Abstract
The landscape of non-coding mutations in cancer progression and immune evasion is largely unexplored. Here, we identify transcrptome-wide somatic and germline 3′ untranslated region (3′-UTR) variants from 375 gastric cancer patients from The Cancer Genome Atlas. By performing gene expression quantitative trait loci (eQTL) and immune landscape QTL (ilQTL) analysis, we discover 3′-UTR variants with cis effects on expression and immune landscape phenotypes, such as immune cell infiltration and T cell receptor diversity. Using a massively parallel reporter assay, we distinguish between causal and correlative effects of 3′-UTR eQTLs in immune-related genes. Our approach identifies numerous 3′-UTR eQTLs and ilQTLs, providing a unique resource for the identification of immunotherapeutic targets and biomarkers. A prioritized ilQTL variant signature predicts response to immunotherapy better than standard-of-care PD-L1 expression in independent patient cohorts, showcasing the untapped potential of non-coding mutations in cancer.
Similar content being viewed by others
Introduction
Immune evasion is a key hallmark of tumorigenesis and cancer progression1. The central role of immune evasion in the development and maintenance of the disease is reflected in the remarkable efficacy of cancer immunotherapies2. The US Food and Drug Administration (FDA) recently approved the use of immune checkpoint inhibitors (ICIs), such as anti-PD-1 or PD-L1 monoclonal antibodies (mAb), as third-line treatments against advanced cancers of different types, including gastric cancer3,4. Despite the celebrated successes of cancer immunotherapy, only a small subset of patients still benefits, while the differences in clinical response even in tumors having similar histopathological types warrant the development of better biomarkers and means for patient stratification5.
During tumor progression, cell populations with immune evasion properties, sometimes acquired through mutations, are clonally expanded6. The importance of coding germline and somatic mutations as drivers of immune escape has been extensively documented and is a domain of intense research7. However, less is known about the contribution of non-coding variants to immune evasion and tumorigenesis8.
Messenger RNA 3′ Untranslated Regions (3′-UTRs) are primary sites for post-transcriptional regulatory events9. These processes account for ~60% of the variation in protein expression, while ~20% of germline expression quantitative trait loci (eQTLs) are located in 3′-UTRs9, which are more conserved than other noncoding loci, suggesting selective pressure10. 3′-UTRs are the most common targets of key regulatory molecules such as RNA binding proteins (RBPs) and microRNAs (miRNAs).
miRNAs are potent post-transcriptional regulators and are implicated in the control of numerous cellular mechanisms11 as well as of all cancer hallmarks12, hence their role in cancer immune surveillance has become a research hotspot12. miRNAs have been found to efficiently regulate Programmed death ligand 1 (PD-L1), other B7 family members, cytokines and numerous immune genes13,14. On the other hand, RBPs have been shown to regulate mRNA processing, localization, interactions, and stability15, while different RBPs such as Mex3B16, Mex3C17, and HNRNPR18 have been shown to regulate key antigen presentation mechanisms.
However, tumors mutate, truncate, or edit their 3’UTRs to escape this tight regulatory control13,19,20,21,22,23. Unfortunately, the current reliance of variant-calling pipelines on whole exome sequencing (WES) data, which do not include probes for 3′-UTR regions, has resulted in a lack of understanding of the role of 3′-UTR variants in cancer progression. Small-scale, targeted studies have identified individual 3′-UTR somatic mutations that associate with changes in cis-gene expression and immune phenotypes, especially for PD-L124,25,26. For instance, a common somatic mutation in the PD-L1 3′-UTR has been shown to disrupt miR-570 binding leading to increased expression26.
RNA sequencing (RNAseq) has been shown to be an alternative variant detection source27. The Pan-Cancer Analysis of Whole Genomes (PCAWG) database contains the largest collection of cancer patient samples with whole genome sequencing (WGS) data to date28. In PCAWG (n = 1188), out of the 87 samples without a driver alteration identified at the DNA level and available RNAseq data, every sample had an RNA-level alteration identified; indicating that driver alterations could have revealed themselves in RNA, rather than DNA29. Until today, no study has leveraged the rich transcriptomic data from The Cancer Genome Atlas (TCGA) to identify mutations across this vast resource.
Here, we perform a comprehensive mutational analysis on raw RNAseq data from hundreds of stomach adenocarcinoma (STAD) samples in TCGA to identify 3′-UTR germline and somatic single-nucleotide variants (SNVs) as well as short insertions-deletions (indels)30. By performing a quantitative trait loci (QTL) analysis, we identify cis-acting gene expression QTLs (cis-eQTLs), as well as variants associated with changes in immune phenotypes, herein termed immune landscape QTLs (ilQTLs). We design and implement a massively parallel reporter assay (MPRA) to validate at scale cis-eQTLs in immune-related genes directly affecting post-transcriptional stability and abundance of respective genes. MPRAs have been utilized successfully in the past for the functional validation of non-coding variants, such as promoter and UTR germline variants31, while this assay is specifically enriched in somatic 3′-UTR mutations. We also investigate the translational potential of the identified 3′-UTR variants and specifically their ability to predict outcomes across diverse cohorts of ICI. Utilizing the prioritized ilQTLs, we establish a polygenic risk score (PRS) that proves more accurate in predicting response to checkpoint inhibition in melanoma and gastric cancer patients than PD-L1 expression, providing direct support of the potential utility of UTR variants in predictive modeling in immunotherapy. In this work, we establish the tools and apply them to unbiasedly identify transcriptome-wide 3′-UTR variants associated with changes in cis-gene expression and immune phenotypes in cancer and lay the foundations for similar 3′-UTR-focused studies in other cancer types.
Results
Characterization of the somatic and germline 3′-UTR variant landscape in gastric adenocarcinoma
Apart from the 1188 PCAWG samples overlapping with TCGA, WGS data are not available for ~90% of TCGA subjects, limiting large scale 3′-UTR variant investigations. Specifically for gastric cancer, only 40 samples comprise WGS data. The only study to date which attempted to analyze 3′-UTR variants in TCGA in non-WGS samples32, mistakenly considered that 3′-UTR regions were captured in the WES probe sets used in the study30,33. We now know that these regions are not covered in the TCGA WES kits30, with only 0.31% of 3’UTR regions being targeted in the TCGA STAD cohort.
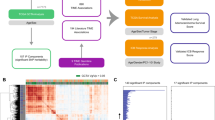
To call variants, we utilized RNAseq data, which has been shown to be a powerful modality for such analyses27,28. Sequencing data from 375 gastric cancer patients (Supplementary Data 1), including 375 primary gastric cancer samples and 40 matched controls, were analyzed following a comprehensive approach using GATK best practices34 (Fig. 1A) and led to the identification of thousands of expressed variants and indels per sample (Supplementary Fig. 1A, B). Analysis of the distribution of called variants along the length of the 3′-UTR revealed that the RNAseq-derived calls matched the distribution of WGS calls from PCAWG (Fig. 1B). As expected, analysis of the distribution of TCGA WES-derived variants along the length of the 3′-UTR showed that most variants fall in the beginning of the 3′-UTR, proximal to coding sequences, likely representing sequences captured by coding region probes (Fig. 1B). The majority of 3′-UTR variants distally to the stop codon are missed by the WES-based variant calling analysis, due to lack of targeting probes.
A Flowchart showing the variant-calling pipeline used for the analysis of TCGA STAD raw RNAseq data. B Distribution of the relative position of single nucleotide variant calls along the 3′-UTR as identified by analysis of TCGA WES, RNAseq and WGS data. C Number of RNAseq-derived somatic 3′-UTR single nucleotide variants per sample, grouped by “Mutation Rate Category” as defined by TCGA WES variant-calling data. D Overlap between transcriptome-wide variant calls in the analysis of RNAseq data with GATK and Strelka2 (GATK = 5,431,118, Strelka2 = 10,175,223, Overlap = 4,692,062). Figure data are provided in the Source Data file.
