Abstract
Many statistical methods are available for genomic selection (GS) through which genetic values of quantitative traits are predicted for plants and animals using whole-genome SNP data. A large number of predictors with much fewer subjects become a major computational challenge in GS. Principal components regression (PCR) and its derivative, i.e., partial least squares regression (PLSR), provide a solution through dimensionality reduction. In this study, we show that PCR can perform better than PLSR in cross validation. PCR often requires extracting more components to achieve the maximum predictive ability than PLSR and thus may be associated with a higher computational cost. However, application of the HAT method (a strategy of describing the relationship between the fitted and observed response variables with a hat matrix) to PCR circumvents conventional cross validation in testing predictive ability, resulting in substantially improved computational efficiency over PLSR where cross validation is mandatory. Advantages of PCR over PLSR are illustrated with a simulated trait of a hypothetical population and four agronomical traits of a rice population. The benefit of using PCR in genomic selection is further demonstrated in an effort to predict 1000 metabolomic traits and 24,973 transcriptomic traits in the same rice population.
Similar content being viewed by others
Introduction
Genomic selection (GS) is a new form of marker-assisted selection (MAS) in modern animal and plant breeding programs (Hayes et al. 2009; Heffner et al. 2009; Xu et al. 2014). It differs from the conventional MAS in that the marker detection step has been skipped and all markers are included in a prediction model regardless their effects (Xu et al. 2014). Many statistical methods are available for GS, including popular Bayesian methods (Bayes B) (Meuwissen et al. 2001), best linear unbiased prediction (BLUP) (VanRaden 2008), kernel-based methods (Gianola and De Los Campos 2008; Gianola et al. 2006), least absolute shrinkage and selection operator (LASSO) (Tibshirani 1996), and partial least squares regression (PLSR) (Colombani et al. 2012). There is no such a method as universally optimal for all traits in all populations. A method performing well for one trait may predict poorly for another trait, depending on the nature of the model and the genetic architecture of the trait. Therefore, all available methods should be tested if convenient software packages are available.
A large number of predictors with much fewer subjects become a major computational challenge in GS (De los Campos et al. 2013). Principal component regression (PCR, Hotelling 1957; Jeffers 1967; Kendall 1957; Wentzell and Montoto 2003) and its derivative, i.e., PLSR (Wold 1966), provide intuitive solutions through dimensionality reduction. Both methods are commonly used in the field of chemometrics (Frank and Friedman, 1993; Wentzell and Montoto 2003). Recently, the two methods have been introduced to breeding and GS studies (Colombani et al. 2012; Coster et al. 2010; Long et al. 2011; Lorenz et al. 2011). PCR is less popular than PLSR due to the unsupervised nature of extracting principal components only from predictor variables. This is in contrast to the supervised counterpart PLSR (Wentzell and Montoto 2003) where extracting components depends also on response variables. Both PCR and PLSR extract independent or orthogonal components from predictor variables to fit one or more response variables, but PLSR also maximizes the covariance between the resultant components and the response variables (Lafaye De Micheaux et al. 2017), which explains why PLSR is called a supervised method. As a result, PLSR is believed to be more efficient than PCR. However, experienced PLSR and PCR users often report that the two prediction methods rarely produce results with noticeable difference, except that PCR always requires extracting substantially more components to achieve the same level of predictive ability as PLSR (Andrew and Worsfold 1994; Hemmateenejad et al. 2007; Solberg et al. 2009; Wentzell and Montoto 2003). More components in PCR mean higher computational cost when samples are large and massive number of predictive variables are considered.
The unsupervised nature of PCR is not necessarily undesirable. We can take advantage of this nature and convert it into a useful tool to improve computational efficiency of prediction. As constructing the principal components (scores) does not involve the response variables, PCR allows an explicit method to assess model predictive ability. Such a method is called the HAT method, which is a strategy of describing the relationship between the fitted and observed response variables with a hat matrix and is routinely used in linear regression for diagnosis of influential observations and outliers (Cook 1977; 1979). The HAT method in linear regression can replace cross validation to evaluate model predictive ability. Considering a linear regression model y = Xβ + e, the HAT method utilizes a hat matrix to describe the linear relationship between the fitted and observed response variables, i.e., \(\hat y = Hy\), where the hat matrix is defined as \(H = X(X^TX)^{ - 1}X^T\). The predicted residual error sum of squares (PRESS) is defined as
where h jj is the jth diagonal element of the hat matrix (Cook 1979; Hoaglin and Welsch 1978). The PRESS calculated from the above HAT method is exactly the same as that of the leave-one-out (LOO) cross validation. In this study, we propose to develop a hat matrix for PCR to avoid time consuming cross validation for model assessment. An approximate HAT method has been developed for BLUP prediction (Xu 2017). In this study, a similar HAT method was developed for PCR. The hat matrix for PCR is not the same as the hat matrix for BLUP. In addition, using the HAT method of PCR for GS has not been reported in previous literatures. As PLSR involves the response variables in construction of the orthogonal components, a comparable HAT method is not available for PLSR. When PCR is implemented with the HAT method, it can outcompete PLSR in both predictive ability and computational efficiency. Another advantage of PCR over PLSR is that one only needs to perform eigenvalue decomposition once on the predictors prior to prediction for all traits in PCR; while PLSR must conduct matrix algebra to extract regression components for every trait if one trait is analyzed at a time as the resultant components from the decomposition are different across traits.
