removal test. Specifically, for a preferred image of a cortical pixel, we identified the K pixels with the highest heatmap values as the key area and generated two versions of the image: one with the key area removed and another with only the key area content preserved (Fig. 7b). As the key area increases, the target cortical pixel’s responses to the key content-removed image will gradually decrease, while the response to the key content-preserved image will increase. We refer to the particular K pixels where these two responses are equal as the critical key area. Feature disparity is defined as the proportion of this critical key area relative to the RF area (Fig. 7c). According to this definition, if the critical features of the preferred images are completely dispersed in the RF, the feature dispersity will be close to 1; while when the critical features only occupy a small part of the RF, the feature dispersity will much smaller than 1. We used the model to calculate the feature dispersity for each cortical pixel. Figure 7d shows topographical maps of feature dispersity in V4 for the two monkeys respectively, revealing clusters of various degrees of feature dispersity distributed between 0 and 1.
a Example of the distribution of critical features within receptive fields (RFs). Left: the preferred images and their corresponding heatmaps of a face-preferring cortical pixel; right: those of a cortical pixel preferring grid textures. The contour outlines the 2 standard deviations (2-std) of the Gaussian envelope of the cortical pixels’ RF. b Illustration of the content removal test. The first row shows the regions containing the K pixels with the highest heatmap values; the second row shows images with only the content in these regions preserved; the third row shows images with these regions’ content removed. See content removal approach in Supplementary Fig. 11 and Methods. The ratio of K pixels to the RF area is indicated at the top. c Model-predicted responses of the two pixels in (a) to Top K removed and Top K preserved stimuli for different K values. Solid lines represent the average response to the top 25 preferred images for each pixel, with the shaded stripes indicating standard errors (salmon: face-preferring, green: grid texture-preferring). We define feature dispersity (FD) as the ratio between the K values and the RF area at the intersect of the Top K removed and Top K preserved curves. Source data are provided as a Source Data file. d Feature dispersity maps for monkey C and monkey B. e Relationship between feature dispersity and frequency selectivity index across domains in monkey C. We averaged the indices of cortical pixels within each domain to obtain the domain index. Colors denote domain categories. Source data are provided as a Source Data file. f Relationship between feature dispersity and color selectivity index across domains in monkey C. Source data are provided as a Source Data file. For better visualization, (e, f) Show the RF-cropped preferred images of the example domains, where the cropped square region encompasses the receptive field (2-std) of the domain. See all the domains’ RF-cropped preferred images in Supplementary Fig. 13 and Supplementary Data 4, 5.
Previous studies have demonstrated that V4 has topographic maps of color selectivity and spatial frequency selectivity. How does the map of feature dispersity relate to these established functional maps? To quantify how cortical responses are tuned to color or high-frequency features, we introduce the color selectivity index (CSI) and frequency selectivity index (FSI) for each cortical pixel. CSI is calculated as the difference in responses to the cortical pixel’s preferred images and the gray images. FSI, on the other hand, is defined as the difference in response to the preferred images, and to the Gaussian blurred version of them (Supplementary Fig. 12a, see Methods). The model shows that both indices form a specific topographic organization in V4 (Supplementary Fig. 12b, c). However, the organization of feature dispersity cannot be fully explained by the topographic organization of color selectivity and frequency selectivity. Although domains with higher FSI tend to be associated with smaller degrees of feature dispersity, and domains with lower FSI tend to be associated with larger degrees of feature dispersity, there are exceptions. For example, domains preferred for high-frequency textures have both high FSI and high feature dispersity (Fig. 7e). Similarly, while most domains with color selectivity have a high feature dispersity, there are also domains with color selectivity but a low feature dispersity (Fig. 7f). Feature dispersity is more closely associated with concepts of shape and texture, rather than spatial frequency or color. Domains with a preference for shape-related features like edges, curvature, and face components tend to have smaller feature diversity. Conversely, domains with a preference for surface or texture features tend to have higher feature diversity (Fig. 7e). These results suggest that feature dispersity is an independent characteristic dimension of V4 functional organization, distinct from color and frequency selectivity. The topographical organization of feature dispersity reflects the distinct processing streams of shape and texture features in V4.
Discussion
Our study aims to identify the specific natural image features that are prominently represented by local subpopulations of V4 neurons. To capture the V4 representation of diverse natural scenes as comprehensively as possible, we used widefield calcium imaging to record cortical responses of large spans of V4 to tens of thousands of natural images. This dataset enables us to train a deep-learning model that can accurately predict the responses of every imaged cortical pixel. This model can be considered a functional digital twin of V412,18,19,20,37. With the help of this digital twin, we systematically investigated the natural image features preferred by the neuronal populations across the cortical surface. We found that V4, remarkably, contains a set of diverse functional domains preferring many non-classical visual features. These domains can be classified into those preferring shape related or texture-related features based on how dispersed the preferred image features are inside their receptive fields. This finding of distinct domains of shape and texture related features suggest a possible functional organization principle of V4 neural codes.
Biological visual systems are designed to process natural senses and hence natural images should be the most effective stimuli for probing the visual systems. However, the complexity and diversity of natural image features pose a challenge for investigating the neural codes of these features when only a limited number of stimuli can be tested. Consequently, many studies have relied on simplified artificial stimuli to explore the neural coding of specific stimulus attributes. Such an approach has revealed that V4 contains maps for encoding color38, orientation39, spatial frequency40,41, and curvature42,43,44. While these findings have enriched our understanding of V4, they are, at best, partial descriptions of the neural codes. Like blind men touching the elephant, each study offers a limited perspective. This leaves us with an incomplete picture of the feature representation of V4. In this study, we establish the feasibility of a paradigm utilizing natural images to characterize the neural codes. This paradigm leverages the testing power of digital twins, modeled by deep neural networks. Using the digital twin to search for the most preferred images for each cortical pixel in a large set of natural images, we could comprehensively capture the visual features that are most salient to the neural population. Through feature attribution analysis, we could then identify the critical regions within the natural stimulus that drive the neural response. This enabled us to effectively estimate the receptive fields of the cortical pixels. Additionally, we could decipher the neural coding of specific features via targeted image transformation (e.g., content removal, image blur, image graying). This paradigm of understanding feature tuning using natural images not only allows us to uncover detailed functional organization that has been previously overlooked but also provides a unified framework for integrating the knowledge gained from past studies. This paradigm can be applied to any visual cortical area, facilitating a more thorough investigation of the neural coding of visual features.
