Abstract
β-catenin-dependent canonical Wnt signaling plays a plethora of roles in neocortex (Ncx) development, but its function in regulating the abundance of intermediate progenitors (IPs) is elusive. Here we identified neCtnnb1, an evolutionarily conserved cis-regulatory element with typical enhancer features in developing Ncx. neCtnnb1 locates 55 kilobase upstream of and spatially close to the promoter of Ctnnb1, the gene encoding β-catenin. CRISPR/Cas9-mediated activation or interference of the neCtnnb1 locus enhanced or inhibited transcription of Ctnnb1. neCtnnb1 drove transcription predominantly in the subventricular zone of developing Ncx. Knock-out of neCtnnb1 in mice resulted in compromised expression of Ctnnb1 and the Wnt reporter in developing Ncx. Importantly, knock-out of neCtnnb1 lead to reduced production and transit-amplification of IPs, which subsequently generated fewer upper-layer Ncx projection neurons (PNs). In contrast, enhancing the canonical Wnt signaling by stabilizing β-catenin in neCtnnb1-active cells promoted the production of IPs and upper-layer Ncx PNs. ASH2L was identified as the key trans-acting factor that associates with neCtnnb1 and Ctnnb1’s promoter to maintain Ctnnb1’s transcription in both mouse and human Ncx progenitors. These findings advance understanding of transcriptional regulation of Ctnnb1, and provide insights into mechanisms underlying Ncx expansion during development.
Similar content being viewed by others
Introduction
The expansion of the neocortex (Ncx), which in humans is the site of our cognitive abilities1, is thought to primarily be due to an increase in neuron production2,3. The excitatory/projection neurons (PNs) of the six-layered mammalian Ncx are born prenatally in an inside-out fashion. Briefly, Ncx radial glial cells (RGCs), which include basal RGCs and outer RGCs (oRGs), give rise to intermediate progenitors (IPs) residing in the subventricular zone. IPs can divide a few times (a.k.a. transit-amplification) before differentiating into PNs. In mice, deep-layer PNs are most generated between E11.5 to E14.5, followed by the production of upper-layer PNs, which migrate radially along the basal/radial fibers of RGCs and reside on top of deep-layer PNs2,4,5,6. IPs are morphologically and molecularly distinct from RGCs: IPs are multipolar and express markers including TBR2, NEUROG2 and BTG2. Notably, IPs first appear in amniotes, with some birds and reptiles having pallial structures similar to the mammalian Ncx7,8. Moreover, the Ncx expansion during development and evolution correlates with the pool size of oRGs and IPs7,9,10,11. Therefore, many recent studies have dissected cellular and molecular mechanisms underlying production, transit-amplification, and neuronal differentiation of neocortical IPs12,13,14.
The canonical Wnt signaling mediated by β-catenin is highly conserved through evolution and is essential for multiple aspects of brain development. It has been reported that the Wnt/β-catenin signaling promotes self-renewal of RGCs and neuronal differentiation of IPs, however, the role of Wnt/β-catenin signaling in production and properties of neocortical IPs remains inclusive, partly due to the involvement of β-catenin in cell cycle progressions, differentiation, and cell adhesion of neural cells15,16,17,18,19,20. Moreover, CTNNB1 is one of the top risk genes associated with neurodevelopmental disorders including autism spectrum disorder (ASD)21,22. Much has been known regarding molecular components and signaling cascade for phosphorylation, degradation/stabilization, nuclear translocation of β-catenin23,24. However, transcriptional regulation of Ctnnb1, the gene that encodes β-catenin, is poorly understood25.
An enhancer is a cis-regulatory DNA element that spatiotemporally controls transcription of a particular gene in specific cell type(s). Many essential fate-specifying genes have multiple enhancers that are respectively responsible for its proper expression and function in distinct cell types26,27. An active enhancer is accessible by trans-acting factors, enriched with chromatin signatures such as H3K27ac and H3K4me1, and spatially proximal to the promoter of its target gene28,29. Enhancer mutations and/or their aberrate activation/inactivation could lead to developmental abnormalities and cancers26,30,31. Thus, characterization of essential enhancers has implications in understanding molecular machineries that govern selective gene expression in developmental and disease scenarios.
Here we identified an evolutionarily conserved enhancer of Ctnnb1, the gene encodes β-catenin. The enhancer, neCtnnb1, exhibits specific activity in developing Ncx and is essential for production and transit-amplification of IPs and subsequent generations of upper-layer Ncx PNs. Finally, ASH2L was identified as the trans-acting factor that associates with neCtnnb1 and Ctnnb1’s promoter to maintain Ctnnb1’s expression in both mouse and human Ncx progenitors.
