Abstract
High-voltage-activated R-type CaV2.3 channel plays pivotal roles in many physiological activities and is implicated in epilepsy, convulsions, and other neurodevelopmental impairments. Here, we determine the high-resolution cryo-electron microscopy (cryo-EM) structure of human CaV2.3 in complex with the α2δ1 and β1 subunits. The VSDII is stabilized in the resting state. Electrophysiological experiments elucidate that the VSDII is not required for channel activation, whereas the other VSDs are essential for channel opening. The intracellular gate is blocked by the W-helix. A pre-W-helix adjacent to the W-helix can significantly regulate closed-state inactivation (CSI) by modulating the association and dissociation of the W-helix with the gate. Electrostatic interactions formed between the negatively charged domain on S6II, which is exclusively conserved in the CaV2 family, and nearby regions at the alpha-interacting domain (AID) and S4-S5II helix are identified. Further functional analyses indicate that these interactions are critical for the open-state inactivation (OSI) of CaV2 channels.
Similar content being viewed by others
Introduction
Voltage-gated calcium (CaV) channels mediate calcium influx to cells in response to changes in membrane potential1,2,3. Their cellular roles have been emphasized for decades in a variety of studies and include hormone secretion4,5, neurotransmitter release6,7, and muscle contraction8,9. CaV channel members are categorized into the CaV1, CaV2, and CaV3 subfamilies based on sequence identity or alternatively classified into T-, L-, P/Q-, N-, and R-types according to their pharmacological and biophysical profiles1. The so-called pharmacoresistant (R-type) CaV2.3 is widely expressed in the brain and enriched in the hippocampus, cerebral cortex, amygdala, and corpus striatum10,11,12. Electrophysiological investigations revealed that currents mediated by CaV2.3 are resistant to common CaV blockers or gating modifiers such as nifedipine, nimodipine, ω-Aga-IVA, etc13. CaV2.3 channels exhibit cumulative inactivation in response to brief and repetitive depolarizations, a process known as preferential closed-state inactivation (CSI)14. Furthermore, CaV2.3 is involved in a broad spectrum of neuronal activities10,12,15. Previous studies have reported that CaV2.3 participates in multiple physiological processes in the central nervous system, such as inducing long-term potentiation (LTP) and post-tetanic potentiation in mossy fiber synapses16, modulating the burst firing mode of action potentials17,18, and regulating synaptic strength in hippocampal CA1 pyramidal neurons19. In recent years, increasing evidence has revealed that dysfunction of CaV2.3 is linked to epilepsy20,21, convulsions22,23, and neurodevelopmental impairments24, suggesting that CaV2.3 is a pivotal player in the pathogenesis of a series of neurological disorders.
The molecular basis of CaV channels has been investigated extensively over the past several decades, including structural studies of L-type CaV1.125,26, N-type CaV2.227,28, and T-type CaV3.129 and CaV3.330 in the apo form or distinct modulator-bound states. These structures provide substantial insights into the architecture, subunit assembly, and modulator actions of the CaV channels. However, the gating mechanism of CaV channels is still far from fully understood. For instance, in the CaV2.2 structure, the VSDII is trapped in a resting state by a PIP2 molecule at a membrane potential of ~0 mV27,28. The functional roles of the VSDII trapped in the resting state by PIP2 remain unknown. A considerable number of pathogenic mutations have been identified in the VSDs of neuronal CaV2 channels, demonstrating that VSD dysfunctions contribute to the genesis of spinocerebellar ataxia (SCA), episodic ataxia (EA), and familial hemiplegic migraine (FHM)31. Moreover, the CaV2.2 and CaV2.3 channels inactivate preferentially from the intermediate closed state along the activation pathway, which is important in controlling the short-term dynamics of synaptic efficacy14,32. In our previous study, we elucidated that residue W768 on the W-helix located within the DII-III linker serves as a key determinant of the CSI of the CaV2.2 channel28. However, CaV2.3 is characterized by a more prominent preferential CSI than CaV2.214. It is also interesting to explore the modulation mechanism of CSI in CaV2.3. Furthermore, the high-voltage-activated (HVA) CaV1 and CaV2 channels harbor a conserved α-helix connecting Domain I and Domain II (alpha-interaction domain, or AID). Previous studies have indicated that AID might contribute to the open-state inactivation (OSI) of the HVA CaV channels33,34. However, the inactivation properties of CaV1 and CaV2 channels are dramatically different33. Mechanistic insight into the inactivation processes of the HVA CaV channels will help us to fully uncover the physiological role of CaV channels and facilitate the development of therapeutic solutions for CaV-related diseases.
In this study, we expressed and purified human CaV2.3 in complex with auxiliary subunits α2δ1 and β1 and unveiled the high-resolution structure of this protein complex. Further mutagenesis and electrophysiological experiments were performed. Our results provide insights into the pharmacological resistance properties of CaV2.3, the asynchronous functional roles of the VSDs, the mechanism by which the pre-W-helix regulates the CSI, and the OSI process modulated by the negatively charged domain on S6II (S6IINCD).
Results and discussion
Architecture of the CaV2.3 complex
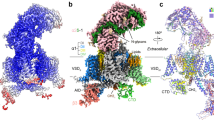
To gain structural insights into the CaV2.3 complex, we expressed and purified full-length wild-type human CaV2.3 α1E subunit (CACNA1E), α2δ1 (CACNA2D1) and β1 (CACB1) using a HEK 293-F expression system. The CaV2.3-α2δ1-β1 complex was solubilized using n-Dodecyl-β-D-maltoside (DDM) and purified using a strep-actin affinity column, followed by further purification by size-exclusion chromatography (SEC) in a running buffer containing glycol-diosgenin (GDN) to remove protein aggregates (Supplementary Fig. 1a, see Method section for details). The peak fractions were subsequently collected and concentrated for cryo-EM sample preparation (Supplementary Fig. 1b). A total of 2096 micrographs were collected. Data processing of the dataset gave rise to a 3.1-Å cryo-EM map, which is rich in high-resolution structural features, including densities for sidechains, lipid molecules, and glycosylations (Fig. 1, Supplementary Fig. 2, and Supplementary Table 1).
a, b Cryo-EM map (a) and model (b) of the CaV2.3-α2δ1−β1 complex. The CaV2.3 α1E pore-forming subunit are colored pink, red, blue, and deep cyan. The C-terminal domain (CTD) of CaV2.3 are colored gray. The auxiliary subunits α2, δ, ανδ β1 are colored green, orange, and white, respectively. c The ion-conducting pathway and pore profiling of the CaV2.3. The ion-conducting pathway is viewed in parallel to the membrane and shown in black dots. The radius of the pore is calculated using the HOLE program. The vertical dashed line marks the 1.0-Å pore radius, which represents the ionic radius of calcium. d Superimposition of the DIII-IV fenestration between CaV2.3 and CaV1.1 complex. The nifedipine molecule bound to the CaV1.1 complex is shown as sticks. Residues stabilizing the nifedipine molecule in CaV1.1, and residues that might form steric clashes in CaV2.3 are shown as sticks. e Superimposition between the ‘P-loops’ (P1 and P2) and extracellular loops (ECLs) of the CaV2.3 and the ziconotide-bound CaV2.2. The CaV2.3 and CaV2.2 are shown as ribbon, overlaid with the ziconotide shown as transparent green surfaces. Potential steric clash between the ECLI of CaV2.3 and the ziconotide is highlighted in red. f Pathogenic mutations of the CaV2.3, shown in blue spheres. The four domains of CaV2.3 are colored pink, red, blue, and deep cyan, respectively. CaV1.1 is colored purple and CaV2.2 is colored gray.