Out of 5,431,118 variants identified across the genome by the RNAseq GATK analysis post-filtering, 3,283,340 (60.5%) overlapped with germline variant calls identified from blood samples from the same patients. The remaining 2,147,778 variants (39.5%) were treated as “likely somatic” calls. Of the likely somatic calls, 1,429,039 variants (66.5%) intersected with common RNA editing events identified in the GTEx database35. Samples deemed as ultramutated by TCGA, based on WES-derived variant calls, exhibited high frequency of RNAseq-derived somatic SNVs (Fig. 1C). We also reanalyzed all TCGA STAD RNAseq samples using Strelka236, an orthogonal variant calling algorithm, and identified a ~90% concordance (Fig. 1D).
High-throughput capture of known functional variants in PD-L1 3′-UTR
We initially evaluated whether our high-throughput approach could capture the functional impact of the small number of 3′-UTR SNVs that have been previously associated with changes in PD-L1 expression in gastric cancer, such as the polymorphisms rs229713625 and rs414381537, or non-small cell lung cancer, such as rs474209838. Indeed, our analysis identified all three variants and when comparing PD-L1 expression in samples with or without the 3′-UTR variants, we observed significantly increased expression levels in patients carrying the alternative allele, as expected based on the literature (Fig. 2).
Normalized PD-L1 expression (log2(read count+1)) in patients carrying the alternative allele (Alt) vs patients homozygous for the reference allele (Ref). For all three SNPs studied, Alt patients show higher levels of PD-L1 expression than Ref patients. Boxplot lines represent the median and upper or lower quartiles, while whiskers define the 1.5x interquartile range. Significance was assessed by two-tailed Student’s t test. rs2297136 (nAlt = 236, nRef = 139, p = 1.42e-8), rs4143815 (nAlt = 163, nRef = 212, p = 1.01e−4), rs4742098 (nAlt = 101, nRef = 274, p = 0.00173). Figure data are provided in the Source Data file. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Prioritization of 3′-UTR germline variants and somatic mutations controlling cis gene expression in gastric adenocarcinoma
We prioritized transcriptome-wide 3′-UTR variants associated with cis-gene expression changes in gastric cancer. We investigated variants that were present in 5 or more samples, corresponding to 1.3% or higher minor allele frequency (MAF) in the tested population (2,917,776 total variants, 68,4%/1,994,516 germline, 31.6%/923,260 somatic, in which 67.2%/620,419 overlapping with editing sites). We performed an eQTL analysis39 with the dosage of each variant as the genotype variable and the inverse quantile-normalized expression of the corresponding gene as the phenotype variable. To remove unwanted variation from the model, we used sex, age, along with the top 5 genetic principal components (PCs, Supplementary Fig. 2A) and expression surrogate variables (SVs)40 as covariates (a simplified version of the formula is captured in Fig. 3A).
A Model used for eQTL analysis. B Waffle plot showing the percentage of eQTLs that mapped to each of the indicated gene biotypes. A waffle plot consists of 100 squares and the number of colored squares represents the percentage of eQTLs mapping to each gene biotype. C Barplot showing the number of protein-coding cis-eQTLs that reside in each genic region (5′UTR, CDS or 3′-UTR). D Number of 3′-UTR cis-eQTLs that overlap with miRNA or RBP binding sites in the indicated databases. TarBase is a database of experimentally validated miRNA binding sites, microT includes predicted miRNA binding sites and POSTAR2 contains predicted RBP binding sites based on CLIP experimental data. E KEGG pathway enrichment analysis of the 500 topmost significant 3′-UTR cis-eGenes. The X axis shows the number of eGenes that belong to the indicated pathway and the color (legend) represents the FDR-adjusted one-sided Fisher’s exact test p value for the enrichment of each gene-set. Figure data are provided in the Source Data file.
With a cutoff of a nominal p value of 1e-7, we identified ~3000 cis-eQTLs in protein-coding genes, accounting for 75% of all eQTLs (Fig. 3B). Out of the 3133 exonic (CDS/UTR) variants in protein coding genes, 1845/58.9% were germline, and 1288/41.1% were somatic (254 overlapping with editing sites). 27.6% of the identified eQTLs overlapped with eQTLs from the Genotype-Tissue Expression project (GTEx), identified using healthy stomach tissue samples (90.6% overlapping germline eQTLs). Interestingly, we found that the highest percentage (60%) of all significant cis-eQTLs in protein-coding genes reside in the 3′-UTR genic region (Fig. 3C), reflecting the importance of 3′-UTR cis-acting elements in controlling gene expression, often post-transcriptionally41. Comparing the number of 3′-UTR variants to the average relative length of the 3′-UTR in protein coding genes42 revealed a significant enrichment (Supplementary Fig. 2B, C) (chi-square p value < 1e-5). Enrichment of eQTLs in the 3′-UTR among exonic sequences has also been reported in other studies, including analyses of normal-tissue eQTL variants from GTEx43,44. Somatic eGenes, where the lead eQTL was somatic, included important gastric cancer oncogenes, such as KRAS, CCDN1, and CCND2 among others45, genes involved in antigen generation, processing, or presentation (e.g., APOBEC3B, CANX, CTSS), and cytokines/chemokines or other key immune regulators (e.g., STAT1, CXCL5, CXCL9, TNFRSF9). Using cell compartment marker sets derived from a gastric cancer single cell atlas46, we observed that the eQTLs of somatic-only variants were enriched in tumor/epithelial cell markers (all somatic: Storey’s q value = 1.19e-13, somatic-only: Storey’s q value = 5.72e-8). This enrichment was diminished for germline-only eQTLs (Storey’s q value = 0.025).
Overlapping the 3′-UTR cis-eQTLs with databases of predicted and experimentally supported miRNA and RBP binding sites showed that around 90% of the variants reside in functionally relevant regulatory elements (Fig. 3D)11,47,48,49. However, by performing a permutation-based test against randomly sampled 3’UTR regions, only predicted miRNA binding sites and not RBPs were identified as significantly enriched in the eQTL loci (p = 0.0002). Finally, gene-set enrichment analysis (GSEA) of the top significant 3′-UTR cis-eQTLs revealed enrichment in immune-related pathways (Fig. 3E), indicating that 3′-UTR variants could have an impact on immune phenotypes in gastric cancer.
Massively parallel reporter assay validation of cis-eQTLs residing in cancer immunoediting genes
Considering the significant enrichment of immune-related pathways in the top 3′-UTR cis-eQTLs as well as the importance of immune escape in cancer progression and as a target for novel therapeutics, we proceeded with a functional validation of prioritized 3′-UTR eQTLs on a compiled list of immune-related genes (Supplementary Data 2, Methods). The manually curated list incorporates immune checkpoint and known or suspected immunomodulatory genes, MHC machinery, genes used in signatures for response to immunotherapy50,51,52,53, and significant hits from hypothesis-free CRISPR–Cas9 screens for CD8+ T-Cell effector function54,55 and in vivo screening of transplantable tumors in mice treated with ICI56.
We selected the top 749 variants (478 somatic, 135 potential editing sites) that resided in 299 prioritized genes based on the curated list described above (Supplementary Data 3). We developed and performed a massive parallel reporter assay (MPRA) in two gastric cancer cell lines to assess the effect of each variant on the post-transcriptional stability of a reporter gene (Fig. 4A, Methods). Briefly, a reporter plasmid library containing barcoded reference and alternative alleles for the 749 eQTL variants was transfected into AGS and SNU719 cells. The effect of the variant on post-transcriptional expression of the reporter was assessed by barcode quantification from amplicon sequencing of RNA extracted from transfected cells.
A Protocol followed for 3′-UTR MPRA assay. Ref: Reference allele, Alt. allele: Alternative allele, Syn. pA terminator: synthetic polyadenylation terminator, oligodT: primer containing 20 T nucleotides. Created with Biorender.com. B Manhattan plot showing the top hits from the MPRA assay in both gastric cancer cell lines. A two-sided Wilcoxon rank sum test was used to calculate p value of enrichment of alternative vs reference alleles and multiple comparison adjustment was performed by FDR. The black line (p. adj. = 0.05) separates significant from non-significant calls. Blue dots represent likely germline variants, while red dots represent likely somatic variants. Figure data are provided in the Source Data file.