Materials and methods
Singular value decomposition and eigenvalue decomposition
Let n be the number of observations and m be the number of variables (features). In GS, m is often many times larger than n. Let X n×m be an n×m feature matrix and y n×1 be an n×1 vector of responses. The PCR requires eigenvalue decomposition for matrix \(X_{m \times n}^TX_{n \times m}\) (Shlens 2014), which is an m×m matrix and may not be decomposed easily when m is very large. However, the common practice in PCR is to use singular value decomposition (SVD) for the feature matrix X n×m as (Bordier et al. 2011; Mandel 1982; Shlens 2014)
where the columns of U are called the left singular vectors, the columns of V are called the right singular vectors and Δ is a rectangular diagonal matrix with non-negative real numbers on the diagonal. These diagonal elements are called singular values. How do we connect singular value decomposition of X with eigenvalue decomposition of XTX?
Let us replace X in XTX by its SVD (denoted by X = UΔVT) and perform the following matrix multiplication,
Clearly, the right singular vector matrix of SVD for X is identical to the eigenvector matrix for XTX. In addition, the eigenvalues of XTX are the squares of singular values of X as indicated by D = ΔTΔ. Therefore, we can simply perform SVD for the feature matrix X to obtain the eigenvectors and eigenvalues for matrix XTX. When m is extremely large, singular value decomposition for X may still be expensive in terms of computational load. Therefore, we propose to perform eigenvalue decomposition for matrix XXT which is an n×n matrix, and then convert its eigenvectors to the eigenvectors of matrix XTX which is an m×m matrix. We first need to show that the two matrices, XTX and XXT, have the same eigenvalues,
where D = ΔΔT. The diagonal non-zero elements of ΔΔT are equal to those of ΔTΔ. Therefore we conclude that the eigenvalue decomposition for XXT is \(XX^T = UDU^T\). It can be shown that the eigenvectors of XTX can be written as \(V = X^TUD^{ - 1/2}\). We can prove this by showing
Therefore, in principal component analysis, it is much more efficient to perform eigenvalue decomposition on XXT (a lower ranking matrix) first and then find the eigenvectors of XTX (a higher ranking matrix) using V = XTUD−1/2. In general, the score matrix can be obtained by the following simple expression
We can treat the scores in principal component analysis as the weighted eigenvector of XXT.
Principal component regression
The eigenvector matrix V is also called X loading and it is an m×m matrix. Let p (p ≤ n) be the number of components chosen by the investigator. The loading matrix is then truncated as an m×p matrix. Define an n×p score matrix by \(T_{n \times p} = X_{n \times m}V_{m \times p}\). The principal component regression is to perform multiple regression of y on the score matrix, y = Tb+e. The estimated coefficients of scores can be expressed as
Because columns of the score matrix are orthogonal, we get \((T^TT)^{ - 1} = (V^TX^TXV)^{ - 1} = D^{ - 1}\). Therefore, \(\widehat b = D^{ - 1}T^Ty\), which is easy to calculate because D is diagonal. Substituting T = XV into Eq. (7), we get
Recall that the original prediction model is y = Xβ + e and we have \(\hat \beta = (X^TX)^{ - 1}X^Ty\) based on the least squares estimation. Instead of estimating β directly using the above model, in the PCR analysis, we estimate b first and then convert b into β using \(\hat \beta {\mathrm{ = }}V\widehat b\) (Greenberg 1975; Næs and Martens 1988).