Our results show that V4 primarily encodes natural image features related to shape and texture. Contour shape and surface texture are important features that support object recognition and scene segmentation45. The encoding of these two types of features in mid-level visual area V4 has thus attracted considerable attention46,47,48,49,50,51,52,53. An earlier study54 on the joint coding of shape and texture found that V4 neurons lie along a continuum from strong tuning for boundary shapes to strong tuning for texture, and among neurons tuned to both attributes, tuning for shape and texture were largely separable. In that study, neural codes for shape and texture were characterized using a set of artificially defined stimulus prototypes. In contrast to their approach, we characterized the feature continuum between shape and texture using a quantitative measure of feature dispersity. This measure relates the concepts of shape and texture to the degree of localization of preferred features within the receptive field, with high dispersity being a characteristic of texture and low dispersity a characteristic of shape. How this dispersity measure can be related to the joint encoding characteristic of shape and texture reported in the earlier study remains unclear. Further deciphering the relationship between feature dispersity and coding of shape and texture may help us understand the nature of shape and texture processing and provide insights into how shape and texture selectivities emerge in the primate brain.
The high-performing V4 model, derived from the large dataset, allows us to systematically characterize the feature preferences of the neuronal populations in V4. Beyond supporting the analysis and findings in this study, our model and data can be useful for various other applications. One interesting possibility is using the model to generate images that evoke responses similar to those of the original reference images, known as ‘metamers’55. This reconstruction analysis can provide insights into the specific image contents represented in the V4 population, enhancing our understanding of information processing in the ventral visual stream. Our widefield imaging dataset can also be a useful resource for benchmarking and evaluating computational models of V4. A popular strategy of brain modeling is to develop task-driven artificial neural networks so that their network features can best explain the relationship between stimuli and neural responses56,57,58. The quality and richness of the neural data available in our dataset will allow a reliable evaluation of the competing computational models in terms of neural response prediction and representational similarity. Furthermore, our calcium imaging dataset provides information about columnar-scale topographic organization, which is not available in existing datasets based on fMRI28 or electrophysiological recordings25,59. This unique feature of our dataset makes it particularly useful for assessing computational models that explore the principles underlying functional organization of the primate visual cortex60,61,62.
Our study focused on the image features most preferred by each cortical pixel. A cortical pixel’s response represents the average response of hundreds of nearby neurons in the superficial layer. Therefore, the preferred image features of a cortical pixel can indicate the features that drive the simultaneous excitation of a local population, which might reflect the functional computation emphasized by local circuits. Based on the similarity of preferred images among cortical pixels, we grouped cortical pixels into distinct functional domains. Our two-photon data showed a relatively abrupt transition in categorical labels at the boundaries of these domains (Supplementary Fig. 8c). However, this does not necessarily imply discrete borders between the domains, as characterizing neuronal selectivity based solely on preferred image categories can by itself introduce discontinuities. To clarify whether the functional domains are discrete or continuous, a more comprehensive investigation of the tuning properties of neurons to natural images, as well as their distribution across the cortex, would be necessary.
Our two-photon experiment establishes the consistency between neuronal tunings and the preference of the cortical pixels. It further reveals the diversity and similarity in neuronal tuning of the local population. The natural image preference maps obtained through widefield imaging can be a useful guide for future studies on neuronal tuning. These detailed maps enable us to apply a ‘divide and conquer’ strategy to study neuronal coding within each functional subpopulation. By integrating information from different scales and neural codes from all the local populations, we can ultimately achieve a comprehensive understanding of the neural representation of natural scenes.
Methods
Animal preparation and surgery
All experimental protocols followed the guidelines provided by the Institutional Animal Care and Use Committee (IACUC) of Peking University Laboratory Animal Center and were approved by the Peking University Animal Care and Use Committee (LSC-TangSM-3). Three adult male rhesus macaques (Macaca mulatta) named A, B, and C, aged between 4 and 6 years, were used in this study.
The details of animal preparation for long-term calcium imaging in awake macaque have been described previously in ref. 34. In summary, each animal underwent three sequential surgeries while under general anesthesia. These surgeries involved the implantation of head posts implant, virus injection, and the installation of imaging window. In the second surgery, a 20 mm craniotomy was made on the skull over the dorsal V4 region, targeting the area encompassing the lunate sulcus (lu) and the terminal portion of the inferior occipital sulcus (io). We then performed pressure injections of AAV1.hSynap.GCaMP5G.WPRE.SV40 (AV-1-PV2478, titer 2.37e13 GC/mL, Penn Vector Core) at 20–30 locations within the V4 cortex. These injections were administered at a depth of approximately 350 μm. To ensure uniform expression of GCaMP, the injection sites were spaced approximately 1 mm apart. Each injection had a volume of 100–150 nL.
Behavioural task
Monkeys were securely restrained using a head fixation apparatus and performed an eye fixation task during image recording. The animal was required to maintain fixation on a small white spot, measuring 0.1°, within a circular window with a diameter of 1°, for 1.5 s to obtain a juice reward. Eye position was monitored with an infrared eye-tracking system (ISCAN) at 120 Hz.