The CaV2.3 complex exhibited a conventional shape that resembles that of the CaV2.2 and CaV1.1 complexes (Fig. 1). The complex is composed of the transmembrane α1E subunit, extracellular α2δ1 subunit, and intracellular β1 subunit (Fig. 1a, b). The α1E subunit is a pseudo-tetrameric pore-forming subunit and can be divided into Domain I (DI) to IV (DIV). Each domain of the α1E subunit is composed of 6 transmembrane helices (S1–S6), comprising the voltage-sensing domain (VSD) (S1–S4) and the pore domain (S5–S6). The P1 and P2 helices located between the S5 and S6 helices formed the selectivity filter (Fig. 1c). Similar to other CaV channels, CaV2.3 harbors four extracellular loops (ECLs) that are also positioned between S5 and P1, as well as between P2 and S6 helices in the pore domain (Fig. 1a, c). The ECLI and ECLII are critical for the association between the α1 subunit and the α2δ1 subunit (Fig. 1a, b). The S6 helices from the four domains converge on the cytoplasmic side and form the intracellular gate of the channel. In our structure, the intracellular gate was determined in its closed state, in line with the observations from other voltage-gated channels (Fig. 1c). Moreover, the closed gate of CaV2.3 is further stabilized by the W-helix from the DII-DIII linker, which is consistent with a previous study on CaV2.2 and indicates that CaV2.3 also adopts the CSI mechanism28.
Most CaV channels serve as pharmaceutical targets of a variety of small-molecule drugs or peptide toxins13. However, previous studies indicate that CaV2.3 is resistant to many CaV modulators, such as nimodipine (L-type), omega-Aga-IVA (P/Q-type), and omega-CTx-GVIA (N-type)13. To clarify the structural basis underlying the pharmacoresistance of CaV2.3, we compared the structures of CaV2.3 with CaV1.1 or CaV2.2 in their ligand-bound states (Fig. 1d, e). In the structure of nifedipine-bound CaV1.1, the nifedipine molecule was located within the DIII-DIV fenestration and stabilized by surrounding residues. However, two critical residues are substituted in CaV2.3, namely, Y1296 and F1708. The bulky sidechains of these two residues in CaV2.3 occupy the DIII-DIV fenestration site and thus hinder the binding of nifedipine (Fig. 1d). Meanwhile, a previous study reported that the Q1010 of CaV1.2 is important for sensitivity to dihydropyridine (DHP) and the Q1010M mutant had a decreased sensitivity to DHP molecules35. The equivalent position in CaV2.3 is occupied by M1300, thus also contributing to the pharmacoresistance of CaV2.3 to DHP molecules. Structural comparison of CaV2.3 and ziconotide-bound CaV2.2 demonstrated that the ECLI loop of CaV2.3 adopts a different conformation, and residues D263 and P264 are placed close to the central axis, giving rise to clashes between the ziconotide and the ECLI of CaV2.3 (Fig. 1e). Moreover, other residues on P-loops and ECLs that are involved in ziconotide binding are also not conserved in CaV2.3 (Supplementary Fig. 3), rendering CaV2.3 insensitive to the ziconotide.
Gain-of-function mutations and polymorphisms of CaV2.3 channels have already been implicated in the pathological process of developmental and epileptic encephalopathy. Thirteen pathogenic mutations have been identified in CaV2.324. Our high-resolution structure provides a structural template to map all of these pathogenic mutations, which are distributed throughout the complex structure (Fig. 1f). Eight of thirteen mutations are located around the intracellular gate, such as I603L, F698S, and I701V, and the result in a hyperpolarizing shift in the half-activation voltage24 (Fig. 1f).
Functional heterogeneity of the VSDs
The voltage-dependent gating characteristics of voltage-gated channels are conferred by their VSDs. The VSDs of CaV are conserved helix bundles consisting of S1, S2, S3, and S4 helices (Supplementary Fig. 4a). S4 was found to be a positively charged 310 helix, harboring approximately five or six arginines or lysines as gating charges lining one side of the helix at intervals of three residues (Supplementary Fig. 4a). The positively charged S4 helices move vertically toward the intracellular or extracellular side of the cell in response to the hyperpolarization or depolarization of the membrane potential. The conformational change of VSD is coupled to the pore domain by a short amphipathic helix S4-S5, which connects the S4 helix of VSD to the S5 helix from the pore domain, thus regulating the transition of the intracellular gate between the open and closed states. Although the four VSDs of CaV channels are considerably similar in terms of sequence and overall structure, they contribute differentially to the opening of pore36.
Superimposition of the structures of CaV2.3 and CaV2.2 revealed that they are comparable overall (r.m.s.d. = 1.46 Å for 2222 Cα atom pairs). The pore domain was fairly superimposable between CaV2.3 and CaV2.2, including the S5 and S6 helices and extracellular loops (ECLs) I, III, and III (Supplementary Fig. 4b). VSDI, VSDIII, and VSDIV in the activated state and VSDII in the resting state were also determined in both CaV2.3 and CaV2.2 (Supplementary Fig. 4a, b). However, a structural discrepancy was visualized at the ECLIV between the two structures. The ECLIV of CaV2.3 extends from the pore domain and lies above the S1-S2III linker, whereas the ECLIV of CaV2.2 is much shorter and wraps around the pore domain before touching the extracellular side of VSDIII (Fig. 2a). Four residues on ECLIV of CaV2.3, namely, P1680, D1681, T1682, and T1683, are involved in the interactions with the residues on the S1-S2III linker, especially V1176, L1177, T1178, and N1179, which consequently stitches the VSD to the pore domain at the extracellular side (Fig. 2a). To explore the functional roles of this interaction, we substituted 1680PDTT1683 on ECLIV with four glycines (CaV2.34G) to disrupt the contacts between ECLIV and S1-S2III loop. Electrophysiological studies indicated that the voltage dependency of the activation curve of CaV2.34G displayed a ~5 mV positive shift (P < 0.0001, two-tailed unpaired t test) compared to that of wild-type CaV2.3 (Fig. 2b and Supplementary Fig. 5). We thus speculate that the interactions between ECLIV and the S1-S2III loop may stabilize the VSDIII in a certain conformation relative to the pore domain that requires less electrical energy to activate the channel, reminiscent of the cholesterol regulation on CaV channels, potentially by stabilizing interactions between the extracellular end of the S1-S2 helix hairpin and the pore domain37,38.
a Superimposition of the VSDs (DIII) and the pore domains (DIV) of the CaV2.3 (red and blue) and CaV2.2 (gray) demonstrating the interaction between the ECLIV and the VSDIII. Residues on ECLIV in close contact with VSDIII (1680PDTT1683) are shown in sticks and colored green. Cholesteryl hemisuccinate (CHS) molecules are shown as sticks and labeled. b Steady-state activation and inactivation curves of the wild-type (WT) CaV2.3 (black), VSDII4Q (pink), and CaV2.34G (green). To examine the voltage dependence of activation, HEK 293-T cells expressing the CaV2.3 complex were tested by 200-ms depolarizing pulses between –60 and 50 mV from a holding potential of –100 mV, in 10-mV increments. To determine inactivation curves, cells were stepped from a holding potential of –100 mV to pre-pulse potentials between –100 and –15 mV in 5-mV increments for 10 s. Activation curve, CaV2.3 WT, n = 14; VSDII4Q, n = 7; CaV2.34G, n = 11. Steady-state inactivation curve, CaV2.3 WT, n = 9; VSDII4Q, n = 7; CaV2.34G, n = 9. c Current density of the wild-type CaV2.3 and VSDII4Q. CaV2.3 WT, n = 8; VSDII4Q, n = 8; CaV2.34G, n = 7. d Voltage-clamp protocol and representative traces of the CaV2.3 WT, VSDI5Q, VSDII4Q, VSDIII5Q, and VSDIV5Q, elicited by a 200-ms test pulse at +10 mV. Data are presented as mean ± SEM. n biological independent cells.