The two cell lines used in the MPRA assay yielded similar outcomes (Supplementary Fig. 3A, B). Approximately 15% of eQTLs (128 variants) showed a significant causative effect on the expression of the reporter (FDR-adjusted p value < 0.05) in at least one of the two cell lines (Supplementary Data 4). A subset of genes with causal regulatory variants (Fig. 4B, Supplementary Data 4) are involved in antigen processing and presentation (e.g., HLA genes, CTSB, CTSS, LGMN, CIITA, TAPBP) as well as RNA-editing enzymes such as ADAR.
Uncovering transcriptome-wide 3′-UTR variants regulating the gastric adenocarcinoma immune landscape
3′-UTR regions not only regulate gene expression, but can also influence mRNA localization, protein-protein interactions and other post-transcriptional, translational and post-translational functions41. Therefore, 3′-UTR variants can affect immune phenotypes in cancer independently of their effect on expression. To unbiasedly associate 3′-UTR variants with changes in the immune landscape of gastric cancer, we performed an immune landscape (il)QTL analysis using a similar model as above (Fig. 3A), focusing on immune phenotypes instead of cis-gene expression as the dependent variable. Immune phenotypes for the TCGA STAD cohort were obtained from the Cancer Research Institute (CRI) iAtlas project and included expression-based immune cell infiltration estimates, TCR/BCR entropy and leukocyte ratio calculated by combined imaging, methylation, and expression-based analyses57.
A total of 1715 ilQTLs were identified, with 159 (9.3%) identified as germline. Among the remaining 90.7%/1,556 “likely somatic” ilQTLs, 370 intersected with common RNA editing events. For almost all immune features, the majority of ilQTLs resided in the 3′-UTR region of protein-coding genes and a large percentage of those were predicted to reside in regions of miRNA/RBP binding (Fig. 5), similarly to eQTLs. By performing the permutation analysis for CD8+ T cell ilQTLs, as in eQTLs, they were found to be enriched in predicted (p = 0.002), and experimentally validated (p = 0.005) miRNA binding sites as well as in experimentally supported RBP binding loci (p = 0.002). CD8+ T cell ilQTLs were also significantly more frequent to be of somatic origin, as compared to germline (Somatic CD8+ T cell ilQTLs: 95.58%, 2.3e-165, two-sided Fisher’s exact test). Significant ilQTLs, the majority of which of somatic origin, were identified in immune-relevant genes, including B2M, HLA genes, CANX, LDHA, PSMB2, and HNRNPR, which are known to affect the tumor immune landscape5,17,58,59,60,61. The top hits also included WARS62 and APOBEC3C63, which were only recently implicated in tumor immunity, showing the potential of this approach for prioritization of novel cancer-specific immunotherapeutic targets.
Left-hand side of dashed black line: distribution of significant ilQTLs along genic regions (5′UTR, CDS or 3′-UTR), showing enrichment in 3′-UTR variants. Right-hand side of dashed line: overlap of 3′-UTR ilQTLs with databases for miRNA response elements (mRE) and RBP binding sites. Figure data are provided in the Source Data file.
We focused on CD8+ T cell ilQTLs since the level of CD8+ T cell infiltration in a tumor is an important determinant of cancer immunotherapy response64. We identified significant CD8+ T cell fraction QTL variants in 467 genes. GSEA analysis showed that the most enriched “cellular component” gene sets in the topmost significant CD8+ T cell infiltration QTL variants are the ribosome and ribosomal subunits (Supplementary Fig. 4). In addition, immunoregulatory CRISPR hits (Supplementary Data 2) were enriched in significant CD8+ T cell ilQTL 3′-UTR variants compared to all 3′-UTR variants (1.42-fold increase, one-sided Fisher’s exact test p value = 0.0057), showing concordance between the two orthogonal approaches of key gene prioritization. In addition, only 5% of those ilQTLs overlapped with eQTLs, showing the ability of ilQTLs to capture associations beyond gene expression regulation.
Functional 3′-UTR cis-eQTL and ilQTL variants in ADAR
Significant functional variants were identified in all our high throughput investigations (eQTLs, ilQTLs, MPRA validated variants) in the Adenosine Deaminase RNA Specific (ADAR) gene. ADAR encodes an enzyme that catalyzes A-to-I editing in RNA and has been implicated in promoting cancer hallmarks in multiple cancer types, including breast, thyroid and gastric malignancies65,66,67. In the CRISPR screen by Manguso et al.56 for genes that sensitize tumors to immunotherapy in mouse models, ADAR is the 4th most enriched out of ~20,000 genes. ADAR downregulation has also been shown to induce inflammatory signaling in gastric cancer specifically68. To investigate the immunoediting role of ADAR in gastric cancer further, we queried the iAtlas portal57, where CNVs on ADAR were reported to exhibit high effect sizes on Leukocyte Fraction (Amp: p = 10−4 to 10−10 (multiple groups)), Lymphocyte Infiltration Score, (Amp: p = 10−6/Del: p = 10−5), and CD8+ T Cell content (Del: p = 10−4). Moreover, ADAR harbored multiple 3′-UTR eQTL and ilQTL variants in gastric cancer (Fig. 6A), while we also found ADAR to be significantly overexpressed in responders (n = 55) compared to non-responders (n = 80) to immune-checkpoint inhibitors, in a cohort consisting of gastric cancer and melanoma patients (Fig. 6B)50,51,69,70. By meta-analyzing additional studies through the tumor immunotherapy gene expression resource (TIGER), we saw that ADAR1 overexpression is a common feature for response to checkpoint inhibition (Supplementary Fig. 5). On the other hand, Manguso and colleagues showed that loss of ADAR1 overcomes resistance to PD-1 checkpoint blockade caused by inactivation of antigen presentation by tumor cells in mouse models of resistance59.
A Plot showing the distribution of CDS and 3′-UTR eQTLs and ilQTLs for the ADAR gene. The y-axis represents the nominal eQTL p value for each variant. The exon where the 3′-UTR is found is colored orange. B A cohort of gastric cancer and melanoma patients was classified into responders (R, n = 80) vs non-responders (NR, n = 55) according to response efficacy with anti-PD-1 immunotherapy. Primary cancer RNAseq data from these patients were analyzed. Differential gene expression analysis revealed increased expression of ADAR in R compared to NR (two-sided Wilcoxon rank sum test, Benjamini Hochberg FDR correction). Boxplot lines represent the median and upper or lower quartiles, while upper whiskers represent the max and min. Gastric and Melanoma cancer types are colored with grey and red, respectively. C Correlation analysis between ADAR and TARDBP log2 TPM normalized expression performed using TIMER v2.0 (timer.cistrome.org). Data from 415 STAD patients are included and the Spearman’s correlation coefficient (rho) and p value are reported. A linear regression line is shown in blue; the gray shaded area represents the standard error of the regression. Figure data are provided in the Source Data file.
Through our QTL and MPRA analysis, we identified a somatic 3′-UTR cis-eQTL variant (chr1:154583325, T-to-C) in ADAR (Fig. 6A) with causal effects on post-transcriptional regulation (Supplementary Fig. 6A, B). ADAR variants including (chr1:154583325, T-to-C) were also found as ilQTLs for multiple immune features (Fig. 6A). Based on a meta-analysis of RNA cross-linking and immunoprecipitation (CLIP) data from the POSTAR2 project49, this ADAR variant is predicted to overlap with multiple RBP (Supplementary Table 1) and miRNA (Supplementary Table 2) binding sites. One of those RBPs, TARDBP (Supplementary Table 3), has been studied for its ability to regulate gene expression, pre-mRNA editing, mRNA localization, and microRNA processing through binding on canonical GU-rich motifs or non-canonical sequences, with 3’UTRs being commonly targeted regions71. TARDBP has been shown to directly regulate ADAR1 expression in liver cancer and leukemia cell line models72. Indeed, correlation analysis in gastric cancer patients from TCGA revealed a strong association in the expression of the two genes (Fig. 6C), suggesting that the regulation axis between TARDBP and ADAR could be functional in gastric cancer as well.
3′-UTR variants as an effective means for immunotherapy response prediction
We next sought to investigate the translational potential of 3′-UTR ilQTL variants and their ability to predict therapeutic outcomes to cancer immunotherapy. To this end, we analyzed the cohort of responders (R) vs non-responders (NR) to ICIs described above50,51,69,70, where tumors were subjected to whole transcriptome sequencing thus enabling the detection of 3′-UTR variants.