Visual stimuli
Visual stimuli were generated using a ViSaGe system (Cambridge Research Systems) and displayed on a 17-inch LCD monitor (Acer V173, 80 Hz refresh rate), positioned 45 cm away from the subject’s eyes. Each stimulus was presented for 0.5 seconds following a pre-fixation period of 0.9 s. For each monkey, the receptive field of the imaged region was first hand-mapped with small gratings. Namely, we manually controlled the presentation of a 0.4-degree diameter grating to determine the receptive field location. In the subsequent experiments, the stimuli were presented over the region’s receptive field (Supplementary Fig. 2b).
Stimuli for V4 large dataset
The natural image stimuli used in the large dataset were sourced from ImageNet23, specifically ILSVRC2012 and 8 synsets from the person subtree. The original images were cropped, resized and masked to create round patches measuring 180 pixels (6 degrees) in diameter with soft fade-off.
Our large-scale V4 dataset compromises a training set consisting of neural responses with each image repeated once, and a validation set consisting of neural responses with each image with ten repetitions in random interleave. Each monkey’s training set contained cortical responses to over 17,000 unique color natural images. Monkey A, B, and C were tested with 20,000, 17,900, and 19,900 images, respectively, over a period of 4 to 5 days. For validation purposes, a separate set of 500 natural images was used. In addition, the validation stimuli included 48 gratings and 8 color patches (Supplementary Fig. 2b). The 48 gratings comprised 8 orientations (22.5° increments), 3 spatial frequencies (1.0, 2.0, and 4.0 cycles/degree), and 2 phases. The 8 color patches consisted of red, orange, yellow, green, blue, purple, white, and black. The validation sets were acquired over one day or two consecutive days. To assess the image quality and consistency of cortical responses across recording days, we generated a fingerprint stimulus set comprising the first 100 pictures from the validation stimulus set. On days when validation data was not collected, we recorded the cortical responses to these 100 fingerprint stimuli with 5 repetitions. This allowed us to evaluate the consistency of the cortical pixels’ tunings as well as their imaging quality.
Stimuli for testing the preference map
The test stimulus set includes preferred images for multiple cortical sites. We manually selected 30 and 50 representative cortical sites with distinct feature preferences for monkey B and monkey C, respectively. For each site, we used the model’s predictions to identify the top 20 images from the 50,000-image set, forming the test stimulus set. We eliminated duplicate images that were selected for different sites, resulting in stimulus sets containing 537 and 905 stimuli for monkey B and monkey C, respectively. Neural responses to these stimuli were recorded using widefield imaging and two-photon imaging techniques. During widefield imaging, the response to each stimulus was measured eight times. In the case of two-photon imaging, the response to each stimulus was measured 6-8 times within each field of view (FOV). To validate the functional domains identified by the model, we selected 16 model-predicted preferred images for each domain as follows. First, we normalize each cortical pixel’s response to the 50 K images to zero-mean and unit variance. Then we averaged the normalized responses of all the pixels in a functional domain for each image, and select the 16 images from the test stimulus set with the highest averaged predicted responses (Supplementary Fig. 5a). These images were then used to test the cortical preferences and assess their alignment with the model’s predictions.
Stimuli for testing the critical feature
From the above stimulus set for testing the preference map, we selected 12 images that are preferred by the face domain of monkey C. For each of these selected images, we computed a heatmap to identify the critical region within the image that drove the response of the face domain. Using Adobe Photoshop, we created two types of images: one in which the critical region was masked and another in which only the critical region was visible while the rest of the image was masked. These additional images, along with the original images, formed a test stimulus set comprising a total of 36 images (12 images × 3 types). We chose a mask color to ensure a smooth transition between the masked and uncovered regions. The cortical responses to these stimuli were recorded using widefield imaging, with each stimulus repeated eight times.
Widefield calcium imaging
Widefield imaging setup
We performed widefield calcium fluorescence imaging with a camera imaging system adapted from Imager 3001/M (Optical Imaging). An excitation blue light was obtained with a LED light source (S3000, Nanjing Hecho Technology Co.) passing through a 470/40 nm filter. The reflected light was collected using a pair of lens (Sigma, 50 mm) and split into green and blue light with a dichroic mirror (525 nm). The green light was further filtered (525/50 nm) and projected onto a green channel camera (Imager 3001/M, Optical Imaging) and was recorded as the fluorescence calcium images at a rate of 33 Hz. The blue light was projected onto a blue channel camera (ZWO ASI533MC Pro, ZWO) and was recorded as the reflectance image at a rate of 20 Hz (Fig. 1a). The reflectance image, which captured the blood vessels well, served as the reference for anatomical registration. The imaging focus was adjusted to 300 μm below the cortical surface. A fast-mechanical shutter was inserted into the blue excitation light pathway to provide intermittent illumination for long-term stable imaging.
Calculating cortical responses
During each stimulus presentation epoch, there were two periods of illumination, each lasting 250 ms. We utilized the fluorescence images recorded during these periods to calculate cortical responses. The first illumination period commenced at 150 ms prior to the onset of the stimulus, while the second illumination period began 350 ms after the stimulus onset. We averaged the frames captured during these two periods to obtain the baseline image (F0) and peak response image (F1) respectively. From these images, we computed maps of fluorescence change (ΔF/F0 map) using the formula (F1-F0)/F0. To eliminate global signal changes unrelated to the stimulus and to reveal local modulation, we performed a subtraction operation, subtracting a Gaussian blurred (σ = 1.0 mm) version of the ΔF/F0 map from the original map, resulting in cortical responses that highlight local changes. We found that the cortical responses obtained by this high-pass filtering matched better the responses measured by two-photon imaging as shown in Supplementary Table 2.