Cross validation (CV) and the HAT prediction
The predicted phenotypic values are \(\hat y = T\widehat b{\mathrm{ = }}TD^{ - 1}T^Ty = Hy\). Therefore, the HAT matrix is defined as\(H = TD^{ - 1}T^T\). Let \(r = \hat y - y = (I - H)y\) be the residual errors. The PRESS is \({\mathrm{PRESS = }}\mathop {\sum}\nolimits_{j = 1}^n {r_j^2/(1 - h_{jj})^2}\), where h jj is the jth diagonal element of the HAT matrix H. Let \({\mathrm{SS = }}\mathop {\sum}\nolimits_{j = 1}^n {(y_j - \bar y)^2}\) be the total sum of squares. If y is already standardized, then SS = n − 1. The predictive ability is \(R_{{\mathrm{HAT}}}^2 = 1 - {\mathrm{PRESS/SS}}\). The residual error sum of squares (RESS) is calculated as \(\mathop {\sum}\nolimits_{j = 1}^n {r_j^2}\). The model goodness of fit is defined as \(R_{{\mathrm{FIT}}}^2 = 1 - {\mathrm{RESS/SS}}\). Note that the model goodness of fit \(R_{{\mathrm{FIT}}}^2\) is not a good measure of model efficiency. Goodness of fit is often close to unity when m is extremely large, a phenomenon called overfitting. The predictive ability \(R_{{\mathrm{HAT}}}^2\) is an appropriate measure of model efficiency. The HAT matrix does not involve y and thus \(R_{{\mathrm{HAT}}}^2\) can replace the predictive ability obtained from the LOO cross validation.
The HAT predictive ability and the predictive ability obtained from 10-fold cross validation for the PCR method will be compared along with the predictive ability obtained from 10-fold cross validation for the PLSR method and the BLUP evaluated from the HAT method (Xu 2017). As variation of predictive ability will occur when the sample is partitioned into ten parts in different ways, we replicated the cross validation experiment ten times by randomly partitioning the sample into ten parts differently each time. The predictive ability reported for each method is the average of predictive ability of ten replicated CV analyses.
Design of a simulation experiment
We simulated 1000 individuals of an F2 family generated from the cross of two inbred lines. Two chromosomes were simulated, each with 2400 centi-Morgan (cM) in length covered by 961 evenly distributed co-dominant markers. The ratio of the three genotypes of each marker, AA, AB, and BB, is 1:2:1, a typical Mendelian ratio. Each genotype was numerically coded as 1, 0, and −1 for AA, AB, and BB, respectively. The distance between two consecutive markers is 2.5 cM. The total number of markers for the entire genome (two chromosomes) is 1922. The numerically coded genotype data are stored in an n×m = 1000×1922 feature matrix X. The first chromosome contains 20 quantitative trait loci (QTL) with effects and positions shown in Fig. 1a as well as in Supplementary Table S1. The second chromosome contains no QTL and this ‘empty’ chromosome was used to control type 1 error in a separate QTL mapping study. Phenotypes of the 1000 individuals were generated using
where β0 = 10 is the intercept, β k is the effect assigned to marker k, and e is the residual error vector following an \(N(0,I\sigma _E^2)\) distribution with \(\sigma _E^2 = 10\). Of the m = 1922 markers, only 20 randomly selected markers have non-zero effects which add up to a predefined constant termed the total genetic effect (Fig. 1a; Table S1). Theoretically, the total genetic variance collectively contributed by the simulated QTL is \(\sigma _G^2 = 59.4303\). The sum of these individual marker variances is \(\Sigma _g = 46.7806\). The difference between \(\sigma _G^2\) and Σ g is C g = 12.6497, which is caused by linkage (covariance) between the simulated QTL. The heritability of the simulated trait is
Although the total contribution of the 20 QTL to the phenotypic variance is high, contribution from each QTL varies from 0.36 to 14.39% with an average contribution of 3.37% (Table S1).
Two additional simulated data sets, which represent low heritability (30%) case and medium heritability (50%) case, respectively, are also analyzed. The effects of the 20 simulated QTL in these two simulation scenarios are shown in Table S2. Details of the simulation experiment and the theoretical analysis are presented in Supplementary Note S1.
Rice data
Four agronomic traits collected from 210 recombinant inbred lines (RILs) were used for prediction; they are yield per plant (YD), 1000-grain weight (KGW), tiller number per plant (TP), and grain number per plant (GN). The experiment was replicated four times in multiple years and multiple locations (MYML) (Hua et al. 2002; 2003; Xing et al. 2002). The average phenotypic values of each trait across the replicates were used as the response variables. The genomic data are represented by 1619 bins inferred from ~270,000 SNPs of the rice genome (Xie et al. 2010; Yu et al. 2011). All SNPs within a bin have exactly the same segregation pattern (perfect linkage disequilibrium or LD) and thus one SNP from a bin is sufficient to represent the entire bin. Bin genotypes of the 210 RILs were coded as 1 for the Zhenshan 97 genotype and 0 for the Minghui 63 genotype, which are the two parental lines of the 210 RILs.
In addition to the agronomic traits, we also predicted 1000 metabolomic traits measured from the 210 RILs using the 1619 bins of the rice genome. The metabolomic data consisted of 683 metabolites measured from flag leaves and 317 metabolites from germinated seeds (Gong et al. 2013). The data were collected in 2009 and 2010 (two replicates). For metabolic profiling, germinated seeds were sampled in one biological replicate in 2009 and one in 2010, and flag leaves were sampled in two biological replicates in 2009. In both tissues, the expression level of each metabolite was log2 transformed. For each line, we took the average of expression levels measured from the two replicates as the measurements of the metabolites.