We separated the samples into a training (n = 68, 39.7% Responders) and a test (n = 67, 40% Responders) set and selected 28 TCGA 3′-UTR ilQTL variants that were enriched in the R vs NR samples of the training set (one-sided Fisher’s exact test q value < 0.05, Supplementary Table 4). The variants were used to devise a Polygenic Risk Score (PRS) for the potential prediction of response to ICI (Methods). The orthogonal test subset was completely insulated from both the variant selection (TCGA STAD) and directionality (training set). When the selected variants were tested on the orthogonal test set (n = 67), the PRS was significantly increased in the responders (Wilcoxon rank sum test, p value = 0.00071, Fig. 7A), and exhibited a higher area under the receiver operating characteristic curve (AUC, ROC) than PD-L1 expression (Fig. 7B), as well as against tumor mutational burden (TMB) or microsatellite instability (MSI) as calculated from WES data (Supplementary Fig. 7). Importantly, the information captured by the PRS score is a predictor independent of PD-L1 expression, and their combination, as well as potentially the integration of the expression or mutational status of additional genes, can be leveraged to further increase the prognostic accuracy of the model (Supplementary Fig. 8). The ilQTL PRS also shows higher generalizability in the test set as compared to standard PRS models generated with the top 3′-UTR or genic variants selected for their ability to predict response in the training set, showing the translational potential of 3′-UTR variants prioritized through the ilQTL analysis in a large discovery cohort (Supplementary Fig. 9). This proof-of-concept application showcases that non-coding variants can be used to predict immunotherapy treatment outcomes in cancer.
A Comparison of the polygenic risk score (PRS) distribution in the non-responder (NR, n = 40) and responder (R, n = 27) groups of the testing population (two-sided, Wilcoxon rank sum test, Benjamini–Hochberg FDR correction). Boxplot lines represent the median and upper or lower quartiles, while upper whiskers represent the max and min. Gastric and Melanoma cancer types are colored with grey and red, respectively. B A receiver operator characteristic (ROC) curve showing the ability of the PRS score and PD-L1 expression classifications to distinguish between R and NR patients in the testing population. An Area Under the Curve (AUC) score is reported for both classifiers. Figure data are provided in the Source Data file.
Discussion
The increasing ease and lower cost of deep genome sequencing technologies will eventually allow the unbiased identification of non-coding variants with high confidence through the analysis of WGS data. However, the small number of currently available cancer samples with combined WGS and RNAseq data prohibits the use of WGS variant calling data for analyses that require high statistical power, such as QTL analysis. By repurposing RNAseq data available for hundreds of gastric adenocarcinoma patients in TCGA30, we deeply investigated 3′-UTR somatic and germline variants in cancer. The use of RNAseq data allowed the identification of variants not only in the coding region of genes, but also in 5′UTR /3′-UTR sequences and non-coding genes like long non-coding RNAs (lncRNAs), which are omitted from standard WES assays, such as those performed for TCGA. To our knowledge, the only study that attempted to characterize 3′-UTR somatic variants from TCGA transcriptome-wide was by Wu et al.32. However, they mistakenly considered that the exome capture used in TCGA was the Illumina TruSeq Exome Enrichment Kit, which also targets 3′-UTRs, instead of the actual assays performed, where UTRs are not included and low coverage statistics are reported if the UTR regions are considered in the metrics33. Our analysis shows that 3′-UTR somatic variants are spread across the 3′-UTR regions as also supported by WGS assays (Fig. 1B) and not proximally to the CDS as previously reported32, which was evidently due the TCGA WES not targeting 3′-UTR regions.
Despite the success of RNAseq data capturing variants in regions not covered by standard WES assays, the ability of this approach is confined to the expression space of the tumor of interest, since low (or no) expression directly inhibits variant calling. To address specificity concerns while maintaining high sensitivity, we followed GATK best practices for RNAseq data by deduplicating identical reads and by following through with variants identified in at least 1.3% of the population tested.
Our study reveals the importance of 3′-UTR variants in driving cis-gene expression in cancer and provides a framework for incorporating 3′-UTR variant-calling data in TCGA and other cohorts. We prioritized functional variants by performing a transcriptome-wide cis-eQTL analysis in the TCGA STAD cohort and identified significant variants across 1117 eGenes. 27.6% of the identified eQTLs overlapped with eQTLs identified from healthy stomach tissue from the GTEx consortium, with 90.6% of the overlapping significant variants being germline. This points not only to the difference between germline variants and somatic mutations but mostly to the regulatory divide between healthy and neoplastic tissue. A recent study comparing eQTLs generated with different combinations of germline/tumor variants and healthy/neoplastic tissue gene expression, concluded that the variation in the eQTLs they observed could be almost entirely attributed to the difference in the source material; highlighting further the genomic, epigenetic, transcriptomic, and regulatory differences observed between healthy and neoplastic tissue73. Around 90% of the 3′-UTR eQTLs overlap with putative or experimentally-supported miRNA and RBP binding sites, providing a potential functional relevance for those variants, with only 1.6% and 1.83% of somatic and germline variants colocalizing on the same microRNA and RBP binding site, respectively. The enrichment in immune-related pathways in the topmost significant 3′-UTR cis-eQTLs indicates the importance of 3′-UTR variants in controlling cancer immunogenicity. Since 3′-UTR regulatory roles go beyond post-transcriptional gene expression regulation and include localization, translation rate control, and even protein-protein interactions and liquid organelle formation41, we performed a transcriptome-wide ilQTL analysis. In this analysis as well, the majority of significant variants resided in 3′-UTR regions. This investigation shows that 3′-UTR variants can be associated with immune phenotype changes in an unbiased hypothesis-free manner. We discovered significant 3′-UTR ilQTL variants in widely studied immunoregulatory genes, such as ADAR and STAT1. One of the ADAR ilQTLs overlapped with multiple RNA binding sites, including TARDBP, an RNA binding protein and known regulator of ADAR expression in liver cancer and leukemia.
In addition to validating previously described 3′-UTR eQTLs, our approach also identified 3′-UTR variants and genes that have not been previously linked to immune-related functions in cancer. We utilized a massively parallel reporter assay (MPRA) to streamline validation across hundreds of candidate 3’UTR variants, with 15% exhibiting functional effects, even though the eQTLs were detected in patient samples and the MPRA assay was performed in gastric cancer cell lines, where the relevant RNA binding proteins, microRNAs and their targets might not exhibit conserved stoichiometry. The validation rate is comparable to the MPRA assay performed in Griesemer et al.43. Interestingly, among the significant hits from the MPRA assay, there were multiple genes encoding ribosomal subunits. Pathway analysis in the top significant CD8+ T cell infiltration QTL variants revealed an overall enrichment in ribosome-related proteins (Supplementary Fig. 4). Previous work has shown that changing the expression of ribosomal subunits can affect MHC class I presentation efficiency and the antigenic profile of a cell in the context of Influenza A virus infection, without altering translation efficiency74, raising the question whether alteration of ribosomal proteins could have a similar phenotypic effect in cancer.
Finally, to investigate the clinical relevance of our ilQTL analysis, we showed that non-coding 3′-UTR ilQTL variants can predict response rates to immunotherapy treatments (Fig. 7A). In this study, we exploit the space of non-coding variants and show that a signature of ilQTL variants has stronger predictive power for drug response than PD-L1 expression in a cohort of melanoma and gastric cancer patients (Fig. 7B). This is a direct application of non-coding mutations to predict response to immunotherapy, providing a potentially strong incentive to start including these important regulatory regions in WES investigations while the community waits to horizontally adopt WGS in somatic samples.
Methods
Cell culture
The AGS (ATCC CRL-1739) cell line was purchased from ATCC and the SNU-719 (KCLB-00719) cell line was purchased from the Korean Cell Line Bank (KCLB). AGS and SNU719 cells were maintained in RPMI-1640 (ThermoFisher Scientific, #11875093) with 10% FBS (ThermoFisher Scientific, A5256801). All cell lines were incubated at 37°C and 5% CO2. The cell lines were verified by the vendors with STR profiling and they were tested for mycoplasma contamination at regular intervals using the MycoAlert Mycoplasma Detection Kit (Lonza, #LT07-318).