Image registration across days
Prior to imaging each day, we carefully adjusted the camera system to ensure alignment with the positions on the first day. Our procedure involves focusing on the plane that exhibits the highest sharpness in the blood vessel image. We then laterally moved the camera system to match the vessel image with the reference image acquired on the first day. To assess alignment accuracy, we developed a custom MATLAB software that evaluates image sharpness and lateral position error. Once the alignment is achieved, we move the depth of focus down by 300 μm for recording.
To correct for minor displacement and distortion of the cortex across multiple imaging days, we incorporated additional image correction during data processing. We first aligned the blood vessel image captured by the blue imaging camera with the fluorescence images acquired by the green imaging camera. This alignment involved rotating, rescaling, and translating the blood vessel images. Then, a transformation matrix was generated between the blood vessel images from each day and the reference image acquired on the first day. This transformation matrix was then used to correct and align the fluorescence images across days. Since the blood vessel images acquired by the blue channel camera have higher spatial resolution than the fluorescence images, this approach allows us to achieve greater registration accuracy.
DNN Modeling
Data preprocessing for modeling
The stimulus images, initially measuring 200 × 200 pixels (30 pixels/degree), were resized to 100 × 100 pixels and input to the model. The raw response map acquired by the camera had a resolution of 512 × 512 pixels and a sampling rates of 45 pixels/mm, which exceeded the intrinsic spatial resolution of the widefield calcium imaging signal. To simplify the modeling analysis, we rescaled the response maps to 128 × 128 pixels using bilinear interpolation. We used validation set responses to identify regions exhibiting significant stimulus-related fluorescence changes. Specifically, we conducted one-way ANOVA across responses of 556 validation stimuli, resulting in regions with P < 10−300, indicating highly significant changes. These regions were designated as regions of interest (ROIs, Supplementary Fig. 2a) for modeling purpose. Responses of regions from outside the ROIs were masked to zero during modeling.
Network architecture
Our feature transfer model consisted of a feature extraction and a two-layer perceptron (Supplementary Fig. 3a). The features are the output maps of Add-6 layer of Xception31 to a stimulus image. We used the Add-6 layer as its outputs have been demonstrated to be a reliable predictor of V4 responses (see Supplementary Fig. 3b). We used a Keras implementation of Xception, which was trained for the ImageNet classification task. The two-layer perceptron was used to map the features to the corresponding cortical responses. The hidden layer of the perceptron consists of 200 units, designed to extract effective features while avoiding overfitting. To enhance the model’s expressive capacity, we incorporate an exponential linear unit (ELU) nonlinearity in the hidden layer. This nonlinearity aids in capturing complex relationships and enhancing the model’s ability to represent the data.
The small Xception-like DNN model (Supplementary Fig. 3c) consisted of a CNN encoder that shared architectural similarities with Xception, including depth-wise separable convolution layers and residual learning blocks. The encoder generated nonlinear feature responses from input images. A readout network was used to map the output of encoder to cortical responses. Sigmoid activation function was used to introduce nonlinearity. The encoder converted the RGB input images to 7 × 7 × 400 feature maps, which were fed into the readout network. Each position within the feature maps corresponded to a distinct retinotopic spatial location, containing a column of features, whereas different positions in the imaged V4 cortex encoded different features with similar spatial receptive fields. In order to transform the spatially organized maps of the encoder into feature-organized maps that resembled the organization in the cortex, the readout network first reorganized the input 7 × 7 × 400 spatial-feature map into a 20 × 20 × 49 feature-spatial map. This reorganization swapped the spatial and feature organization. The 400 feature channels were now organized as a 20 × 20 spatial map, with each column containing that feature channel’s responses across various retinotopic locations. This feature-spatial map was then passed through sequences of convolutional and locally connected layers to generate the final response output.
To prevent overfitting, dropout layers with a dropout ratio of 0.1 were introduced to all of the above models, meaning that during training, 10% of the responses were randomly dropped to encourage the model to generalize better to unseen data. Additionally, the feature transfer model utilized an L1 penalty regularization on the connection weights between the hidden layer and Xception features. This regularization encourages sparsity and promotes the selection of more relevant and informative features, reducing the risk of overfitting and improving generalization.
Model training
All the models were optimized using stochastic gradient descent with the Adam optimizer63 and a batch size of 20. When fitting the neural data, models are trained to minimize the mean square error (MSE) on the training set. To prevent overfitting and ensure the best generalization performance, we used early stopping based on the MSE between predicted and measured neural responses on the validation set. If the MSE failed to decrease during any consecutive 50 passes through the entire training set (50 epochs), the training process would be halted. The model that achieved the best performance on the validation set during the training phase would be saved as the final model. This approach allows us to capture the model’s optimal performance while avoiding unnecessary training iteration.
For knowledge distillation, we used the results generated by the feature transfer model as data to train the small DNN model using supervised training. This generated data consisted of image-response pairs derived from 100,000 ImageNet images. The responses were predicted by the feature transfer model that was trained on the neural data. The generated data was split into a training set containing 90,000 images and a validation set containing 10,000 images. We trained the small DNN on this data by minimizing the training set MSE. We also used early stopping based on the MSE on the validation set. The training would be halted if the MSE on the validation set did not decrease in ten consecutive epochs.
Predictivity evaluation
The models were assessed by evaluating the correlation between the predicted responses and the measured responses of each cortical pixel to the validation stimuli. There is an inherent limit to the maximum achievable correlation due to response variability within the same day and across days. To estimate this limit, or noise ceiling, we calculated the correlation (see Supplementary Fig. 2f for statistics) between the responses to the fingerprint stimuli in the validation set and the responses to the fingerprint stimuli averaged across days when the training set were collected. This allowed us to determine the upper bound of correlation that can be achieved considering the inherent variability in the neural responses. The models’ achieved correlation is then normalized by the noise ceiling to provide a performance measure.