We also used transcriptomic data as traits to evaluate the performances of different models in the prediction. The transcriptomic data contained 24,973 gene expression traits measured in tissues sampled from flag leaves for all the 210 RILs (Wang et al. 2014). For each RIL, the RNA samples used for expression profiling were the mixture (1:1) of two replicates of that line. The RNA samples were hybridized with Affymetrix Rice Genome Arrays, and the expression data were preprocessed using Robust Multiarray Average (RMA) suite of Affy package and log2 transformed.
Software
All analyses were performed in R. We developed an R pipeline to implement the genomic prediction using PCR, PLSR, and BLUP. The source code for the pipeline is available at github (https://github.com/JulongWei/GS_PCR). Supplementary Note S2 also provides a detailed instruction for implementing the R pipeline to reproduce the results in our study.
Results
Computing time of eigenvalue decomposition for a hypothetical large data
We used a large simulated data to investigate the computational efficiency of the new eigenvalue decomposition method in comparison to the singular value decomposition algorithm from the svd() function of R and the principal component analysis method from the prcomp() function of R. The simulated sample has n = 5000 observations and m = 100,000 variables stored in matrix X n×m . This data matrix was generated via random draws of n×m independent standardized normal distributions using the following R statement
X = matrix(rnorm(n × m),n,m)
.
The three methods of eigenvalue decomposition are (1) SVD(X), (2) EIGEN(XXT), and (3) PRCOMP(X), where the second method is the one developed in this study. On a desktop computer DELL with a single core of Intel i7-6700K 4.00 GHz CPU, the new method EIGEN(XXT) spent significantly less computational time than the other two methods, i.e., SVD(X) and PRCOMP(X), to complete the eigenvalue decomposition (Table 1).
Analysis of a simulated data set
For a simulated data set generated from the method described in the Methods section, we performed PCR and PLSR predictions and also estimated the marker effects. Figure 1a–c shows the estimated effects of 961 markers on the first chromosome from the two methods along with the true effects. In addition, we also present the estimated effects from the BLUP method (ridge regression) (see Fig. 1d). It is well-known that PCR, PLSR, and BLUP are not suitable for detection of associations (Frank and Friedman 1993). In contrast to the true QTL effects, we do observe severe shrinkages of the effects for all three methods. PCR and PLSR show stronger shrinkage than BLUP. It is surprising that the shrunk estimates of the QTL effects for each method appear to follow a similar pattern to the true effects. Two additional simulations also show similar shrinkage patterns for the three methods (Figure S1 and S2). This phenomenon implies that the three shrinkage methods may be able to detect QTL, which is contrary to the common belief that they are not suitable for association studies.
We now compare the predictive abilities of PCR-HAT, PCR-CV, and PLSR-CV along with BLUP-HAT, where HAT and CV indicate the HAT method and the 10-fold cross validation method, respectively. From the simulated sample, we plotted the predictive ability against the number of components for the three methods (Fig. 2). The maximum predictive ability of the PCR-HAT method is 0.7568 with 204 extracted components. The maximum predictive ability and the number of components needed to achieve this predictive ability are 0.7598 and 225 for the PCR-CV method and are 0.7598 and 7 for the PLSR method, respectively. The three methods have virtually the same predictive ability, but achieved it with quite different numbers of components. The PLSR method only required seven components, in contrast to 204 and 225 for the PCR-HAT and PCR-CV methods, respectively. Of the two PCR-based methods, predictive ability of the HAT method decreased more rapidly than the CV method after reaching the maximum value of predictive ability. Surprisingly, predictive ability of the PLSR method reached a constant value of 0.60 after 100 components are extracted.
Predictive abilities plotted against the number of extracted components for three methods. (1) Principal component regression implemented via the HAT method (PCR-HAT), (2) principal component regression implemented via the 10-fold cross validation method (PCR-CV), and (3) partial least squares regression via 10-fold cross validation (PLSR-CV)
Predictive ability drawn from a 10-fold cross validation varies slightly depending on how the 10 parts of the sample are partitioned. Therefore, we replicated the 10-fold CV ten times with a different way of partitioning the sample each time. The average predictive ability and the average numbers of extracted components for methods PCR-CV and PLSR-CV are presented in Table 2 along with the corresponding numbers for the PCR-HAT method. The conclusion from the replicated CV analysis remains the same as that from one 10-fold CV analysis shown in Fig. 2. Table 2 also shows that the BLUP method has a higher predictive ability than PCR and PLSR for the simulated data. The standard deviation of the 10 replicates appears to be small for both the PCR and PLSR methods. Two-sample t-test indicated there is no significant difference in predictive ability between PCR-CV and PLSR-CV (p = 0.3867). The predictive ability plots for all replicates of the two methods (PCR-CV and PLSR-CV) are shown in Fig. 3. The two methods were further compared with two additional sets of β values, with 30% (low heritability) and 50% (medium heritability) of phenotypic variation being explained by the simulated QTL in two scenarios, respectively. The results showed that PCR consistently had higher predictive ability than PLSR (Table S3).