The VSDIIs of CaV2.3 and CaV2.2 were determined in the resting (S4 down) state, while most other VSDs in the voltage-gated channels, such as CaV1.1, CaV3.1, and NaVs, were determined in the activated (S4 up) state (Supplementary Fig. 4c, d). This finding suggests that VSDII plays a unique role in the gating mechanism among CaV2 members. Structural analyses of CaV2.2 have suggested that the VSDII is trapped in the resting state by a PIP2 molecule27,28. The cryo-EM map of CaV2.3 is of high quality around S4-S5II (Supplementary Fig. 2e), and we identified a single strip-shaped density that does not look like a PIP2 molecule. However, another recent structural investigation of CaV2.3 channel showed that PIP2 could bind to this site but is not responsible for the resting state of VSDII39. To shed light on the functional roles of VSDII in the gating mechanism of CaV2.3, we constructed a gating charge neutralization mutant on the VSDII (VSDII4Q, R572Q/R575Q/R578Q/K581Q). Interestingly, the neutralization mutation of the VSDII (VSDII4Q) exhibits ~9-mV left shifts in the voltage dependency of both activation and steady-state inactivation compared to the wild-type (Fig. 2b and Supplementary Fig. 5a, b). Consequently, the current density-voltage curve of the VSDII4Q mutant was also left-shifted (Fig. 2c). Moreover, we tested whether the VSDII4Q mutant has a distinct CSI profile. The cumulative inactivation of this mutant in response to action potential (AP) trains is markedly enhanced (Supplementary Fig. 5c, e). These results suggested that the conformation of VSDII influences the gating of CaV2.3. However, its OSI kinetics remain unaltered (Supplementary Fig. 5d). In contrast, the gating charge-neutralized mutation in the VSDI, VSDIII, and VSDIV resulted in failure to mediate inward current (Fig. 2d), in line with previous results showing that the VSDI, VSDIII, and VSDIV are important for gating of the closely-related CaV2.240,41,42, while the VSDII is not necessary for channel activation by sensing the depolarization of membrane potential; instead, the VSDII is crucial to modulating channel properties, such as CSI and voltage dependency of channel activation and inactivation.
Molecular mechanism of closed-state inactivation
Preferential closed-state inactivation is a featured kinetic characteristic of neuronal CaV channels14,28. During the state-transition pathway in the activation of CaV channels, CSI occurs preferentially in a specific pre-open closed state and in a voltage-dependent manner. CSI can be visualized by the cumulative inactivation in response to action potential (AP) trains, as reported in previous studies, which demonstrated that the peak current triggered by each AP shrank sequentially, suggesting that a substantial amount of the channels turn inactivated after the repolarization of an AP. CSI plays an important role in the orchestrated modulation mechanism of CaV channels and is of vital importance for the precise regulation of physiological processes such as neurotransmitter release and synapse plasticity. CSI is detected in all neuronal CaV2 channels at distinct levels14. The R-type CaV2.3 displayed a more prominent CSI than the N-type CaV2.2, and both showed far more prominent CSI than the P/Q-type CaV2.114. Previous structural investigations on the N-type CaV2.2 channel have revealed that a conserved W-helix is the structural determinant of CSI, and W768 from the W-helix on the DII-DIII linker functions as a lid blocking the pore and stabilizes the intracellular gate in its closed states by hydrophobic interactions. However, the molecular mechanisms underlying the CSI of CaVs are still not fully understood, as the conserved W-helix is unable to explain the disparity of CSI mechanisms among the CaV2.1, CaV2.2, and CaV2.3 channels.
The W-helix determined in CaV2.2 is also conserved and well-resolved in CaV2.3 (Fig. 3a, b). The W-helix in CaV2.3 (772RHHMSVWEQRTSQLRKH788) is a positively charged short helix and positioned underneath the intracellular gate, with W778 inserting into the gate and forming extensive interactions with residues from surrounding gating helices (Fig. 3a, b). These structural observations are consistent with the W-helix of CaV2.2. First, we designed the CaV2.3 W/Q (W778Q) to disrupt interactions between the W-helix and intracellular gate (Fig. 3c–g and Supplementary Fig. 6). It turns out that the CaV2.3 W/Q exhibited a ~8-mV positive shift on the steady-state inactivation curve (Fig. 3d) and an alleviated cumulative inactivation in response to AP trains (Fig. 3e, g) without affecting the voltage dependence of channel activation (Fig. 3c), consistent with the observations in CaV2.228, suggesting that the W778 is important for CSI process of CaV2.3 channel. Moreover, we speculate that the positively charged residues on the W-helix could putatively respond to the membrane potential change and may be important for the initiation of the CSI process. To evaluate our speculations, we constructed the CaV2.3RK/A mutant by substituting the R781, R786, and K787 with alanine (R781A/R786A/K787A). The activation curve of the CaV2.3RK/A mutant remains unaltered (Fig. 3c). However, the CSI of CaV2.3RK/A mutant was significantly suppressed, exhibiting a ~9 mV right shift on the steady-state inactivation curve (Fig. 3d), an alleviated cumulative inactivation to AP trains (Fig. 3e, g), and an accelerated recovery rate from CSI (Fig. 3f and Supplementary Fig. 6b). These alterations on the CSI profile suggested that the positively charged R781, R786, and K787 are critical for the CSI mechanism of CaV2.3.
a Binding pocket of the W-helix. The W-helix is accommodated in a negatively charged pocket on the intracellular side of CaV2.3. The CaV2.3 α1E subunit is shown in the cartoon, and the negatively charged pocket accommodating W-helix is shown as red surface. b Zoomed-in view of the W-helix, which is stabilized in the intracellular gate at a closed state. The W-helix is shown in cartoon and overlaid with an electrostatic surface. W778 and other residues involved in the hydrophobic or charge interactions are shown in sticks. c Activation curves of the wild-type (WT) CaV2.3 and the mutants. CaV2.3 WT, n = 14; Δw-helix, n = 7; CaV2.3RK/A, n = 7. d Steady-state inactivation curves of CaV2.3 WT and the mutants. CaV2.3 WT, n = 9; Δw-helix, n = 8; CaV2.3RK/A, n = 10. e Inactivation ratio quantified using the current density (I) elicited by each spike of AP trains divided by the maximum current (IPeak) elicited by the first spike. CaV2.3 WT, n = 13; CaV2.3 W/Q, n = 15; Δw-helix, n = 9; CaV2.3RK/A, n = 14. f Recovery rate from the CSI, quantified by the inactivation ratio between the two peak currents (I and I2,048ms) obtained in the two-pulse protocol. HEK 293-T cells were held at –40 mV for 1500 ms and were then stepped to –100 mV for a series of time intervals (4–2,048 ms) before a +10 mV test pulse (35 ms). CaV2.3 WT, n = 6; CaV2.3 W/Q, n = 6; Δw-helix, n = 6; CaV2.3RK/A, n = 7. g Representative current responses to the AP trains. The AP trains used to stimulate the HEK 293-T cells were recorded from a mouse hippocampal CA1 pyramidal neuron after current injection in whole-cell current-clamp mode. See Supplementary Figure 5 for voltage-clamp protocols and Method section for literature reference. CaV2.3 WT, black; CaV2.3 W/Q, orange; CaV2.3RK/A, blue. Data are presented as mean ± SEM. n biological independent cells.