RNAseq variant calling
Raw RNAseq data for 375 TCGA STAD primary cancer and 40 matched normal samples were obtained from Genomic Data Commons (GDC) following NIH dbGAP approval75. Reads were mapped against the human genome (hg38) using STAR76. Mapped reads were deduplicated and short variants/indels were called using Mutect2 following GATK (v4.1.4.0) best practices34. The Mutect2 output was converted to a gVCF format by using region coverage statistics. Since Mutect2 cannot perform genotype calling and does not distinguish between homozygous reference and no-call regions, HaplotypeCaller was also run in parallel by following GATK best practices for RNA77. For samples lacking a Mutect2 call at a specific variant position, HaplotypeCaller was used to distinguish whether the lack of a Mutect2 call was because of no coverage in that region or a homozygous reference genotype. A mutation was characterized as likely somatic by calculating the posterior probability of the event, while using variant call statistics, clonality in tumor samples, matched healthy tissues, and gnomAD variants78 as priors. Calls were filtered using the FilterMutect2 tags “base_qual”, “map_qual”, “n_ratio” and “slippage”. Only biallelic variants present in at least 5 out of the 375 (>1.3%) samples were pursued further. Variants that were present in at least 2 out of the 40 matched normal samples comprised the Panel of Normals (PoN). Strelka236 was also run following the same preprocessing steps as for GATK callers.
Germline variant calling
Affymetrix SNP array 6.0 data from blood samples for all TCGA STAD patients analyzed in this study were downloaded from GDC. The SNP array data were converted to a VCF format using birdseed2vcf (https://github.com/ding-lab/birdseed2vcf), and then whole-genome variant calls were imputed using Minimac4 and the 1000 Genomes Phase 3 project as a reference on the Michigan Imputation Server79. Somatic variants were further intersected against GTEx RNA editing events from REDIportal V2.035.
eQTL analysis
Gene-level expression in TCGA STAD samples was calculated using Salmon v0.91 and Ensembl genome annotation v7780,81. A linear model was utilized to call eQTLs with FastQTL39 following best practices82. In brief, gene expression across libraries was normalized using trimmed mean of m-values as implemented in edgeR83. Genes were selected based on an expression threshold of 1 read in at least 80% of the samples. An inverse quantile normal transformation was performed on the expression values prior to their inclusion into the linear model. Mutect2/HaplotypeCaller alternative allele dosage was utilized as genotype input, while age at diagnosis, sex and the top 5 genetic principal components (gPCs) and expression surrogate variables (SVs) were included as covariates39,84, as follows: gene expression ~Alt Dosage+age+sex+gPC1+gPC2+gPC3+gPC4+gPC5+SV1+SV2+SV3+SV4+SV5. Genetic PCs were calculated using SmartPCA85 on WES-derived germline variants from the same TCGA STAD patients, obtained from Huang et al.86.
Following the nominal run of FastQTL39, for each gene we selected variants that mapped in cis (within the genomic coordinates of the gene) and calculated a per-gene FDR-adjusted q value for each variant. Genes that contained at least one variant with a q value lower than 0.05 were defined as eGenes. A nominal threshold was defined for each eGene based on the highest nominal p value that corresponded to a q value < 0.05. The distribution of the threshold nominal p value in all eGenes is shown in Supplementary Fig. 10. The median threshold nominal p value (1e-7) was applied horizontally, across all eGenes, to describe significant calls (final cutoff: q value < 0.05 and nominal p value < 1e-7). eQTL calls were mapped to transcript annotations (Gencode v32) and relative genomic locations (5′UTR, CDS, 3′-UTR) were assigned using annotatr87. Only eQTLs with significant cis effects were retained for further analysis. Variant annotation for potential overlap with post-transcriptional regulatory regions was performed using the GenomicRanges package in R (v1.38.0)88. Experimentally-supported miRNA binding site coordinates were obtained from TarBase47, predicted miRNA binding sites were acquired from microT-CDS48, and CLIP-based predictions of RBP binding sites were obtained from the POSTAR2 database49.
ilQTL analysis
Immune data per sample were obtained from the CRI iAtlas project57. QTL analysis was performed with FastQTL, using the same genotype and covariate data as above, while using quantile-normalized immune profile estimates as phenotypes. ilQTL selection as well as genomic and regulatory annotation were performed as for eQTLs.
Enrichment and over-representation analyses
For pathway enrichment/over-representation analyses, the top 500 eGenes, ranked based on their lowest nominal p value, were investigated by pathway enrichment analysis using ClusterProfiler (v3.12.0)89. Pathway information was obtained from the Gene Ontology Resource90 and the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway database91. Plots were generated in R using ggplot2 (v3.3.3). One-sided Fisher’s exact test was utilized to evaluate cell type-specific enrichment of eQTL genes against the reference marker gene sets from the gastric cancer single-cell atlas established by Sun et al.46, corresponding to individual cell types or subtypes.
Locus permutation analyses were performed with RegioneR92 utilizing a resampling (n = 5000) permutation test. As a sampling space, all 3’UTR regions were split into 50 bp segments using a walking window of step = 1 (n = 39,142,309 windows). All evaluated regulatory regions from microT, TarBase, and POSTAR were included in the sampling space.
Cancer immunology-related genes
A collection of more than 2500 immune-related genes was manually curated from the literature and experimental resources (Supplementary Data 2). Specifically, the list includes immune checkpoint and immunomodulatory genes, genes involved in the MHC machinery and microsatellite instability, cytokines and chemokines93,94,95, gene markers for metabolic reprogramming93,96 and oncogenes or complexes that can affect the tumor transcriptional and immune landscape, such as EZH2–PRC2 chromatin remodeling complex members and BAF/PBAF complex members97,98. We also incorporated significant genes from hypothesis-free CRISPR–Cas9 screens for CD8+ T-Cell effector function54,55 and in vivo screening of transplantable tumors in mice treated with immunotherapy56. In addition, we included the Urea cycle (GO:0000050) and Mismatch repair (GO:0006298) Gene Ontology terms, the list of Human DNA repair genes from Lange et al.99, and the following entries from the Kyoto Encyclopedia of Genes and Genomes (KEGG): MAPK signaling pathway (hsa04010), PI3K-Akt signaling pathway (hsa04151), Wnt signaling pathway (hsa04310), JAK-STAT signaling pathway (hsa04630), Antigen processing and presentation (hsa04612). Finally, the list comprises more than 250 genes from signatures associated with response to ICI50,51,52,53.
MPRA pool design
A MPRA was performed with eQTL variants in eGenes included within the curated cancer-immunology gene list that had a nominal p-value lower than 1e-5 (Supplementary Data 3). The nominal p value threshold was higher than that for other downstream eQTL analyses to allow the capture of causative effects by less common variants, such as rare somatic eQTLs. A pool of 19220 150 bp-long oligonucleotides (oligos) was synthesized commercially (Twist Biosciences). Each oligo contained the reference or alternative allele of the eQTL variant in the middle flanked by 50 bp of reference transcriptomic sequence on either side. In the case of eQTLs residing in alternative transcript isoforms, oligos were synthesized for all possible transcripts. For eQTLs that were close to the end of the transcript a random sequence was added to bring up the length of the sequence to 101 bp. The random sequence was the same in all oligos and was selected to lack any predicted 7 or 8 bp-long RBP and miRNA-seed binding sites.
An 8 bp-long barcode was added at the 3′ end of the 101 bp-long sequence. Each allele was represented by 10 unique barcodes. All 8 bp barcodes that matched RBP or miRNA seed binding sites were removed100,101. Finally, 20 bp sequences were added on either side of the oligo that matched the 5′ and 3′-end sequences of the XhoI-/NotI-digested psicheck2 vector (Promega, C8021) to allow cloning with the NEBuilder HiFi DNA Assembly kit (NEB, E2621S).