Predictive abilities of ten replicated cross validations plotted against the number of extracted components for two methods. (1) Principal component regression implemented via the 10-fold cross validation method (PCR-CV) and (2) Partial least squares regression via 10-fold cross validation (PLSR-CV)
Analysis of rice data
The four methods (PCR-HAT, PCR-CV, PLSR-CV, and BLUP-HAT) were also used to analyze four agronomic traits of 210 lines of an RIL rice population. The results are summarized in Table 3. The conclusions in terms of predictive ability are slightly different from that of the simulated data analysis. In the real data analysis, the PCR-HAT method often has a slightly lower predictive ability than the PCR-CV method. However, both PCR-HAT and PCR-CV have higher predictive abilities than the PLSR-CV method. Two-sample t-test was performed to compare the predictive abilities between PCR-CV and PLSR-CV when four agronomic traits were considered, respectively. In the YD and KGW traits, PCR-CV has significantly higher predictive ability than PLSR-CV (p < 0.05). No significant difference in predictive ability was detected between PCR-CV and PLSR-CV (p > 0.05) for traits TP and GN, although visual inspection showed that PCR-CV tends to predict better than PLSR. The BLUP-HAT method again shows higher predictive ability than the PCR and PLSR methods. Does this mean that BLUP-HAT is always better than PCR and PLSR? These four agronomic traits are a small number of traits under evaluation. In the next two sections, we predicted 1000 metabolomic traits and 24,994 transcriptomic traits to compare the performances of the four methods.
Prediction of 1000 metabolomic traits of the rice data
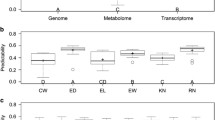
We now compare the predictive abilities of 1000 metabolomic traits with the 1619 bin genotypes of the rice population using the four methods (PCR-HAT, PCR-CV, PLSR-CV, and BLUP-HAT). Six pair-wise plots are shown in Fig. 4, where the diagonal lines of each panel indicates the equality line between the two methods under the comparison. For each panel (comparison), if more points are above the diagonal line, the method on the y-axis, on average, predicts better than the method on the x-axis. The mean predictive abilities (mean ± stdev) and pair-wise comparisons of the four methods are summarized in Table 4. Note that the standard deviations of these predictive abilities appear to be large because these represent the summary scores for 1000 metabolomic traits, which is similar to the values in Table 5 where many thousands of transcriptomic traits are considered. Overall, BLUP-HAT is the best method, followed by PCR-HAT and PCR-CV, and PLSR-CV is the worst one. Two-sample t-test suggested that PCR-CV had significantly higher predictive ability than PLSR-CV (p < 0.05). The two PCR-based methods performed equally well. The PCR-HAT method, however, has 211 metabolites predicted better than the BLUP-HAT method. We were not able to show this result when using only four agronomic traits. The big data (1000 metabolomic traits) indeed help achieve more than what can be achieved in traditional field experiments.
Regarding the computational time of the four methods for prediction of the 1000 metabolites, PCR-HAT and BLUP-HAT only took less than 2 min to complete the analysis, while PLSR and PCR-CV took about 6.3 and 2.9 h, respectively. The gain in computational efficiency mainly came with the HAT prediction that avoids lengthy cross validations. One may wonder whether a similar HAT method can be used for PLSR prediction. The answer is NO because PLSR requires response variables to construct the scores. Once the response variables are involved in generating the predictor components (the scores), the HAT method will exaggerate the predictive ability. In PCR, the scores are constructed using only the predictors and thus the HAT method works well, just like the HAT prediction in ordinary least squares regression. Why do PCR-HAT and PCR-CV generate slightly different predictions? The answer is that we used the entire data to construct the scores in PCR-HAT, while the PCR-CV constructs the scores only based on portion of the data (training set). Simulation studies and real data analysis both showed that the two PCR-based methods perform equally well.
Prediction of transcriptomic traits of the rice data
Finally, we used a much larger data set of 24,973 transcriptomic traits (big data) to compare the prediction behaviors of the four methods (PCR-HAT, PCR-CV, PLSR-CV, and BLUP-HAT). Six pair-wise plots are shown in Fig. 5. The mean predictive ability and pair-wide comparisons are summarized in Table 5. The conclusion remains the same as that obtained from the metabolomic trait prediction. The BLUP-HAT method gave the best prediction, followed by PCR-HAT, PCR-CV, and PLSR-CV is the worst one. Two-sample t-test suggested that PCR-CV had significantly higher predictive ability than PLSR-CV (p < 0.001). Again, the two PCR-based methods performed equally well with PCR-HAT having 15,301 (out of 24,973) gene expressions predicted better than PCR-CV, and the average predictive ability of PCR-HAT was slightly higher than that of PCR-CV. PCR-HAT had 21,954 gene expressions (~90% of all expression traits) with higher predictive ability than PLSR-CV, and 5474 expression traits predicted better than BLUP-HAT.