Intriguingly, sequence alignment among the neuronal CaVs revealed that a peptide segment (pre-W-helix) that resembled the W-helix is located adjacent to the W-helix of CaV2.3 (Fig. 4a). The sequence of pre-W-helix (753RHHMSMWEPRSSHLRER769) is nearly identical to that of the W-helix (~65% identity), including the conserved tryptophan plug (W759) and positively charged residues (R762, R767, and R769) (Fig. 4a). However, we noticed that a non-conserved proline (P761) was in the middle of the pre-W-helix, which may undermine the stability of both the pre-W-helix itself and its interactions with the gate. Moreover, in our cryo-EM map of the CaV2.3 complex, the helical density beneath the gate perfectly fits the atomic model of the W-helix, enabling us to unambiguously determine that the W-helix, instead of the pre-W-helix, exists in our structure (Supplementary Fig. 2e). Considering that the pre-W-helix is located immediately before the W-helix and that they share high sequence identity, we speculate that the pre-W-helix participates in the CSI event of CaV2.3. To evaluate the contribution of the pre-W-helix to the CSI of CaV2.3, we constructed two mutants by deleting the W-helix (Δw-helix) and pre-W-helix (Δpre-w-helix) (Fig. 4b–e, 4k, and Supplementary Fig. 6). The Δw-helix exhibited a ~4-mV positive shift on the steady-state inactivation curve (Fig. 4c) and alleviated cumulative inactivation in response to AP trains (Fig. 4d, k) without affecting the voltage dependence of channel activation (Fig. 4b), indicating that the W-helix plays pivotal roles in the CSI of CaV2.3. However, compared with the CSI of CaV2.2, which was almost abolished by deleting the W-helix, a substantial portion of the CSI in the Δw-helix mutant remained unaltered (Fig. 4d, k), suggesting that the CSI modulation of CaV2.3 is distinct from that of CaV2.2 and that other elements may also contribute to the CSI of CaV2.3. We also used a two-pulse protocol to assess the recovery rate from CSI, i.e., the release process of CSI (Supplementary Fig. 6a). Consistent with the electrophysiological results above, an accelerated recovery rate from CSI was observed in the Δw-helix compared to the WT (Fig. 4e and Supplementary Fig. 6b). Strikingly, a negative-shift of ~8 mV was detected on the inactivation curve of the Δpre-w-helix mutant (Fig. 4c), and its cumulative inactivation to AP trains was surprisingly enhanced (Fig. 4d, k), demonstrating that the development process of CSI in the Δpre-w-helix was significantly boosted. Considering that the pre-W and W helices are close to each other and share high sequence identity, we speculate that the pre-W-helix may serve as a competitive negative regulator to interfere with the binding of the intracellular gate to the W-helix (Fig. 4f). In the absence of the pre-W-helix, the CSI is consequently enhanced (Fig. 4c, d, k). Paradoxically, compared with the WT, the recovery rate from CSI of the Δpre-w-helix mutant was substantially accelerated (Fig. 4e and Supplementary Fig. 6b). This suggests that once CSI occurs, the pre-W-helix appears to stabilize the W-helix for interaction with the gate during membrane potential repolarization, thereby slowing the recovery of CaV2.3 from the CSI. We also designed a mutant by deleting both the pre-W-helix and the W-helix (Δpre-w/Δw-helix). This mutant displayed a ~5-mV positive shift on the inactivation curve (Fig. 4c), reduced cumulative inactivation to AP trains (Fig. 4d, k), and an accelerated recovery rate from CSI (Fig. 4e and Supplementary Fig. 6b). These effects of this double deletion construct are essentially identical to those of the Δw-helix, demonstrating that the modulatory role of the pre-W-helix on CSI is largely dependent on the W-helix, probably by regulating the binding or dissociation of the W-helix with the intracellular gate. To further investigate the regulatory mechanism on the CSI of CaV2.3, we constructed two mutants by neutralizing the arginines (CaV2.3preR/Q, R753Q/R762Q/R767Q/R769Q) or substituting the tryptophan on the pre-W-helix with glutamine (CaV2.3preW/Q, W759Q) (Fig. 4g–k and Supplementary Fig. 6). Impressively, the mutants CaV2.3preR/Q and CaV2.3preW/Q exhibit similar gating kinetics and voltage dependence of channel activation and inactivation, but significantly different from that of the WT CaV2.3 channel (Fig. 4g–k, Supplementary Fig. 5a, b and Supplementary Fig. 6b). In particular, they displayed a ~7-mV negative shift on inactivation curve (Fig. 4h), enhanced cumulative inactivation to AP trains (Fig. 4i, k), and an accelerated recovery rate from CSI (Fig. 4j and Supplementary Fig. 6b), highly identical to the CSI profile of the Δpre-w-helix. The kinetic characteristics of these mutants indicated that the positively charged residues and W759 are important for the regulatory effect of the pre-W-helix. Our data also show that the development and release of CSI are two independent processes. In particular, the Δpre-w-helix, CaV2.3preR/Q, and CaV2.3preW/Q exhibited enhanced cumulative inactivation during AP trains (Fig. 4d, i, k) but a faster recovery rate from CSI (Fig. 4e, j and Supplementary Fig. 6b) at a membrane potential of –100 mV, suggesting that the channel may have distinct conformational states along the activation pathway. The W-helix may bind preferentially to the channel in the intermediate closed state(s) near the open state, thus leading to the largest inactivation before the channel opens (Fig. 4f). At strongly hyperpolarized potentials (~100 mV), the channel is probably stabilized in a different closed state, exhibiting a lower affinity to the W-helix, causing the W-helix to detach from the gate and the channel to recover from the CSI (Fig. 4f). Taken together, the pre-W-helix in CaV2.3 plays significant roles in regulating the association or dissociation of the W-helix with the intracellular gate and thus exerts a regulatory effect on the development and release of the CSI.
a Sequence alignment of the DII-III linker among the CaV2 members. Residues on the pre-W-helix and W-helix are underlined and labeled, respectively. Exon 19 on the genomic DNA of CaV2.3 encoding the pre-W-helix is also labeled. b, g Activation curves of the wild-type CaV2.3 and the mutants. CaV2.3 WT, n = 14; Δw-helix, n = 7; Δpre-w-helix, n = 7; Δpre-w/Δw-helix, n = 7; CaV2.3preR/Q, n = 7; CaV2.3preW/Q, n = 8. c, h Steady-state inactivation curves of wild-type CaV2.3 and the mutants. CaV2.3 WT, n = 9; Δw-helix, n = 8; Δpre-w-helix, n = 7; Δpre-w/Δw-helix, n = 7; CaV2.3preR/Q, n = 8; CaV2.3preW/Q, n = 6. d, i Inactivation ratio quantified using I/IPeak elicited by the AP trains. CaV2.3 WT, n = 13; Δw-helix, n = 9; Δpre-w-helix, n = 13; Δpre-w/Δw-helix, n = 14; CaV2.3preR/Q, n = 9; CaV2.3preW/Q, n = 13. e, j Recovery rate from the CSI, quantified using the previously described two-pulse protocol. CaV2.3 WT, n = 6; Δw-helix, n = 6; Δpre-w-helix, n = 7; Δpre-w/Δw-helix, n = 6; CaV2.3preR/Q, n = 8; CaV2.3preW/Q, n = 7. f Proposed model of the putative regulatory role of the pre-W-helix in the CSI of CaV2.3. Membrane planes are indicated using gray lines. Pore domains are depicted as cartoon and colored purple. Positively charged S4 helices are shown as gray rounded bars. Pre-W-helix and the W-helix are shown as cartoon. In the development process of CSI (left), the intracellular gate might have a high affinity to W-helix. As a consequence, W-helix and pre-W-helix competitively bind to the gate. In the release process of CSI (right), the gate might adopt an alternative conformation and the W-helix is likely to disassociate from the gate. k Representative current responses stimulated by AP trains. See Supplementary Figure 5 for voltage-clamp protocols and Method section for literature reference. CaV2.3 WT, black; Δw-helix, purple; Δpre-w-helix, red; Δpre-w/Δw-helix, green-cyan; CaV2.3preR/Q, green; CaV2.3preW/Q, yellow. Data are presented as mean ± SEM. n biological independent cells.