Preference map analysis
Preference map synthesis
We gathered a set of 50,000 images from ImageNet and prepared them in the same way as we did for generating the stimulus sets used in the experiment. We used the KD Transfer model to predict the cortical responses of these 50,000 images. We then organized the nine most responsive images for each cortical pixel into a 3 × 3 grid and display it at the corresponding cortical location in the acquired image to derive the preference map.
Hierarchical clustering on the preference map
In the preference map, the preference of each cortical pixel is represented by its top nine images. To cluster these cortical pixels based on the similarities of their preferred images, we employed a method illustrated in Supplementary Fig. 4. First, we computed the model’s prediction of each cortical pixel’s response to the entire set of 50,000 images, and normalized them to range between 0 and 1. Then, we combined all the pixels’ normalized responses to generate a predicted cortical activation pattern associated with each image. We then averaged the activation patterns of the top nine images preferred by each pixel to create the ‘cortical response vector’ of that pixel. The similarity between two cortical pixels was determined by computing the Pearson correlation between their respective cortical response vectors. This similarity metric enabled us to identify groups of cortical pixels that exhibited similar activation patterns when exposed to their preferred images. To identify functional domains within the V4 cortex based on shared image preferences, we employed hierarchical clustering based on average-linkage, and grouped cortical pixels within a distance threshold of 0.4 (computed as one minus the similarity) into a cluster. Any cluster that included connected regions larger than 40 pixels was considered a functional domain. This approach allowed us to distinguish distinct functional domains within the V4 cortex, based on their collective preference for specific image features.
Two-photon calcium imaging
Two-photon imaging setup
We performed two-photon calcium imaging on monkeys B and C with a Bruker two-photon imaging system (Prairie Ultima IV, Bruker Nano). The wavelength of the femtosecond laser (Insight X3, Spetra-Physics) was set to 1000 nm. Field of views (FOVs) of 600 μm × 600 μm were imaged under 1.4× zoom with a 16× objective (0.8-N.A., Nikon) at a resolution of 1.2 μm/pixel. A fast-resonant scan (30 frames per second) was used to obtain images of neuronal activity. We averaged every two frames, resulting in an effective frame rate of 15 fps. In total, we recorded 20 FOVs, 10 from monkey B and 10 from monkey C, with recording depths ranging from 100 μm to 300 μm. To determine the precise position of each FOV relative to the widefield imaging map, we recorded the blood vessel image directly above each FOV as a reference to align the two-photon imaging data with the widefield imaging map.
Data processing for two-photon imaging
We used customized MATLAB code to process the data obtained from the experiments. First, we associated the two-photon image series with the corresponding visual stimuli using the time sequence information recorded by Neural Signal Processor (Cerebus system, Blackrock Microsystem). Then, the images were motion corrected using a normalized cross-correlation-based translation algorithm34. This step helped to align the images and mitigate any image shifts caused by motion during recording. For the response to each stimulus, we computed the F0 image by averaging the five frames preceding the onset of the stimulus. Similarly, the F1 image was obtained by averaging the frames from the fifth to the tenth frames after stimulus onset. These F0 and F1 images provided baseline and peak response information associated with a stimulus, respectively. An additional non-rigid motion correction64 was applied to the F0 and F1 images to correct for the cortical deformation during the long recording session.
We used the differential image (ΔF) obtained by F1-F0 to extract regions of interest (ROIs). We averaged the differential images across all repeated trials of the same stimulus. A band-pass Difference of Gaussian filtering (standard deviations of positive and negative Gaussians are 1 and 30 pixels respectively) was then applied to the averaged differential images. The connected subsets of pixels ( > 30 pixels) with pixel values > 3.5 standard deviations of the mean brightness were selected as ROIs. We further refined the shape of the ROI by calculating the correlation between the ΔF values of the ROI and its neighboring pixels. Pixels with a correlation greater than 0.3 will be assigned to the ROI. Using the above methods, we obtained many overlapping ROIs. To determine whether these ROIs should be merged, we perform hierarchical clustering on the responses of pixels within these ROIs. The pixel response was calculated by ΔF/F0 of the ROI to which it belonged. In cases where a pixel belonged to the multiple ROIs, the response was computed as the average of ΔF/F0 of those ROIs. We calculated the response of each selected ROI using ΔF/F0 and identified visually responsive ROIs that exhibit significant response selectivity (P < 10−5, tested with a one-way ANOVA) for any of the 537 and 905 test stimuli for monkey B and monkey C, respectively.
Feature attribution analysis
Heatmap synthesis
We used the SmoothGrad-Square algorithm35,36 to produce heatmap of the input image for any output unit of the model. This algorithm relies on computing the gradient map of the unit’s response with respect to the input image. The SmoothGrad-Square algorithm operates by introducing Gaussian noise to the image of interest, generating a set of similar images. For each generated image, the unit’s response is backpropagated to the input of the deep learning model to generate a gradient map, which captures the sensitivity of the unit’s response to changes in the input. These gradient maps are then squared and aggregated to produce the final heatmap. SmoothGrad-Square involves two hyper-parameters: σ, the standard deviation of the Gaussian noise, and n, the number of samples to sum over. Here we used σ = 0.2 (image value \(\in \)[0,1]) and n = 20. To obtain the heatmap of a specific cortical pixel, we applied the SmoothGrad-Square algorithm to the corresponding model output unit. When generating heatmap for a particular cortical region (e.g., the face domain in Supplementary Fig. 6b), we first added a linear connection unit to the model that summed the outputs of the relevant pixels. We then applied the SmoothGrad-Square algorithm to this unit to produce the final heatmap.