Considering the expensive computational time of PCR-CV and PLSR-CV, we performed the prediction using the HPCC cluster at University of California Riverside (UCR) with 32 CPUs. The two HAT methods, PCR-HAT and BLUP-HAT, took about 2 and 7 min to complete the analysis, respectively. The two CV-based methods, PCR-CV and PLSR-CV, however, required substantially more computational time with about 2.5 and 6 h, respectively.
Discussion
We developed a new method of eigenvalue decomposition for PCR. The new method requires eigenvalue decomposition for an n×n matrix rather than an m×m matrix, where m (number of variables) is often substantially larger than n (number of observations) in GS. In addition to PCR, principal component analysis (PCA) is often used in genome-wide association studies (GWAS) to control population structure. The eigenvalue decomposition applied to GWAS is an m×m variance-covariance matrix. However, with the new method, we can decompose the n×n kinship matrix and convert its eigenvectors into those of an m×m covariance matrix, which is further used to calculate scores as input variables to control potential population structure.
This study does not intend to compare efficiencies of all possible prediction methods (see Xu et al. (2017) for comprehensive comparison of different methods). Rather, we hope to demonstrate the advantage of the PCR method over the PLSR method. It has been commonly believed that PLSR is more efficient than PCR because PLSR takes response variable information into account when constructing scores components. Moreover, PLSR can be used to model relationship between multivariate response variables and multidimensional features, and it can be easily implemented, for example, by ‘pls’ package in R (Wehrens and Mevik 2007). Nevertheless, we observed that PCR is often more efficient with a slight higher predictive ability than PLSR in our GS analyses. This may be ascribed to the distinction between two component-extraction processes used in PCR and PLSR. Two optimization steps are involved in PCR: (1) derivation of sorted components from predictor variables without using any information from the response variable(s), and (2) identification of a sub-regression model, which best explains the response variable(s) with the selected components. In PLSR, the only optimization step is extraction of the regression components from predictors by maximizing the covariance between these components and the response variables; therefore, the following regression step does not provide new contribution to the model optimization. It seems that PCR has advantage over PLSR by feeding the regression step with improved data. The component-extraction step in PCR may be able to distill better candidate regression components by scrutinizing the covariance structure among predictive variables. Such a structure, if exists, may be overlooked by PLSR in which the maximization of component-response covariance is the priority. However, the performances of different methods depend on the nature of data which are analyzed. This explains why PCR had higher predictive ability than PLSR for some metabolomic and transcriptomic traits while the results were opposite when some other metabolomics and transcriptomic traits were considered, with PCR being generally preferred to PLSR in terms of predictive ability. Although PCR requires extracting more components to achieve the maximum predictive ability, application of the HAT method to PCR is computationally more efficient than PLSR because of the avoidance of lengthy cross validation. Therefore, PCR-HAT is preferred to PLSR in GS. On the basis of our results, neither PCR nor PLSR is as efficient as BLUP in general. However, the metabolomic prediction studies showed that 211 metabolites (out of 1000) have higher predictive ability with PCR than with BLUP. Out of all 24,973 expression traits, 6074 of them were predicted better by PCR-HAT than BLUP-HAT. In reality, all available prediction methods should be used in GS because different methods vary in predictive ability across different traits over different populations (De los Campos et al. 2013).
Neither PCR nor PLSR was developed for association studies. In fact, the original developers already warned that such a method is only suitable for prediction (Tobias 1995). However, with a RIL population we showed that the method heavily shrunk the effects (Frank and Friedman 1993), just like ridge regression (BLUP), but the shrinkage appears to be proportional to the true QTL effects. The shrinkage estimates and the true effects vary with similar patterns, which implies that the shrinkage methods may be used for association studies. This conjecture needs to be verified using other populations, for example, a natural population of less related genotypes. Duarte et al. (2014) actually recognized the similarity of the pattern between shrunk effects and estimated effects under the fixed model GWAS. They developed a simple method to bring the shrunk test statistics back to the levels comparable to the unshrunk test statistics. There must be a similar method that can be applied to PCR and PLSR to make them suitable for association studies. Further investigation is necessary to unshrink the PCR and PLSR estimated effects and their test statistics.