Interestingly, on the genomic DNA of CaV2.3, the pre-W-helix is encoded by exon 19 (residues R748–R769) (Fig. 4a). Early studies have reported that alternative splicing events might occur at this site, as exon 19 is spliced out in the mature mRNA encoding CaV2.3e, an isoform of CaV2.3 that displays decreased Ca2+ sensitivity in its calcium-dependent modulation mechanisms43. Gene expression profiling has revealed that CaV2.3e is enriched in endocrine tissues, including the kidney and pancreas, and the majority of CaV2.3 in the brain contains the pre-W-helix44. This implies that the kinetic variability of CaV2.3 mediated by the pre-W-helix is an important regulatory mechanism to fine-tune the properties of the CaV2.3 channel to adapt to the distinct physiological needs of neuronal and endocrinal excitable cells43.
Modulation of open-state inactivation
Upon the opening of the pore, CaV channels undergo an inactivation process called open-state inactivation (OSI), which describes the mechanism by which the intracellular gate shifts swiftly from the open state into the inactivation state33,45. This process is an intrinsic mechanism that precisely regulates calcium influx into cells during depolarization. Neuronal (P/Q-, N- and R-type) CaV2 channels bear a stronger OSI and mediate a rapidly inactivated current, while cardiac (L-type) CaV channels display a much weaker OSI, mediating a long-lasting current46. Dysfunction of OSI, i.e., the gain-of-function of neuronal CaVs, is linked to a series of neurological disorders, including trigeminal neuralgia47, myoclonus-dystonia-like syndrome48, and epileptic encephalopathies21,24. Although the structures of L-type CaV1.125 and N-type CaV2.227,28 complexes have been elucidated at high resolution, the structural basis for their distinct OSI properties remains elusive. Intriguingly, when comparing the EM density maps of the CaV1.1, CaV2.2, and CaV2.3 complexes, we observed that AID displayed a blurred density in CaV1.1 but was clearly resolved in CaV2.2 and CaV2.3 (Supplementary Fig. 7). Previous studies suggested that the AID helix is able to regulate the OSI in CaV channels33,34.
In the structure of CaV2.3, the S6II helix is much longer than that of CaV1.1 and extends into the cytosol. Interestingly, we identified a negatively charged domain 715DEQEEEE721 in the intracellular juxtamembrane region of the S6II helix (S6IINCD) (Fig. 5a, b). Taking a closer look at the structure, this negatively charged region forms electrostatic interactions with the positively charged R590 on S4-S5II, as well as R371 and R378 on the AID (Fig. 5b). Strikingly, the S6IINCD is conserved among P/Q-, N- and R-type channels (Fig. 5c). The equivalent segment of L-type CaV channels contains fewer negatively charged residues, interspersed with some positively charged residues, indicating that the electrostatic interactions between S6IINCD and AID are not present in the L-type CaV channels, which is consistent with structural observations that the AID of CaV1.1 has high motility (Fig. 5c and Supplementary Fig. 7). Additionally, the positively charged R590 and R378 are conserved only in CaV2 channels. R378 is further reverted to a negatively charged glutamate in the CaV1 subfamily, reflecting the coevolutionary linkages within this interaction site (Fig. 5c). We speculate that the charge interactions centered on S6IINCD is critical to regulating the OSI of CaV2 channels.
a Interaction of the AID between the negatively charged domains on S6II (S6IINCD) and S4-S5II. The CaV2.3 α1E subunit is shown as cartoon. The S6IINCD is overlaid as red spheres. b Zoomed-in view of the side-chain interactions among S6IINCD, AID, and S4-S5II. The S6IINCD (715DEQEEEE721) is shown as sticks. Residues involved in charge interactions on AID and S4-S5II are also shown as sticks. Negatively charged residues within the S6IINCD are highlighted using red labels. c Sequence alignment of the interaction sites among CaV1 and CaV2 members. Secondary structures are labeled over the sequences. Mutation sites are indicated by arrows. Residues with positively- and negatively charged sidechains are highlighted by blue and red, respectively. d Ratio of open-state inactivation (R200) at 10-mV test pulses, measured using the mean current at the end of the 200-ms test pulse divided by the peak amplitude. WT, n = 6; CaV2.3NQ, n = 10; R590E, n = 6; R590Q, n = 10; R378E, n = 6; R378Q, n = 9; R371E, n = 6; R371Q, n = 9. Data are plotted as box plots. The box encompasses the interquartile range (25th–75th percentile). Whiskers illustrates the minima and maxima of the values. Mean values and medians are indicated using plus signs and dashes, respectively. Significances were determined using two-sided, unpaired t test. P values, CaV2.3 WT vs. mutants; 0.01 (CaV2.3NQ), 0.005 (R590E), 0.02 (R590Q), 0.004 (R371E), and 0.004 (R378Q). n biological independent cells.
To validate our hypothesis, we mutated the key residues to disrupt electrostatic interactions cross-linking the S6II, S4-S5, and AID helices, including replacing 715DEQEEEE721 with 715NNQNNNN721 (CaV2.3NQ), R590Q, R378Q/E, and R371Q/E (Fig. 5d and Supplementary Fig. 8). We employed Ba2+ as the charge carrier in our whole-cell patch-clamp analysis to exclude the effects of calcium-dependent inactivation (CDI) of the CaV2.3 channels. The CaV2.3NQ mutant mediates a current that decays much more slowly than the wild-type CaV2.3 during a 200-ms test pulse (Supplementary Fig. 8a, b), suggesting that OSI is remarkably decreased. To quantify the OSI of the CaV2.3 mutants, we employed the R200 value as an indicator, which is calculated by the mean current density at the end of the 200-ms test pulse divided by the peak amplitude (Fig. 5d and Supplementary Fig. 8c). Specifically, the mean value of R200 increased from 0.17 ± 0.01 in the wild-type CaV2.3 to 0.31 ± 0.03 in CaV2.3NQ under the test pulse holding at 10 mV (Fig. 5d and Supplementary Fig. 8c). Moreover, R590E and R590Q also exhibited remarkably suppressed OSI, with increased R200 values of 0.35 ± 0.04 and 0.27 ± 0.03, respectively (10-mV test pulse) (Fig. 5d and Supplementary Fig. 8c), suggesting that the R590-S6IINCD interaction plays an essential role in the development of OSI in CaV2 channels. In contrast, mutants on the AID side show complicated effects on OSI kinetics (Fig. 5d and Supplementary Fig. 8c). In particular, R378Q displayed an enhanced OSI, with a decreased R200 value of 0.11 ± 0.02 (10-mV test pulse). R371E, which is located in the adjacent region of R378, exhibited an enhanced OSI as well, displaying an R200 value of 0.09 ± 0.01 (10-mV test pulses). Nevertheless, OSI of the R371Q and R378E mutants do not show a significant difference with that of the WT CaV2.3 channel (Fig. 5d and Supplementary Fig. 8c). Moreover, the time course of channel inactivation could be well fitted by a single exponential. Conclusions drawn using time constants are nearly identical to those using R200 values (Supplementary Fig. 8d), further supporting that S6IINCD, R371 (AID), R378 (AID), and R590 (S4-S5II) play important roles in OSI modulation. However, the interaction between the AID and S6IINCD could go beyond the current structures of CaV channels and is worth investigating in future studies.
Methods
Expression and protein purification of the human CaV2.3 complex
Full-length CaV2.3 α1E (CACNA1E), α2δ1 (CACNA2D1), and β1 (CACB1) were amplified from a human cDNA library and subcloned into pEG BacMam vectors. To detect the expression and assembly levels of the CaV2.3 complex, a superfolder GFP, an mCherry, and an mKalama tag were fused to the C-terminal, N-terminal, and N-terminal regions of the CaV2.3 α1, α2δ1, and β1 subunits, respectively. Twin-Strep tags were tandemly inserted into the C-terminal and N-terminal of the α1 and α2δ1 subunits, respectively. Primers are provided in Supplementary Table 2. The Bac-to-Bac baculovirus system (Invitrogen, USA) was used to conduct protein expression in HEK 293-F cells (Thermofisher, 11625019) following the manufacturer’s protocol. The bacmids were prepared using DH10Bac competent cells, and P1 viruses were generated from Sf9 cells (Thermofisher, 10902096) after bacmid transfection. P2 viruses (1%, v/v) were used to infect HEK 293-F cells supplemented with 1% (v/v) fetal bovine serum. The cells were cultured at 37 °C and 5% CO2 for 12 h before the addition of 10 mM sodium butyrate to the medium. The cells were cultured at 30 °C and 5% CO2 for another 48 h before harvest. No authentication was performed for the HEK 293-F or the Sf9 cell line. No Mycoplasma contamination was observed.