Estimating receptive field
To estimate the receptive field of a target cortical pixel/region, we averaged the heatmaps generated from a large set of natural images. Namely, we first calculated the heatmaps for the top 1,000 images in the 50,000-image set for the target cortical pixel/region. Next, we normalized the heatmap for each image to ensure that the sum of values on the heatmap equals the cortical response elicited by the image. Finally, we fitted the average of these normalized heatmaps with an elliptical Gaussian:
where \(A\) is the amplitude of the Gaussian, \(B\) is the offset, \({\sigma }_{a}\) and \({\sigma }_{b}\) are the standard deviations of the elliptical Gaussian along its two principal axes, and \({x}^{{\prime} }\) and \({y}^{{\prime} }\) are transformations of the coordinates \(x\) and \(y\), taking into account the angle \(\theta\) and the offset (\({x}_{c}\) and \({y}_{c}\)) of the ellipse. In total, there were seven free parameters in the fitting procedure: \(A,\, B,\,{\sigma }_{a},\, {\sigma }_{b},\, \theta,\;{x}_{c}\) and \({y}_{c}\). We define the RF area as the area within the half-maximum contour of the \(G\left(x,\, {y}\right)\), and the receptive field size as the square root of the RF area (i.e. \(\sqrt{2\pi \, {\mathrm{ln}}(2)\, {\sigma }_{a}{\sigma }_{b}}\)). This definition of the RF area will ensure that \({\int }_{{\rm{within}}\; {\rm{RF}}\; {\rm{area}}}G\left(x,\, {y}\right)\,{\rm{d}}x{\rm{d}}y={\int }_{{\rm{outside}}\; {\rm{RF}}\; {\rm{area}}}G\left(x,\, {y}\right)\,{\rm{d}}x{\rm{d}}y\).
Content removal test
To quantify the dispersion of preferred features within the receptive field, we define a metric called feature dispersity based on the content removal test. For a given cortical pixel, we select its top 25 preferred images from the 50,000-image set. For each preferred image, we first calculate the heatmap of the target cortical pixel and identify the key area with the highest heatmap value. We then generate a series of images, with the key area removed or preserved at various key area sizes. We used the model to predict the target cortical pixel’s responses to the 25 preferred images with and without the key content. We call the area when the cortical pixel’s average response curves to these two types of images intersect the critical key area. The ratio of this critical key area relative to the RF area is defined as the feature dispersity for that cortical pixel.
To smoothly remove the image content (inside or outside the key area), we adopted a method that performs information removal separately in the low-frequency and high-frequency domains (Supplementary Fig. 11). We obtain the low-frequency information (X1) of the original image by applying a Gaussian filter with a sigma value equal to half of σRF (σRF = \(\sqrt{{\sigma }_{a}{\sigma }_{b}}\)). The Gaussian filter size was chosen to ensure the receptive field does not contain sufficient information to resolve period signals, in accordance with the Shannon sampling theorem. The high-frequency information (X2) was then defined as the difference between the original image and the low-frequency image. To remove content in the low-frequency domain, we set the target pixels’ RGB values to 0.5 and applied the same low-pass filter as before, obtaining the content removed low-frequency image (X1’). For high frequencies, we apply Gaussian smoothing (sigma = 2 pixels) to the target pixels’ spatial mask and then mask the X2 with the smoothed mask to obtain the content removed high-frequency image (X2’). The final content-removed image is obtained by adding X1’ and X2’.
Calculating frequency selectivity index and color selectivity index
We used the frequency selectivity index (FSI) and color selectivity index (CSI) to measure how cortical responses are tuned to high-frequency and color information. For a given cortical pixel, we selected its top 25 preferred images from the 50,000-image set. FSI is calculated for each image as the response difference between the original image and its Gaussian blurred version (sigma = σRF/2), normalized by the original image response. CSI is calculated as the difference in response between the original image and its grayscale counterpart, also normalized by the original image response. The grayscale conversion is performed using the formula Gray = 0.299×R + 0.587×G + 0.114×B. The results from the 25 preferred images are averaged to obtain the FSI and CSI for the target cortical pixel.
Statistics & Reproducibility
Statistical analyses were performed using MATLAB (R2021a). We used the one-sided Wilcoxon signed-ranked test to compare the performances of the models. The one-sided paired t-test followed by Bonferroni correction was used to compare responses to 3 types of stimuli for testing critical features. One-way ANOVA analyses were used to identify significant differences in the means of multiple groups. Three male macaque monkeys participated in the experiments. No statistical method was used to predetermine sample size.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The widefield calcium imaging dataset and the two-photon imaging data used in this study are available at Zenodo (https://doi.org/10.5281/zenodo.10972034). Natural images used in the stimulus set were taken from the ImageNet database23 (https://image-net.org/download-images.php). Source data are provided with this paper.
Code availability
Custom code for model training and related analysis are available at Zenodo (https://doi.org/10.5281/zenodo.10972034).
References
Simoncelli, E. P. & Olshausen, B. A. Natural image statistics and neural representation. Annu Rev. Neurosci. 24, 1193–1216 (2001).
DiCarlo, J. J. & Cox, D. D. Untangling invariant object recognition. Trends Cogn. Sci. 11, 333–341 (2007).
Olshausen, B. A., Mangun, G. & Gazzaniga, M. Perception as an inference problem (MIT Press, 2014).
DiCarlo, J. J., Zoccolan, D. & Rust, N. C. How does the brain solve visual object recognition? Neuron 73, 415–434 (2012).
Collins, C. E., Airey, D. C., Young, N. A., Leitch, D. B. & Kaas, J. H. Neuron densities vary across and within cortical areas in primates. Proc. Natl Acad. Sci. 107, 15927–15932 (2010).
Douglas, R. J. & Martin, K. A. C. A Functional Microcircuit for Cat Visual-Cortex. J. Physiol.-Lond. 440, 735–769 (1991).
Bastos, A. M. et al. Canonical Microcircuits for Predictive Coding. Neuron 76, 695–711 (2012).
Hubel, D. H. Laminar and Columnar Distribution of Geniculo-Cortical Fibers in Macaque Monkey. J. Comp. Neurol. 146, 421–450 (1972).