References
Andrew KN, Worsfold PJ (1994) Comparison of multivariate calibration techniques for the quantification of model process streams using diode-array spectrophotometry. Analyst 119:1541–1546
Bordier C, Dojat M, Lafaye De Micheaux P (2011) Temporal and spatial independent component analysis for fMRI data sets embedded in the AnalyzeFMRI R package. J Stat Softw 44:1–24
Colombani C, Croiseau P, Fritz S, Guillaume F, Legarra A, Ducrocq V et al. (2012) A comparison of partial least squares (PLS) and sparse PLS regressions in genomic selection in French dairy cattle. J Dairy Sci 95:2120–2131
Cook RD (1977) Detection of influential observation in linear regression. Technometrics 19:15–18
Cook RD (1979) Influential observations in linear regression. J Am Stat Assoc 74:169–174
Coster A, Bastiaansen JW, Calus MP, van Arendonk JA, Bovenhuis H (2010) Sensitivity of methods for estimating breeding values using genetic markers to the number of QTL and distribution of QTL variance. Genet Sel Evol 42:9
De los Campos G, Hickey JM, Pong-Wong R, Daetwyler HD, Calus MP (2013) Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics 193:327–345
Duarte JLG, Cantet RJ, Bates RO, Ernst CW, Raney NE, Steibel JP (2014) Rapid screening for phenotype-genotype associations by linear transformations of genomic evaluations. BMC Bioinformatics 15:246
Frank LE, Friedman JH (1993) A statistical view of some chemometrics regression tools. Technometrics 35:109–135
Gianola D, De Los Campos G (2008) Inferring genetic values for quantitative traits non-parametrically. Genet Res 90:525–540
Gianola D, Fernando RL, Stella A (2006) Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics 173:1761–1776
Gong L, Chen W, Gao Y, Liu X, Zhang H, Xu C et al. (2013) Genetic analysis of the metabolome exemplified using a rice population. Proc Natl Acad Sci USA 110:20320–20325
Greenberg E (1975) Minimum variance properties of principal component regression. J Am Stat Assoc 70:194–197
Hayes BJ, Bowman PJ, Chamberlain A, Goddard M (2009) Invited review: genomic selection in dairy cattle: progress and challenges. J Dairy Sci 92:433–443
Heffner EL, Sorrells ME, Jannink J-L (2009) Genomic selection for crop improvement. Crop Sci 49:1–12
Hemmateenejad B, Akhond M, Samari F (2007) A comparative study between PCR and PLS in simultaneous spectrophotometric determination of diphenylamine, aniline, and phenol: effect of wavelength selection. Spectrochim Acta A 67:958–965
Hoaglin DC, Welsch RE (1978) The hat matrix in regression and ANOVA. Am Stat 32:17–22
Hotelling H (1957) The relations of the newer multivariate statistical methods to factor analysis. Br J Math Stat Psychol 10:67–79
Hua J, Xing Y, Wu W, Xu C, Sun X, Yu S et al. (2003) Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 100:2574–2579
Hua J, Xing Y, Xu C, Sun X, Yu S, Zhang Q (2002) Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 162:1885–1895
Jeffers J (1967). Two case studies in the application of principal component analysis. Appl Stat 16:225–236.
Kendall MG (1957) A course in multivariate analysis. Griffin’s statistical monographs & courses. no. 2. Hafner Publishing Company, New York
Lafaye De Micheaux P, Liquet B, Sutton M (2017). A unified parallel algorithm for regularized group PLS scalable to big data. Preprint at arXiv:1702.07066
Long N, Gianola D, Rosa G, Weigel K (2011) Dimension reduction and variable selection for genomic selection: application to predicting milk yield in Holsteins. J Anim Breed Genet 128:247–257
Lorenz AJ, Chao S, Asoro FG, Heffner EL, Hayashi T, Iwata H et al. (2011) genomic selection in plant breeding: knowledge and prospects. Adv Agron 110(C):77–123
Mandel J (1982) Use of the singular value decomposition in regression analysis. Am Stat 36:15–24
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Næs T, Martens H (1988) Principal component regression in NIR analysis: viewpoints, background details and selection of components. J Chemom 2:155–167
Shlens J (2014). A tutorial on principal component analysis. Preprint at arXiv:14041100
Solberg TR, Sonesson AK, Woolliams JA, Meuwissen TH (2009) Reducing dimensionality for prediction of genome-wide breeding values. Genet Sel Evol 41:29
Tibshirani R (1996). Regression shrinkage and selection via the lasso. J R Stat Soc B 58:267–288
Tobias RD (1995) Proceedings of the twentieth annual SAS users group international conference. SAS Institute, Cary, NC, pp 1250–1257
VanRaden P (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Wang J, Yu H, Weng X, Xie W, Xu C, Li X et al. (2014) An expression quantitative trait loci-guided co-expression analysis for constructing regulatory network using a rice recombinant inbred line population. J Exp Bot 65:1069–1079
Wehrens R, Mevik B-H (2007). The pls package: principal component and partial least squares regression in R. J Stat Softw 18:1–24