The cell pellets were resuspended at 4 °C using Buffer W containing 20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol (β-ME), 2 μg/mL aprotinin, 1.4 μg/mL leupeptin, and 0.5 μg/mL pepstatin A (MedChemExpress, USA) by a Dounce homogenizer, followed by centrifugation at 110,000 × g for 1 h to collect the membrane. The membrane was resuspended again using Buffer W and solubilized by the addition of 1% (w/v) n-dodecyl-β-D-maltoside (DDM) (Anatrace, USA), 0.15% (w/v) cholesteryl hemisuccinate (CHS) (Anatrace, USA), 2 mM adenosine triphosphate (ATP) and 5 mM MgCl2 on a rotating mixer at 4 °C for 2 h. The addition of ATP and MgCl2 is to remove associated heat shock proteins. The insoluble debris of cells was removed by another centrifugation at 110,000 × g for 1 h. The supernatant was passed through a 0.22 μm filter (Millipore, USA) before being loaded into 6 mL Streptactin Beads 4FF (Smart-Lifesciences, China). The resin was washed using 6 column volumes of Buffer W1 (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-ME, 0.03% (w/v) glycol-diosgenin (GDN) (Anatrace, USA), 2 mM ATP, and 5 mM MgCl2). The purified CaV2.3 complex was eluted using 15 mL elution buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-ME, 0.03% (w/v) GDN, and 5 mM d-Desthiobiotin (Sigma-Aldrich, USA)) and concentrated to 1 mL using a 100 kDa MWCO Amicon (Millipore, USA). The concentrated protein sample was subjected to further size-exclusion chromatography (SEC) by a Superose 6 Increase 10/300 GL gel filtration column (GE Healthcare, USA) using a flow rate of 0.3 mL/min and a running buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-ME, and 0.01% (w/v) GDN (Anatrace, USA). The monodispersed peak fraction within 12–13.5 mL was pooled and concentrated to 5 mg/mL before preparing the cryo-EM grids.
Cryo-EM sample preparation and data collection
Holey carbon grids (Au R1.2/1.3 300 mesh) (Quantifoil Micro Tools, Germany) were glow-discharged using H2 and O2 for 60 s before being loaded with 2.5 μL purified CaV2.3 complex. The grids were automatically blotted for 4 s at 4 °C and 100% humidity and flash-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific, USA). Cryo-EM data were collected using a 300 kV Titan Krios G2 (Thermo Fisher Scientific, USA) equipped with a K2 Summit direct electron detector (Gatan, USA) and a GIF Quantum LS energy filter (Gatan, USA). The dose rate was set to ~9.2 e−/(pixel*s), and the energy filter slit width was set to 20 eV. A total exposure time of 6.72 s was dose-fractioned into 32 frames. A nominal magnification of ×130,000 was used, resulting in a calibrated super-resolution pixel size of 0.52 Å on images. SerialEM49 was used to automatically acquire the movie stacks. The nominal defocus range was set from –1.2 μm to –2.2 μm.
Cryo-EM data processing
Motion correction was performed on 2096 movie stacks using MotionCor250 with 5 × 5 patches, generating dose-weighted micrographs. The parameters of the contrast transfer function (CTF) were estimated using Gctf51. Particles were initially picked using the blob picker in cryoSPARC52, followed by 2D classifications to produce 2D templates and ab initio reconstruction to generate an initial reference map. Another round of particle picking was conducted using Template Picker in cryoSPARC, generating a dataset of 787,518 particles that were used for further processing. All data processing steps were performed in RELION-3.153 unless otherwise specified. A round of multi-reference 3D classification was conducted against one good and 4 biased references, generating 5 classes. Class 5 (75.2%), which was calculated using the good reference, displayed a classical shape of CaV complexes featuring a transmembrane subunit and two soluble subunits residing on both sides of the micelle. Particles from class 5 were re-extracted and subjected to another round of 3D classification, resulting in 6 classes. Classes 1, 4, and 5 (43.5%) displayed well-resolved structural features, including continuous transmembrane helices of the α1E subunit and secondary structures within the α2δ1 and β1 subunits. To improve the map quality, Bayesian polish and CTF refinement were then conducted. The following 3D auto refinement generated a 3.1-Å map. The particle dataset was then imported back to cryoSPARC, where the final map was generated by Non-uniform (NU) refinement, which was reported at 3.1 Å according to the golden-standard Fourier shell correlation (GSFSC) criterion.
Model building
The cryo-EM map of CaV2.3 was reported at near-atomic resolution, which enabled us to reliably build and refine the model. The structure of the CaV2.2-α2δ1-β1 complex (PDB ID: 7VFS)28 was selected as the starting model because of the high sequence identity and was docked into the map of CaV2.3 complexes using UCSF Chimera54. Sidechains of the a1 subunit were manually mutated according to the sequence alignment between CaV2.3 and CaV2.2 and adjusted according to the EM density using Coot55. Sidechains of α2δ1 were also manually adjusted according to the EM density. β1 was initially fit into the EM maps as a rigid body and manually refined against a low-resolution map of the CaV2.3 complex in Coot due to local structural heterogeneity. The manually adjusted models were then automatically refined against the cryo-EM maps using the integrated Real Space Refinement program within the PHENIX software package56. Model stereochemistry was also evaluated using the Comprehensive validation (cryo-EM) tool in PHENIX.
All the figures were prepared using Open-Source PyMOL (Schrödinger, USA), UCSF Chimera54, or UCSF ChimeraX57.
Whole-cell voltage-clamp recordings of CaV2.3 channels in HEK 293-T cells
HEK 293-T cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) supplemented with 15% (v/v) fetal bovine serum (FBS) (PAN-Biotech, Germany) at 37 °C with 5% CO2. The cells were grown in culture dishes (d = 3.5 cm) (Thermo Fisher Scientific, USA) for 24 h and then transiently transfected with 2 μg of control or mutant plasmid expressing the human R-type CaV2.3 calcium channel complex (CaV2.3 α1E, β1, α2δ1) using 1.2 μg of Lipofectamine 2000 Reagent (Thermo Fisher Scientific, USA). Patch-clamp experiments were performed 12 to 24 h post-transfection at room temperature (21~25 °C) as described previously. Briefly, cells were placed on a glass chamber containing 105 mM NaCl, 10 mM BaCl2, 10 mM HEPES, 10 mM D-glucose, 30 mM TEA-Cl, 1 mM MgCl2, and 5 mM CsCl (pH = 7.3 with NaOH and an osmolarity of ~310 mosmol−1). Whole-cell voltage-clamp recordings were made from isolated, GFP-positive cells using 1.5~2.5 MΩ fire-polished pipettes (Sutter Instrument, USA) filled with standard internal solution containing 135 mM K-gluconate, 10 mM HEPES, 5 mM EGTA, 2 mM MgCl2, 5 mM NaCl, and 4 mM Mg-ATP (pH = 7.2 with CsOH and osmolarity of ~295 mosmol−1). Whole-cell currents were recorded using an EPC-10 amplifier (HEKA Electronik, Germany) at a 20 kHz sample rate and were low-pass filtered at 5 kHz. The series resistance was 2~4.5 MΩ and was compensated 80~90%. The data were acquired by the PatchMaster program (HEKA Electronik, Germany).