Pool amplification and cloning
Amplification of the pool, prior to cloning, was performed using 0.5 μM of each of the PCR_lib_fwd and PCR_lib_rev primer pair (Supplementary Data 5) with the NEB Next High-Fidelity 2x PCR Master Mix (NEB, M0541L). The following PCR conditions were used: 98 °C for 30 sec, 20 cycles (98 °C for 10 sec, 63 °C for 10 sec, 72 °C for 15 sec), 72 °C for 2 min. The amplified oligo pool was introduced into a XhoI-/NotI-digested psicheck2 vector using NEBuilder HiFi DNA Assembly kit (NEB, E2621S), as per the manufacturer’s protocol. The assembly reaction product was purified following a standard isopropanol precipitation protocol, as described in Joung et al.102. The purified plasmid pool was transformed into Endura ElectroCompetent cells (Lucigen, #60242-1) at 50 ng plasmid per 25 μl of bacteria ratio, following the provider’s protocol. A total of 8 transformation reactions were pooled together and plated onto large 15 cm LB Agar plates at 37 °C for 12 h. A large enough number of colonies to ensure at least 500 colonies/oligo representation was harvested directly from the LB Agar plates and the plasmid pool was purified by performing at least 2 midipreps per 15 cm LB Agar plate, using the Qiagen Plasmid Plus Midi kit (Qiagen, #12943).
MPRA transfection and library prep
SNU719 and AGS cells were seeded in 15 cm plates to achieve 80% confluence the next day. Cells were transfected with 10 μg of the MPRA plasmid library using TransIT-X2 reagent (Mirus Bio, MIR 6004) as per the manufacturer’s protocol, aiming for a transfection efficiency of 50-80%. Total RNA was collected 48 hr post-transfection using the miRNeasy mini kit (Qiagen, #217004). Genomic DNA was removed using the Turbo DNA-free kit (ThermoFisher Scientific, AM1907) following the manufacturer’s “Rigorous DNase treatment” protocol. Per replicate, 15 μg total RNA was reverse transcribed with SuperScript IV Reverse Transcriptase (ThermoFisher Scientific, #18090010) using oligo-dT primers. Amplicon sequencing libraries from cDNA or plasmid pool DNA were constructed through two PCR reactions, adapted from Pinto et al.103. In the first PCR round, 1:10 diluted cDNA was amplified using 0.2 μM of the DT_barcodePE_Fv2 and 0.2 μM of an equimolar mix of the DT_barcodePE_Rv2 primers (0 to 6 random Ns, Supplementary Data 5). The PCR reaction was performed with the NEB Next High-Fidelity 2x PCR Master Mix (NEB, M0541L) and the following conditions: 98 °C for 30 sec, 10 cycles (98 °C for 10 sec, 63 °C for 10 sec, 72 °C for 15 sec), 72 °C for 2 min. Enough PCR reactions were run to ensure that all the cDNA from each replicate was amplified. In the second PCR reaction, 1:10 diluted PCR round 1 product was amplified using 0.5 μM of a unique pair of multiplexing Illumina primers (PE_i5 and PE_index in Supplementary Data 5). The following PCR conditions were used: 98 °C for 30 sec, 10 cycles (98 °C for 10 sec, 62 °C for 10 sec, 72 °C for 15 sec), 72 °C for 2 min. For each replicate, the second round PCR product was purified through gel extraction using the Monarch Gel Extraction kit (NEB, T1020S). The quality of each library was assessed by an Agilent Tapestation D1000 assay (Agilent). An equimolar mix of all libraries was sent for single-end 150 bp sequencing on an Illumina sequencer, with 20% PhiX spike-in to increase library complexity. The mixed library was sequenced at a depth to ensure at least 10 M reads per replicate (>500 reads per oligo).
MPRA analysis
MPRA analysis was performed similarly to Mattioli et al.104. Briefly, barcode counts were calculated from raw reads and then normalized per sample based on sequencing depth. For each sample, a barcode RNA to DNA ratio was calculated by dividing the barcode counts in each replicate to that in the plasmid pool library. The RNA to DNA ratios were then log-transformed and quantile normalized across samples. A two-sided Wilcoxon test was performed to compare barcode count ratios between reference and alternative allele oligos in each replicate. To combine replicate p-values, the Stouffer’s method was used, and Benjamini-Hochberg false discovery rate (FDR) correction was applied. Each oligo was represented by 10 barcodes, so to obtain a per-oligo activity in each sample, the median activity was calculated. Fold-change was defined as the ratio of the alternative to the reference allele median activity.
ICI cohort analysis
Pre-treatment tumor RNAseq data were retrieved from four published ICI studies (anti-PD1 or anti-CTLA4 treatment), of which three addressed melanoma patients (n = 90)50,51,69 and one addressed gastric cancer patients (n = 45)70. The combined cohort (n = 135) included 55 responders and 80 non-responders to immunotherapy. Gene expression of pre-treatment tumor samples was quantified from RNAseq reads using Salmon v0.9180. To calculate differential expression of ADAR between responders and non-responders, a Wilcoxon rank sum test with continuity correction was performed. The ICI cohort was also randomly split into a training (n = 68) and a testing set (n = 67) and the Variant Calling and ilQTL pipelines were run on the pre-treatment tumor RNAseq data. The 3′-UTR ilQTL variants enriched in ICI responders in the training set (n = 28, FDR-adjusted p value < 0.05) were selected to comprise the PRS. The score is calculated as the number of variants detected in the patient’s tumor sample, therefore ranging from 0 to 28. All variants were present across the cohort in 1 or more individuals, with 15 being present in the smaller gastric cancer sub-population (n = 45).
PD-L1 expression and the ilQTL signature were combined with a multivariate linear model, and its performance was assessed in the test set (n = 67). For the PRS calculation of the 3′-UTR and CDS + UTR regions, a similar approach was followed as described above. The top 28 enriched variants based on the odds ratio per gene in 3′-UTR or CDS + UTR in the R vs NR samples of the training set were selected accordingly (one-sided Fisher’s exact test q value < 0.05). Only the most highly enriched variant per gene was included in the final models. All PRS models (ilQTL, ilQTL + PD-L1, 3′-UTR, Genic Variants) were trained and tested on identical patient sets.
Whole exome sequencing analysis
WES data for melanoma51 and gastric cancer70 were downloaded from SRA using the sra-toolkit and processed according to GATK best practices using GATK 4.4105. Briefly, FASTQ files were checked for presence of contamination using FastQC 0.12.1106 and MultiQC 1.17107, and following inspection, were aligned using the Burrows-Wheeler Aligner (BWA) 0.7.17108 using the BWA-MEM algorithm against the hg38 genome distributed by the GATK team (https://console.cloud.google.com/storage/browser/genomics-public-data/resources/broad/hg38/v0). The resulting SAM files were sorted and indexed using samtools 1.18109. The files were then post-processed, marking duplicates and running Base Quality Score Recalibration (BQSR). A panel of normals was generated for each study using the healthy patient samples, which was then used along with each tumor-normal WES pair to call mutations using Mutect2, with gnomAD as a germline resource. To minimize artifact calls and contamination, the read orientation artifact workflow was followed before filtering the Mutect2 calls. To accelerate runtime, intervals were used where available, utilizing the capture kit information for each study. SnpEff v5.2110, with the GRCh38.105 database, was used to annotate the resulting VCF files, and then the TMB was calculated with pyTMB v1.3111, using a variant allele fraction of 0.05, a MAF of 0.001, minimum depth of 20 and minimum alternative depth of 2 to minimize noise, while filtering out low quality, non-coding, synonymous and polymorphic calls against gnomAD78. For the MSI calculation, MSIsensor-pro v1.2.0112 was used by first scanning the reference genome for microsatellite information and then running in tumor-normal mode on the aligned reads with default parameters.