Fujita, I., Tanaka, K., Ito, M. & Cheng, K. Columns for Visual Features of Objects in Monkey Inferotemporal Cortex. Nature 360, 343–346 (1992).
Mountcastle, V. B. The columnar organization of the neocortex. Brain 120, 701–722 (1997).
Horton, J. C. & Adams, D. L. The cortical column: a structure without a function. Philos. Trans. R. Soc. B: Biol. Sci. 360, 837–862 (2005).
Willeke, K. F. et al. Deep learning-driven characterization of single cell tuning in primate visual area V4 unveils topological organization. bioRxiv, 2023.2005. 2012.540591 (2023).
Ts’o, D. Y., Frostig, R. D., Lieke, E. E. & Grinvald, A. Functional organization of primate visual cortex revealed by high resolution optical imaging. Science 249, 417–420 (1990).
Seidemann, E. et al. Calcium imaging with genetically encoded indicators in behaving primates. Elife 5, https://doi.org/10.7554/eLife.16178 (2016).
Rust, N. C. & Movshon, J. A. In praise of artifice. Nat. Neurosci. 8, 1647–1650 (2005).
Rousselet, G. A., Thorpe, S. J. & Fabre-Thorpe, M. How parallel is visual processing in the ventral pathway. Trends Cogn. Sci. 8, 363–370 (2004).
Naselaris, T., Allen, E. & Kay, K. Extensive sampling for complete models of individual brains. Curr. Opin. Behav. Sci. 40, 45–51 (2021).
Bashivan, P., Kar, K. & DiCarlo, J. J. Neural population control via deep image synthesis. Science 364, https://doi.org/10.1126/science.aav9436 (2019).
Walker, E. Y. et al. Inception loops discover what excites neurons most using deep predictive models. Nat. Neurosci. 22, 2060–2065 (2019).
Ratan Murty, N. A., Bashivan, P., Abate, A., DiCarlo, J. J. & Kanwisher, N. Computational models of category-selective brain regions enable high-throughput tests of selectivity. Nat. Commun. 12, 5540 (2021).
Richards, B., Tsao, D. & Zador, A. The application of artificial intelligence to biology and neuroscience. Cell 185, 2640–2643 (2022).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Deng, J. et al. ImageNet: A Large-Scale Hierarchical Image Database. Proc Cvpr Ieee, 248-255, https://doi.org/10.1109/cvpr.2009.5206848 (2009).
Schrimpf, M. et al. Brain-score: which artificial neural network for object recognition is most brain-like? Preprint at bioRxiv https://doi.org/10.1101/407007 (2020).
Cadena, S. A. et al. Deep convolutional models improve predictions of macaque V1 responses to natural images. PLoS Comput Biol. 15, e1006897 (2019).
Yamins, D. L. & DiCarlo, J. J. Using goal-driven deep learning models to understand sensory cortex. Nat. Neurosci. 19, 356–365 (2016).
Yamins, D. L. et al. Performance-optimized hierarchical models predict neural responses in higher visual cortex. Proc. Natl Acad. Sci. USA 111, 8619–8624 (2014).
Allen, E. J. et al. A massive 7T fMRI dataset to bridge cognitive neuroscience and artificial intelligence. Nat. Neurosci. 25, 116–126 (2022).
Wang, L. & Yoon, K.-J. Knowledge distillation and student-teacher learning for visual intelligence: A review and new outlooks. IEEE Transac. Pattern Anal. Machine Intell. 44, 3048–3068 (2021).
Gou, J., Yu, B., Maybank, S. J. & Tao, D. Knowledge distillation: A survey. Int. J. Computer Vis. 129, 1789–1819 (2021).
Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. 30th Ieee Conference on Computer Vision and Pattern Recognition (Cvpr 2017), 1800-1807, https://doi.org/10.1109/Cvpr.2017.195 (2017).
Ukita, J., Yoshida, T. & Ohki, K. Characterisation of nonlinear receptive fields of visual neurons by convolutional neural network. Scientific Reports 9, https://doi.org/10.1038/s41598-019-40535-4 (2019).
Abbasi-Asl, R. et al. The DeepTune framework for modeling and characterizing neurons in visual cortex area V4. Preprint at bioRxiv, https://doi.org/10.1101/465534 (2018).
Li, M., Liu, F., Jiang, H., Lee, T. S. & Tang, S. Long-Term Two-Photon Imaging in Awake Macaque Monkey. Neuron 93, 1049–1057.e1043 (2017).
Smilkov, D., Thorat, N., Kim, B., Viégas, F. & Wattenberg, M. Smoothgrad: removing noise by adding noise. In ICML Workshop on Visualization for Deep Learning (ICML, 2017).
Hooker, S., Erhan, D., Kindermans, P.-J. & Kim, B. A benchmark for interpretability methods in deep neural networks. In Conference on Neural Information Processing Systems (NIPS, 2019).
Franke, K. et al. State-dependent pupil dilation rapidly shifts visual feature selectivity. Nature 610, 128–134 (2022).
Liu, Y. et al. Hierarchical Representation for Chromatic Processing across Macaque V1, V2, and V4. Neuron 108, 538–550.e535 (2020).
Tanigawa, H., Lu, H. D. & Roe, A. W. Functional organization for color and orientation in macaque V4. Nat. Neurosci. 13, 1542–1548 (2010).
Zhang, Y., Schriver, K. E., Hu, J. M. & Roe, A. W. Spatial frequency representation in V2 and V4 of macaque monkey. Elife 12, e81794 (2023).
Lu, Y. et al. Revealing Detail along the Visual Hierarchy: Neural Clustering Preserves Acuity from V1 to V4. Neuron 98, 417–428.e413 (2018).