To obtain activation curves of CaV2.3 channels, cells were held at –100 mV, and then a series of 200-ms voltage steps from –60 mV to +50 mV in 5-mV increments were applied. The steady-state inactivation properties of CaV2.3 channels were assessed with 10-s holding voltages ranging from –100 mV to –15 mV (5-mV increments) followed by a 135-ms test pulse at +10 mV. To assess the time-dependent recovery from CSI, cells were depolarized to –40 mV (pre-pulse) for 1500 ms to allow CaV2.3 channels to enter CSI, and recovery hyperpolarization steps to –100 mV were applied for the indicated period (4 ms–2048 ms), followed by a 35-ms test pulse at +10 mV. To assess the cumulative inactivation of CaV2.3 channels in response to AP trains, the cells were held at –100 mV, and then the physiologically relevant AP trains was applied. The AP trains used to stimulate the HEK 293-T cells were recorded from a mouse hippocampal CA1 pyramidal neuron after current injection in the whole-cell current-clamp mode58. The spike pattern contained 13 action potentials in 2 s (mean frequency = 6.5 Hz). The percentage inactivation of CaV2.3 channels was calculated from the first spike eliciting maximal current to the other spikes in the AP trains. To analyze the extent of OSI, the ratio of remaining currents at 200 ms post-depolarization and the peak currents was calculated.
Electrophysiological data analysis
All data are reported as the mean ± SEM. Data analyses were performed using Origin 2019b (OriginLab, USA) and Prism 9 (GraphPad, USA).
Steady-state activation curves were generated using a Boltzmann Eq. (1).
where G is the conductance, calculated by G = I/(V−Vrev), where I is the current at the test potential and Vrev is the reversal potential; Gmax is the maximal conductance of the CaV2.3 channel during the test pulse; V is the test potential; V0.5 is the half-maximal activation potential; and k is the slope factor.
Steady-state activation and inactivation curves were generated using a Boltzmann Eq. (2).
where I is the current at the indicated test pulse; Imax is the maximal current of CaV2.3 activation during the test pulse; V is the test potential; V0.5 is the half-maximal inactivation potential; and k is the slope factor.
Recovery curves from CSI were calculated from the results of 7–9 independent experiments where a series of recovery traces from inactivation time points were acquired. The data were fit using a single exponential of the following Eq. (3).
where I is the current at the indicated intervals; Imax is the current at 2048 ms; y0 is the remaining current at –40 mV for 1500 ms; t is the indicated hyperpolarization time; and τ is the time constant of recovery from CSI.
Statistical significance (p < 0.05) was determined using unpaired Student’s t tests or one-way ANOVA with Tukey’s post hoc test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding authors upon reasonable request. The cryo-EM density map of the CaV2.3-α2δ1-β1 complex has been deposited in the Electron Microscopy Data Bank (EMDB) under the accession code EMD-33285. The coordinate for the CaV2.3 complex have been deposited in the Protein Data Bank (PDB) under the PDB ID 7XLQ. The starting model used to build CaV2.3-α2δ1-β1 is available in the PDB under the PDB ID 7VFS (CaV2.2-α2δ1-β1). DNA sequences of human CaV2.3 α1E (CACNA1E isoform 1), human α2δ1 (CACNA2D1), and human β1 (CACB1) are available in the Universal Protein Resource (UniProt) databases under accession codes Q15878-1 [https://www.uniprot.org/uniprotkb/Q15878/entry], P54289, and Q02641, respectively. Source data including an uncropped scan of the gel image and values of all electrophysiological experiment graphs are provided as Source Data Files. Source data are provided in this paper.
References
Catterall, W. A. Voltage-gated calcium channels. Cold Spring Harb. Perspect. Biol. 3, a003947 (2011).
Zamponi, G. W., Striessnig, J., Koschak, A. & Dolphin, A. C. The physiology, pathology, and pharmacology of voltage-gated calcium channels and their future therapeutic potential. Pharm. Rev. 67, 821–870 (2015).
Dolphin, A. C. A short history of voltage-gated calcium channels. Br. J. Pharm. 147, S56–S62 (2006).
Yang, S. N. & Berggren, P. O. The role of voltage-gated calcium channels in pancreatic beta-cell physiology and pathophysiology. Endocr. Rev. 27, 621–676 (2006).
Van Goor, F., Zivadinovic, D., Martinez-Fuentes, A. J. & Stojilkovic, S. S. Dependence of pituitary hormone secretion on the pattern of spontaneous voltage-gated calcium influx. Cell type-specific action potential secretion coupling. J. Biol. Chem. 276, 33840–33846 (2001).
Tsien, R. W., Lipscombe, D., Madison, D. V., Bley, K. R. & Fox, A. P. Multiple types of neuronal calcium channels and their selective modulation. Trends Neurosci. 11, 431–438 (1988).
Spafford, J. D. & Zamponi, G. W. Functional interactions between presynaptic calcium channels and the neurotransmitter release machinery. Curr. Opin. Neurobiol. 13, 308–314 (2003).
Tsien, R. W. Calcium channels in excitable cell membranes. Annu Rev. Physiol. 45, 341–358 (1983).
Takahashi, M., Seagar, M. J., Jones, J. F., Reber, B. F. & Catterall, W. A. Subunit structure of dihydropyridine-sensitive calcium channels from skeletal muscle. Proc. Natl Acad. Sci. USA 84, 5478–5482 (1987).
Wormuth, C. et al. Review: Cav2.3 R-type voltage-gated Ca(2+) channels - functional implications in convulsive and non-convulsive seizure activity. Open Neurol. J. 10, 99–126 (2016).
Lee, S. C. et al. Molecular basis of R-type calcium channels in central amygdala neurons of the mouse. Proc. Natl Acad. Sci. USA 99, 3276–3281 (2002).
Schneider, T., Neumaier, F., Hescheler, J. & Alpdogan, S. Cav2.3 R-type calcium channels: from its discovery to pathogenic de novo CACNA1E variants: a historical perspective. Pflug. Arch. 472, 811–816 (2020).
Randall, A. & Tsien, R. W. Pharmacological dissection of multiple types of Ca2+ channel currents in rat cerebellar granule neurons. J. Neurosci. 15, 2995–3012 (1995).
Patil, P. G., Brody, D. L. & Yue, D. T. Preferential closed-state inactivation of neuronal calcium channels. Neuron 20, 1027–1038 (1998).
Huang, C. et al. Proteomic analysis of olfactory bulb suggests CACNA1E as a promoter of CREB signaling in microbiota-induced depression. J. Proteom. 194, 132–147 (2019).
Dietrich, D. et al. Functional specialization of presynaptic Cav2.3 Ca2+ channels. Neuron 39, 483–496 (2003).
Zaman, T. et al. Cav2.3 channels are critical for oscillatory burst discharges in the reticular thalamus and absence epilepsy. Neuron 70, 95–108 (2011).
Benkert, J. et al. Cav2.3 channels contribute to dopaminergic neuron loss in a model of Parkinson’s disease. Nat. Commun. 10, 5094 (2019).
Bloodgood, B. L. & Sabatini, B. L. Nonlinear regulation of unitary synaptic signals by CaV(2.3) voltage-sensitive calcium channels located in dendritic spines. Neuron 53, 249–260 (2007).
Kuzmiski, J. B., Barr, W., Zamponi, G. W. & MacVicar, B. A. Topiramate inhibits the initiation of plateau potentials in CA1 neurons by depressing R-type calcium channels. Epilepsia 46, 481–489 (2005).
Carvill, G. L. Calcium channel dysfunction in epilepsy: gain of CACNA1E. Epilepsy Curr. 19, 199–201 (2019).
Hainsworth, A. H., McNaughton, N. C. L., Pereverzev, A., Schneider, T. & Randall, A. D. Actions of sipatrigine, 202W92 and lamotrigine on R-type and T-type Ca2+ channel currents. Eur. J. Pharmacol. 467, 77–80 (2003).