Statistics and reproducibility
All statistical analyses were performed using R or GraphPad Prism 8. The QTL analysis was performed with 375 STAD patients for which TCGA contained primary cancer RNAseq data. The MPRA assay (Fig. 4A, B and Supplementary Figs. 3A, B and 6A, B) was independently repeated three times in two gastric cancer cell lines. No statistical method was used to predetermine sample size. No data were excluded from the analyses. Randomization was utilized in the selection of the training/testing sets for the predictive models as described in the Methods section. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are derived from publicly available datasets. Raw RNAseq, WES, and SNP array data for gastric adenocarcinoma patients were obtained from The Cancer Genome Atlas (TCGA) database at gdc.cancer.gov. The TCGA barcodes of the STAD patients included in the study are provided in the Supplementary Information. The PRS and predictive model analyses were performed on a combined cohort of publically available melanoma and gastric cancer pre-treatment RNAseq data obtained from the Gene Expression Omnibus (GEO) database under accession codes GSE11582150, GSE7822051, and GSE9106169, and the European Nucleotide Archive (ENA) under accession PRJEB2578070. Matched WES data for patients included in the PRS and predictive model analyses were obtained from the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih. gov/sra) under accessions SRP067938 and SRP09029451 and from ENA under accession ERP10773470. The raw MPRA amplicon sequencing data generated in this study have been deposited in the GEO database under accession code GSE261709. The remaining data are available within the Article, Supplementary Information, or Source Data file. Source data are provided with this paper.
Code availability
Computational analyses are described in detail in the methods section. The majority of analyses have been performed utilizing publicly available tools. Custom code utilized for the MPRA analysis has been deposited at https://github.com/ivlachos/3UTR.
References
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Waldman, A. D., Fritz, J. M. & Lenardo, M. J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668 (2020).
Brar, G. & Shah, M. A. The role of pembrolizumab in the treatment of PD-L1 expressing gastric and gastroesophageal junction adenocarcinoma. Ther. Adv. Gastroenterol. 12, 1756284819869767 (2019).
Fashoyin-Aje, L. et al. FDA approval summary: pembrolizumab for recurrent locally advanced or metastatic gastric or gastroesophageal junction adenocarcinoma expressing PD-L1. Oncologist 24, 103–109 (2019).
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
He, Q. et al. Genome-wide prediction of cancer driver genes based on SNP and cancer SNV data. Am. J. Cancer Res. 4, 394–410 (2014).
Slack, F. J. & Chinnaiyan, A. M. The role of non-coding RNAs in oncology. Cell 179, 1033–1055 (2019).
Weinhold, N., Jacobsen, A., Schultz, N., Sander, C. & Lee, W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 46, 1160–1165 (2014).
Schuster, S. L. & Hsieh, A. C. The untranslated regions of mRNAs in cancer. Trends Cancer 5, 245–262 (2019).
Litterman, A. J. et al. A massively parallel 3′ UTR reporter assay reveals relationships between nucleotide content, sequence conservation, and mRNA destabilization. Genome Res. 29, 896–906 (2019).
Vlachos, I. S. & Hatzigeorgiou, A. G. Online resources for miRNA analysis. Clin. Biochem. 46, 879–900 (2013).
Cortez, M. A. et al. Role of miRNAs in immune responses and immunotherapy in cancer. Genes. Chromosom. Cancer 58, 244–253 (2019).
Kumar, S. & Sharawat, S. K. Epigenetic regulators of programmed death-ligand 1 expression in human cancers. Transl. Res.: J. Lab. Clin. Med. 202, 129–145 (2018).
Yang, Q., Cao, W., Wang, Z., Zhang, B. & Liu, J. Regulation of cancer immune escape: the roles of miRNAs in immune checkpoint proteins. Cancer Lett. 431, 73–84 (2018).
Friedrich, M. et al. Methods in Enzymology, Vol. 636 (eds. Lorenzo Galluzzi & Nils-Petter Rudqvist) 339–350 (Academic Press, 2020).
Huang, L. et al. The RNA-binding protein MEX3B mediates resistance to cancer immunotherapy by downregulating HLA-A expression. Clin. Cancer Res. 24, 3366–3376 (2018).
Friedrich, M. et al. Tumor-induced escape mechanisms and their association with resistance to checkpoint inhibitor therapy. Cancer Immunol. Immunother. 68, 1689–1700 (2019).
Reches, A. et al. HNRNPR regulates the expression of classical and nonclassical MHC class I proteins. J. Immunol. 196, 4967–4976 (2016).
Arthur, S. E. et al. Non-coding NFKBIZ 3′ UTR mutations promote cell growth and resistance to targeted therapeutics in diffuse large B-cell lymphoma. bioRxiv https://doi.org/10.1101/2021.05.22.445261 (2021).
Arthur, S. E. et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat. Commun. 9, 4001 (2018).
Supek, F., Miñana, B., Valcárcel, J., Gabaldón, T. & Lehner, B. Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335 (2014).
Kurkowiak, M. et al. The effects of RNA editing in cancer tissue at different stages in carcinogenesis. RNA Biol. 18, 1524–1539 (2021).
Liu, J., Wang, F., Zhang, Y., Liu, J. & Zhao, B. ADAR1-mediated RNA editing and its role in cancer. Front. Cell. Dev. Biol. 10, 956649 (2022).
Kataoka, K. et al. Aberrant PD-L1 expression through 3’-UTR disruption in multiple cancers. Nature 534, 402–406 (2016).
Wu, Y. et al. Polymorphism of the programmed death-ligand 1 gene is associated with its protein expression and prognosis in gastric cancer. J. Gastroenterol. Hepatol. 34, 1201–1207 (2019).
Wang, W. et al. A frequent somatic mutation in CD274 3’-UTR leads to protein over-expression in gastric cancer by disrupting miR-570 binding. Hum. Mutat. 33, 480–484 (2012).
Coudray, A., Battenhouse, A. M., Bucher, P. & Iyer, V. R. Detection and benchmarking of somatic mutations in cancer genomes using RNA-seq data. PeerJ 6, e5362–e5362 (2018).
Rheinbay, E. et al. Analyses of non-coding somatic drivers in 2658 cancer whole genomes. Nature 578, 102–111 (2020).
Calabrese, C. et al. Genomic basis for RNA alterations in cancer. Nature 578, 129–136 (2020).
Network, C. G. A. R. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209, (2014).
Kinney, J. B. & McCandlish, D. M. Massively parallel assays and quantitative sequence–function relationships. Annu. Rev. Genomics Hum. Genet. 20, 99–127 (2019).
Wu, W. et al. miRNA mediated noise making of 3’UTR mutations in cancer. Genes. (Basel) 9, 545 (2018).
Wang, V. G., Kim, H. & Chuang, J. H. Whole-exome sequencing capture kit biases yield false negative mutation calls in TCGA cohorts. PloS One 13, e0204912 (2018).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Schaffer, A. A. et al. The cell line A-to-I RNA editing catalogue. Nucl. Acids Res. 48, 5849–5858 (2020).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Wang, W. et al. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum. Genet. 132, 641–648 (2013).
Du, W. et al. Variant SNPs at the microRNA complementary site in the B7‑H1 3’‑untranslated region increase the risk of non‑small cell lung cancer. Mol. Med. Rep. 16, 2682–2690 (2017).
Ongen, H., Buil, A., Brown, A. A., Dermitzakis, E. T. & Delaneau, O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32, 1479–1485 (2016).
Leek, J. T. & Storey, J. D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3, 1724–1735 (2007).
Mayr, C. What are 3’ UTRs doing? Cold Spring Harb. Perspect. Biol. 11, https://doi.org/10.1101/cshperspect.a034728 (2019).
Piovesan, A., Caracausi, M., Antonaros, F., Pelleri, M. C. & Vitale, L. GeneBase 1.1: a tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics. Database (Oxford) 2016, https://doi.org/10.1093/database/baw153 (2016).
Griesemer, D. et al. Genome-wide functional screen of 3’UTR variants uncovers causal variants for human disease and evolution. Cell 184, 5247–5260.e5219 (2021).
Wang, Q. S. et al. Leveraging supervised learning for functionally informed fine-mapping of cis-eQTLs identifies an additional 20,913 putative causal eQTLs. Nat. Commun. 12, 3394 (2021).
Pappou, E. P. & Ahuja, N. The role of oncogenes in gastrointestinal cancer. Gastrointest Cancer Res. S2–S15 (2010).
Sun, K. et al. scRNA-seq of gastric tumor shows complex intercellular interaction with an alternative T cell exhaustion trajectory. Nat. Commun. 13, 4943 (2022).
Vlachos, I. S. et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucl. Acids Res. 43, D153–159, (2015).
Paraskevopoulou, M. D. et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucl. Acids Res. 41, W169–173 (2013).