Jiang, R., Andolina, I. M., Li, M. & Tang, S. Clustered functional domains for curves and corners in cortical area V4. Elife 10, https://doi.org/10.7554/eLife.63798 (2021).
Tang, R. et al. Curvature-processing domains in primate V4. Elife 9, https://doi.org/10.7554/eLife.57502 (2020).
Hu, J. M., Song, X. M., Wang, Q. & Roe, A. W. Curvature domains in V4 of macaque monkey. Elife 9, https://doi.org/10.7554/eLife.57261 (2020).
Pasupathy, A., Kim, T. & Popovkina, D. V. Object shape and surface properties are jointly encoded in mid-level ventral visual cortex. Curr. Opin. Neurobiol. 58, 199–208 (2019).
Gallant, J. L., Braun, J. & Vanessen, D. C. Selectivity for Polar, Hyperbolic, and Cartesian Gratings in Macaque Visual-Cortex. Science 259, 100–103 (1993).
Nandy, A. S., Sharpee, T. O., Reynolds, J. H. & Mitchell, J. F. The Fine Structure of Shape Tuning in Area V4. Neuron 78, 1102–1115 (2013).
Pasupathy, A. & Connor, C. E. Population coding of shape in area V4. Nat. Neurosci. 5, 1332–1338 (2002).
Carlson, E. T., Rasquinha, R. J., Zhang, K. & Connor, C. E. A Sparse Object Coding Scheme in Area V4. Curr. Biol. 21, 288–293 (2011).
Okazawa, G., Tajima, S. & Komatsu, H. Image statistics underlying natural texture selectivity of neurons in macaque V4. Proceedings of the National Academy of Sciences 112, https://doi.org/10.1073/pnas.1415146112 (2014).
Kim, T., Bair, W. & Pasupathy, A. Perceptual Texture Dimensions Modulate Neuronal Response Dynamics in Visual Cortical Area V4. J. Neurosci. 42, 631–642 (2022).
Bushnell, B. N., Harding, P. J., Kosai, Y., Bair, W. & Pasupathy, A. Equiluminance Cells in Visual Cortical Area V4. J. Neurosci. 31, 12398–12412 (2011).
Zeki, S. M. Color Coding in Rhesus-Monkey Prestriate Cortex. Brain Res 53, 422–427 (1973).
Kim, T., Bair, W. & Pasupathy, A. Neural Coding for Shape and Texture in Macaque Area V4. J. Neurosci. 39, 4760–4774 (2019).
Feather, J., Leclerc, G., Mądry, A. & McDermott, J. H. Model metamers reveal divergent invariances between biological and artificial neural networks. Nat. Neurosci. 26, 2017–2034 (2023).
Schrimpf, M. et al. Integrative Benchmarking to Advance Neurally Mechanistic Models of Human Intelligence. Neuron 108, 413–423 (2020).
Chang, L., Egger, B., Vetter, T. & Tsao, D. Y. Explaining face representation in the primate brain using different computational models. Curr. Biol. 31, 2785–2795.e2784 (2021).
Zhuang, C. et al. Unsupervised neural network models of the ventral visual stream. Proc. Natl. Acad. Sci. USA. 118, https://doi.org/10.1073/pnas.2014196118 (2021).
Majaj, N. J., Hong, H., Solomon, E. A. & DiCarlo, J. J. Simple Learned Weighted Sums of Inferior Temporal Neuronal Firing Rates Accurately Predict Human Core Object Recognition Performance. J. Neurosci. 35, 13402–13418 (2015).
Blauch, N. M., Behrmann, M. & Plaut, D. C. A connectivity-constrained computational account of topographic organization in primate high-level visual cortex. Proc Natl Acad Sci USA 119, https://doi.org/10.1073/pnas.2112566119 (2022).
Margalit, E. et al. A unifying framework for functional organization in early and higher ventral visual cortex. Neuron, https://doi.org/10.1016/j.neuron.2024.04.018 (2024).
Durbin, R. & Mitchison, G. A dimension reduction framework for understanding cortical maps. Nature 343, 644–647 (1990).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. Preprint at arXiv https://doi.org/10.48550/arXiv.1412.6980 (2014).
Giovannucci, A. et al. CaImAn an open source tool for scalable calcium imaging data analysis. eLife 8, https://doi.org/10.7554/eLife.38173 (2019).
Acknowledgements
We would like to thank Doris Y. Tsao and Winrich A. Freiwald for their helpful comments and suggestion on the manuscript. We thank the Peking University Laboratory Animal Center for excellent animal care. This work was supported by STI2030-Major Projects, grant no. 2022ZD0204600 (to S.T.), National Natural Science Foundation of China, grant no. U1909205 (to S.T.) and funds from the Peking-Tsinghua Center for Life Sciences (to S.T.). T.S.L. was supported by NSF CISE RI 1816568 and NIH R01 EY030226-01A1. This work was also supported by High-performance Computing Platform of Peking University.
Author information
Authors and Affiliations
Contributions
Conceptualization, T.W., T.S.L., and S.T.; Methodology, T.W., H.Y., and S.T.; Software, T.W., H.Y., J.H., and S.T.; Formal Analysis, T.W., H.Y., J.H., T.S.L., and S.T.; Investigation, T.W. and H.Y.; Resources, Y.L., H.J., and S.T.; Writing-Original Draft, T.W., T.S.L., H.Y., and S.T.; Writing-Review & Editing, T.S.L., I.M.A, T.W., and S.T.; Visualization, T.W., H.Y., J.H., and T.S.L.; Supervision, S.T.; Funding Acquisition, S.T.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Peichao Li, Zhe Li and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, T., Lee, T.S., Yao, H. et al. Large-scale calcium imaging reveals a systematic V4 map for encoding natural scenes. Nat Commun 15, 6401 (2024). https://doi.org/10.1038/s41467-024-50821-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-50821-z
- Springer Nature Limited