Dibue-Adjei, M. et al. Cav2.3 (R-Type) calcium channels are critical for mediating anticonvulsive and neuroprotective properties of lamotrigine in vivo. Cell Physiol. Biochem. 44, 935–947 (2017).
Helbig, K. L. et al. De novo pathogenic variants in CACNA1E cause developmental and epileptic encephalopathy with contractures, macrocephaly, and dyskinesias. Am. J. Hum. Genet. 103, 666–678 (2018).
Wu, J. et al. Structure of the voltage-gated calcium channel Ca(v)1.1 at 3.6 A resolution. Nature 537, 191–196 (2016).
Zhao, Y. et al. Molecular basis for ligand modulation of a mammalian voltage-gated Ca(2+) channel. Cell 177, 1495–1506 e1412 (2019).
Gao, S., Yao, X. & Yan, N. Structure of human Cav2.2 channel blocked by the painkiller ziconotide. Nature 596, 143–147 (2021).
Dong, Y. et al. Closed-state inactivation and pore-blocker modulation mechanisms of human CaV2.2. Cell Rep. 37, 109931 (2021).
Zhao, Y. et al. Cryo-EM structures of apo and antagonist-bound human Cav3.1. Nature 576, 492–497 (2019).
He, L. et al. Structure, gating, and pharmacology of human CaV3.3 channel. Nat. Commun. 13, https://doi.org/10.1038/s41467-022-29728-0 (2022).
Striessnig, J. Voltage-gated ca2+-channel alpha 1-subunit de novo missense mutations: gain or loss of function - implications for potential therapies. Front. Synaptic Neurosci. 13, 634760 (2021).
McDavid, S. & Currie, K. P. G-proteins modulate cumulative inactivation of N-type (Cav2.2) calcium channels. J. Neurosci. 26, 13373–13383 (2006).
Stotz, S. C., Hamid, J., Spaetgens, R. L., Jarvis, S. E. & Zamponi, G. W. Fast inactivation of voltage-dependent calcium channels. A hinged-lid mechanism? J. Biol. Chem. 275, 24575–24582 (2000).
Berrou, L., Bernatchez, G. & Parent, L. Molecular determinants of inactivation within the I-II linker of α1E (CaV2.3) calcium channels. Biophys. J. 80, 215–228 (2001).
He, M., Bodi, I., Mikala, G. & Schwartz, A. Motif III S5 of L-type calcium channels is involved in the dihydropyridine binding site - a combined radioligand binding and electrophysiological study. J. Biol. Chem. 272, 2629–2633 (1997).
Pantazis, A., Savalli, N., Sigg, D., Neely, A. & Olcese, R. Functional heterogeneity of the four voltage sensors of a human L-type calcium channel. Proc. Natl Acad. Sci. USA 111, 18381–18386 (2014).
Xia, F. et al. Inhibition of cholesterol biosynthesis impairs insulin secretion and voltage-gated calcium channel function in pancreatic beta-cells. Endocrinology 149, 5136–5145 (2008).
Purcell, E. K., Liu, L., Thomas, P. V. & Duncan, R. K. Cholesterol influences voltage-gated calcium channels and BK-type potassium channels in auditory hair cells. PLoS One 6, e26289 (2011).
Yao, X. et al. Structures of the R-type human Ca(v)2.3 channel reveal conformational crosstalk of the intracellular segments. Nat. Commun. 13, 7358 (2022).
Lin, Z., Haus, S., Edgerton, J. & Lipscombe, D. Identification of functionally distinct isoforms of the N-type Ca2+ channel in rat sympathetic ganglia and brain. Neuron 18, 153–166 (1997).
Zhong, H., Li, B., Scheuer, T. & Catterall, W. A. Control of gating mode by a single amino acid residue in transmembrane segment IS3 of the N-type Ca2+ channel. Proc. Natl Acad. Sci. USA 98, 4705–4709 (2001).
Lin, Y., McDonough, S. I. & Lipscombe, D. Alternative splicing in the voltage-sensing region of N-Type CaV2.2 channels modulates channel kinetics. J. Neurophysiol. 92, 2820–2830 (2004).
Pereverzev, A. et al. Alternate splicing in the cytosolic II-III loop and the carboxy terminus of human E-type voltage-gated Ca(2+) channels: electrophysiological characterization of isoforms. Mol. Cell Neurosci. 21, 352–365 (2002).
Vajna, R. et al. New isoform of the neuronal Ca2+ channel alpha1E subunit in islets of Langerhans and kidney–distribution of voltage-gated Ca2+ channel alpha1 subunits in cell lines and tissues. Eur. J. Biochem. 257, 274–285 (1998).
Armstrong, C. M. Na channel inactivation from open and closed states. Proc. Natl Acad. Sci. USA 103, 17991–17996 (2006).
Lipscombe, D., Helton, T. D. & Xu, W. L-type calcium channels: the low down. J. Neurophysiol. 92, 2633–2641 (2004).
Gambeta, E., Gandini, M. A., Souza, I. A., Ferron, L. & Zamponi, G. W. A CACNA1A variant associated with trigeminal neuralgia alters the gating of Cav2.1 channels. Mol. Brain 14, 4 (2021).
Groen, J. L. et al. CACNA1B mutation is linked to unique myoclonus-dystonia syndrome. Hum. Mol. Genet. 24, 987–993 (2015).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Scheres, S. H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Pettersen, E. F. et al. UCSF chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr. Sect. D. Struct. Biol. 75, 861–877 (2019).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Liu, Y. Q. et al. CDYL suppresses epileptogenesis in mice through repression of axonal Nav1.6 sodium channel expression. Nat. Commun. 8, 355 (2017).
Acknowledgements
We thank Xiaojun Huang, Xujing Li, Lihong Chen, and other staff members at the Center for Biological Imaging (CBI), Core Facilities for Protein Science at the Institute of Biophysics, Chinese Academy of Science for their support in cryo-EM data collection. We thank Yan Wu for his research assistance. This work is funded by the National Key Research and Development Program of China (Grant No. 2021YFA1301501 to Y.Z.), the Chinese Academy of Sciences Strategic Priority Research Program (Grant No. XDB37030304 to Y.Z.; Grant No. XDB37030301 to X.C.Z.), the National Natural Science Foundation of China (Grant No. 92157102 to Y.Z.; Grant No. 31971134 to X.C.Z.; Grant No. 81371432 to Z.H., and U20A6005 to J.S.), Chinese National Programs for Brain Science and Brain-like Intelligence Technology (Grant No. 2022ZD0205800 to Y.Z.; Grant No. 2021ZD0202102 to Z.H.), and National Science and Technology Major Project (2021ZD0202501 to J.S.).
Author information
Authors and Affiliations
Contributions
Y.Z. conceived the project and supervised the research. Y.G. and Y.Q. carried out molecular cloning experiments. Y.G. expressed and purified protein samples. Y.D. prepared samples for cryo-EM study. Y.G. and B.Z. carried out cryo-EM data collection. Y.G. processed the cryo-EM data. Y.G. built and refined the atomic model. Y.Z., Y.G., X.C.Z., and Y.W. analyzed the structure. Y.Z., Z.H., Y.G., S.X., and J.S. designed the electrophysiological experiments. S.X., X.C., H.X., C.P., and S.L. conducted the whole-cell voltage patch-clamp analysis. Y.G. wrote the original draft of the manuscript and prepared the figures. Y.Z., Y.G., and X.C.Z. edited the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks SeCheol Oh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gao, Y., Xu, S., Cui, X. et al. Molecular insights into the gating mechanisms of voltage-gated calcium channel CaV2.3. Nat Commun 14, 516 (2023). https://doi.org/10.1038/s41467-023-36260-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-023-36260-2
- Springer Nature Limited