Wentzell PD, Montoto LV (2003) Comparison of principal components regression and partial least squares regression through generic simulations of complex mixtures. Chemom Intell Lab Syst 65:257–279
Wold H (1966). Estimation of principal components and related models by iterative least squares. In: Krishnaiaah PR (ed) Multivariate analysis. Academic Press, New York, pp. 391–420
Xie W, Feng Q, Yu H, Huang X, Zhao Q, Xing Y et al. (2010) Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci USA 107:10578–10583
Xing Y, Tan Y, Hua J, Sun X, Xu C, Zhang Q (2002) Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet 105:248–257
Xu S (2017) Predicted residual error sum of squares of mixed models: an application for genomic prediction. G3 7:895–909
Xu S, Zhu D, Zhang Q (2014) Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proc Natl Acad Sci USA 111:12456–12461
Xu Y, Xu C, Xu S (2017) Prediction and association mapping of agronomic traits in maize using multiple omic data. Heredity 119:174–184
Yu H, Xie W, Wang J, Xing Y, Xu C, Li X et al. (2011) Gains in QTL detection using an ultra-high density SNP map based on population sequencing relative to traditional RFLP/SSR markers. PLoS ONE 6:e17595. https://doi.org/17510.11371/journal.pone.0017595
Acknowledgements
We truly appreciate Dr. Weibo Xie from Huazhong Agricultural University for providing the rice data.
Funding
This work was supported for start-up funding of UCR to Zhenyu Jia.
Author contributions
ZJ and JW conceived and designed the experiments. CD, JW, and SW conducted the experiments and analyzed data. CD and JW developed the R program. CD, JW, SW, and ZJ wrote the manuscript. All authors have read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Appendix
Appendix
A toy example for eigenvalue demonstration
A toy data with n = 3 observations and m = 5 variables are used to demonstrate the new method of eigenvalue decomposition. This matrix is stored in X n×m ,
The ultimate goal is to perform eigenvalue decomposition on XTX, an m×m = 5×5 symmetric matrix,
Eigenvalue decomposition of this matrix using the eigen(XTX) function in R produces the following eigenvalues (converted into a diagonal matrix),
and eigenvectors,
Note that the rank of matrix XTX is \(p = \min (n,m) = \min (3,5) = 3\) in this case. Therefore, there are only three non-zero eigenvalues in this example. It is a common practice to delete the zero eigenvalues and also delete all columns of the eigenvectors corresponding to the zero eigenvalues because the values of those columns are irrelevant to the principal component analysis. After this manipulation, we have eigenvalues of
and eigenvectors of
We now use the svd(X) function in R to decompose X so that X = U ΔVT, where
and
The svd() function deletes zero eigenvalues and columns of eigenvectors corresponding the zero eigenvalues. Note that the right eigenvectors are the same as the eigenvectors of matrix XTX (some columns differ by signs, but irrelevant to principal components analysis). Note that
where
Therefore, singular value decomposition on X will produce information for both eigenvalues and eigenvectors of matrix XTX.
When the number of variables (m) is substantially larger than the sample size (n), an alternative but more efficient way to do eigenvalue decomposition for XTX (an m×m matrix) is through eigenvalue decomposition on XXT (an n×n matrix). This matrix from the toy data is
Eigenvalue decomposition for XXT=UDUT produces
and
Let us define B as
This matrix is exactly the same as V, the eigenvectors of matrix XTX after deleting columns corresponding to zero eigenvalues. Therefore, the eigenvectors of XTX can be obtained via eigenvectors of matrix XXT.
Principal components (also called scores) are constructed based on
One can verify that TTT = D. If we have a response variable y, a principal component regression can be performed using y as the dependent variable and T as the predictor matrix.
Rights and permissions
About this article
Cite this article
Du, C., Wei, J., Wang, S. et al. Genomic selection using principal component regression. Heredity 121, 12–23 (2018). https://doi.org/10.1038/s41437-018-0078-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-018-0078-x
- Springer Nature Switzerland AG
This article is cited by
-
Optimization and prediction of the cotton fabric dyeing process using Taguchi design-integrated machine learning approach
Scientific Reports (2023)
-
Haplotype-tagged SNPs improve genomic prediction accuracy for Fusarium head blight resistance and yield-related traits in wheat
Theoretical and Applied Genetics (2023)
-
A dimensionality-reduction genomic prediction method without direct inverse of the genomic relationship matrix for large genomic data
Plant Cell Reports (2023)
-
PCA outperforms popular hidden variable inference methods for molecular QTL mapping
Genome Biology (2022)
-
Comparison of artificial intelligence algorithms and their ranking for the prediction of genetic merit in sheep
Scientific Reports (2022)