Hu, B., Yang, Y. T., Huang, Y., Zhu, Y. & Lu, Z. J. POSTAR: a platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucl. Acids Res. 45, D104–D114 (2017).
Auslander, N. et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 24, 1545–1549 (2018).
Hugo, W. et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018).
Van Allen, E. M. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015).
Patel, S. J. et al. Identification of essential genes for cancer immunotherapy. Nature 548, 537–542 (2017).
Dong, M. B. et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 178, 1189–1204.e1123 (2019).
Manguso, R. T. et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017).
Eddy, J. A. et al. CRI iAtlas: an interactive portal for immuno-oncology research. F1000Res 9, 1028 (2020).
Herbert, A. ADAR and immune silencing in cancer. Trends Cancer 5, 272–282 (2019).
Ishizuka, J. J. et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48 (2019).
Brand, A. et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell. Metab. 24, 657–671 (2016).
Chen, Y. et al. Calnexin impairs the antitumor immunity of CD4+ and CD8+ T Cells. Cancer Immunol. Res. 7, 123–135 (2019).
Bartok, O. et al. Anti-tumour immunity induces aberrant peptide presentation in melanoma. Nature 590, 332–337 (2021).
Asaoka, M., Ishikawa, T., Takabe, K. & Patnaik, S. K. APOBEC3-mediated RNA editing in breast cancer is associated with heightened immune activity and improved survival. Int. J. Mol. Sci. 20, 5621 (2019).
Farhood, B., Najafi, M. & Mortezaee, K. CD8. J. Cell. Physiol. 234, 8509–8521 (2019).
Ramírez-Moya, J. et al. An ADAR1-dependent RNA editing event in the cyclin-dependent kinase CDK13 promotes thyroid cancer hallmarks. Mol. Cancer 20, 115 (2021).
Kung, C. P. et al. Evaluating the therapeutic potential of ADAR1 inhibition for triple-negative breast cancer. Oncogene 40, 189–202 (2021).
Chan, T. H. et al. ADAR-mediated RNA editing predicts progression and prognosis of gastric cancer. Gastroenterology 151, 637–650.e610 (2016).
Jiang, L. et al. ADAR1 suppresses interferon signaling in gastric cancer cells by microRNA-302a-mediated IRF9/STAT1 regulation. Int. J. Mol. Sci. 21, https://doi.org/10.3390/ijms21176195 (2020).
Riaz, N. et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 171, 934–949.e916 (2017).
Kim, S. T. et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 24, 1449–1458 (2018).
Zacco, E., Martin, S. R., Thorogate, R. & Pastore, A. The RNA-recognition motifs of TAR DNA-binding protein 43 may play a role in the aberrant self-assembly of the protein. Front. Mol. Neurosci. 11, 372 (2018).
Quinones-Valdez, G. et al. Regulation of RNA editing by RNA-binding proteins in human cells. Commun. Biol. 2, 19 (2019).
Sheng, Q. et al. Cancer-specific expression quantitative loci are affected by expression dysregulation. Brief. Bioinform. 21, 338–347 (2020).
Wei, J. et al. Ribosomal proteins regulate MHC class I peptide generation for immunosurveillance. Mol. Cell. 73, 1162–1173.e1165 (2019).
Tryka, K. A. et al. NCBI’s database of genotypes and phenotypes: dbGaP. Nucl. Acids Res. 42, D975–979 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Lescai, F. et al. Identification and validation of loss of function variants in clinical contexts. Mol. Genet. Genomic Med. 2, 58–63 (2014).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Yates, A. D. et al. Ensembl 2020. Nucl. Acids Res. 48, D682–d688 (2020).
Consortium, G. T. et al. Genetic effects on gene expression across human tissues. Nature 550, 204 https://www.nature.com/articles/nature24277#supplementary-information (2017).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Leek, J. T. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 42, https://doi.org/10.1093/nar/gku864 (2014).
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Huang, K. L. et al. Pathogenic germline variants in 10,389 adult cancers. Cell 173, 355–370.e314 (2018).
Cavalcante, R. G. & Sartor, M. A. annotatr: genomic regions in context. Bioinformatics 33, 2381–2383 (2017).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 (2013).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29 (2000).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucl. Acids Res. 44, D457–462, (2016).
Gel, B. et al. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32, 289–291 (2015).
O’Donnell, J. S., Teng, M. W. L. & Smyth, M. J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 16, 151–167 (2019).
Havel, J. J., Chowell, D. & Chan, T. A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 19, 133–150 (2019).
Nagarsheth, N., Wicha, M. S. & Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 17, 559–572 (2017).
Conway, J. R., Kofman, E., Mo, S. S., Elmarakeby, H. & Van Allen, E. Genomics of response to immune checkpoint therapies for cancer: implications for precision medicine. Genome Med. 10, 93 (2018).
Teng, F., Meng, X., Kong, L. & Yu, J. Progress and challenges of predictive biomarkers of anti PD-1/PD-L1 immunotherapy: a systematic review. Cancer Lett. 414, 166–173 (2018).
Otoshi, T., Nagano, T., Tachihara, M. & Nishimura, Y. Possible biomarkers for cancer immunotherapy. Cancers (Basel) 11, https://doi.org/10.3390/cancers11070935 (2019).
Lange, S. S., Takata, K. & Wood, R. D. DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110 (2011).
Dominguez, D. et al. Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell. 70, 854–867.e859 (2018).
Griffiths-Jones, S. The microRNA Registry. Nucl. Acids Res. 32, D109–D111 (2004).
Joung, J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Pinto, B. S. et al. Impeding transcription of expanded microsatellite repeats by deactivated Cas9. Mol. Cell. 68, 479–490.e475 (2017).
Mattioli, K. et al. High-throughput functional analysis of lncRNA core promoters elucidates rules governing tissue specificity. Genome Res. 29, 344–355 (2019).
Van der Auwera, G. A. & O’Connor, B. D. Genomics in the cloud: using Docker, GATK, and WDL in Terra. (O’Reilly Media, 2020).
Andrews, S. (Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom, 2010).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, https://doi.org/10.1093/gigascience/giab008 (2021).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (Austin) 6, 80–92 (2012).
Curie, I. pyTMB,† Tumor mutational burden score. https://github.com/bioinfo-pf-curie/TMB.
Niu, B. et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 30, 1015–1016 (2014).
Acknowledgements
We acknowledge the NCI award R01CA258776 to ISV and the NCI Outstanding Investigator Award R35CA232105 to FJS. The results presented here are based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. TCGA data were accessed through dbGaP phs000178.v11.p8. Portions of this research were conducted on the Ithaca High-Performance Computing system, Department of Pathology, BIDMC, and the O2 High-Performance Compute Cluster at Harvard Medical School.
Author information
Authors and Affiliations
Contributions
I.S.V., F.J.S., and C.M. conceived and designed the study. C.M. performed the variant calling, e/ilQTL analysis, and MPRA assay and analysis. Y.M. performed the single-cell enrichment analysis and contributed to the 3’UTR variant analyses; X.L.K. and F.B. performed functional enrichment analyses. E.K. and D.K. developed and applied the PRS and predictive models. C.M. and D.K. designed the MPRA pool. K.M. contributed to the MPRA assay analysis. S.N. analyzed the ADAR expression in ICI response cohorts. N.K. performed the WES analysis. Y.P.J. and V.R.R. reviewed the data and provided significant intellectual input. I.S.V., F.J.S., and C.M. wrote the manuscript with input from all authors. I.S.V. and F.J.S. provided oversight for the study.
Corresponding authors
Ethics declarations
Competing interests
I.S.V. consults for Guidepoint Global, Cowen, Mosaic, and NextRNA. Beth Israel Deaconess Medical Center has filed a patent application based on this work for “Methods and compositions related to non-coding variants for the prediction of response to cancer immunotherapy” under 63/378,392, where I.S.V., F.J.S., C.M., and E.K. are named as co-inventors. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miliotis, C., Ma, Y., Katopodi, XL. et al. Determinants of gastric cancer immune escape identified from non-coding immune-landscape quantitative trait loci. Nat Commun 15, 4319 (2024). https://doi.org/10.1038/s41467-024-48436-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-48436-5
- Springer Nature Limited