Results
neCtnnb1 is a conserved cis-regulatory element with typical enhancer features in developing neocortices
We examined cis-regulatory elements that potentially regulate expressions of components of canonical Wnt signaling during Ncx development. We noticed that a genomic region — neCtnnb1, located 55 kilobase (kb) upstream of the transcription start site (TSS) of Ctnnb1, bears essential enhancer features, namely, prominent enrichment of DNase I hypersensitivity (HS), H3K4me1, H3K27ac, and P300 with low H3K4me3 and H3K36me3 signals in mid-late dorsal forebrains (shaded region in Fig. 1a, Supplementary Fig. S1a). The 2.28 kb genomic region of neCtnnb1 (neocortical enhancer of Ctnnb1) contains a 696 bp evolutionarily conserved fragment and matches peaks of H3K27ac, DNase I HS and P300. However, these enhancer features are much less obvious in developing midbrains, hindbrains and in other non-neural tissues (Supplementary Fig. S1b–d). Moreover, analyses of Hi-C data revealed neCtnnb1 and the gene body of Ctnnb1 are in the same topologically associating domain (TAD) of in-vitro differentiated neural progenitor cells (NPCs) (Fig. 1b).
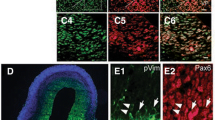
a Schematic representation of the upstream region of mouse Ctnnb1 gene and the location of putative enhancer neCtnnb1 (2.28 kb, orange shading), which is evolutionarily conserved among amniotes and enriched with H3K27ac, DNase I hypersensitivity and P300 in developing forebrains. Data were obtained from ENCODE. Locations of ChIP sites in (h) were indicated. b Hi-C data of in-vitro differentiated NPCs were obtained from the 3D genome browser. Boundaries of the TADs and locations of Ctnnb1 and neCtnnb1 (red bar) were marked below. c Top: a schematic illustration showing the transgenic construct carries three tandem neCtnnb1 sequences, the hsp68 mini-promoter, the LacZ reporter gene and inducible CreERT2 (iCre). Bottom: X-Gal staining (blue) in E11.5 and E15.5 neCtnnb1-LacZ-iCre embryos and brains. Signals at E15.5 dorsal forebrains (fb) were indicated with red arrows and enlarged. d Immunofluorescence of PAX6 (gray), TBR2 (green), and β-Gal (red) on coronal sections of E15.5 neCtnnb1-LacZ-iCre Ncx. e Bar plots showing TBR2 + β-gal+ cells relative of TBR2 + cells (left) and PAX6 + TBR2 + β-gal+ cells relative of PAX6 + TBR2 + cells (right) in E15.5 neCtnnb1-LacZ-iCre Ncx. n = 2 for neCtnnb1-LacZ-iCre brains. Each point represents an individual brain. f The 3C assay performed using Neuro-2a neuroblastoma cells. The Ctnnb1 promoter (pCtnnb1) is the anchor point from which long-range DNA interactions across the neCtnnb1 interval were measured. Numbers over the data points represent fragment locations. g Schematics show the ChIP-qPCR evaluating the physical association between the pCtnnb1 and neCtnnb1. h ChIP-qPCR measuring enrichment of pCtnnb1 and neCtnnb1 after Flag-tagged dCas9 were targeted to neCtnnb1 and pCtnnb1 respectively. Each point represents an independent experiment. I, k Schematics showing the CRISPR activation (CRISPRa, i) and CRISPR interference (CRISPRi, k) experiments. j, l RNA levels of Ctnnb1 in Neuro-2a cells transfected with indicated CRISPRa (j) or CRISPRi (l) vectors for two days. In h quantification data are shown as means ± SEM, statistical significance was determined using two-way ANOVA followed by Tukey’s multiple comparisons test. In j and l, quantification data are shown as means ± SD, statistical significance was determined using one-way ANOVA analysis, *P < 0.05, **P < 0.01, and ***P < 0.001. ns, not significant. Scale bars, 1 mm (c), 50 μm (d). fb, forebrain; CP, cortical plate; VZ, ventricular zone; SVZ, subventricular zone.
neCtnnb1 drives expression in intermediate progenitors (IPs) and newborn neurons of developing neocortices
Enhancers spatiotemporally control gene expression in the process of cell-fate determination during development27,32,33. We thus generated neCtnnb1 transgenic mice, in which three 2.28-kb-long neCtnnb1 sequences were tandemly engineered upstream of the Hsp68 mini-promoter followed by a LacZ reporter and a 2A-CreERT2 cassette (neCtnnb1-LacZ-iCre) (Fig. 1c, top). Five F0 neCtnnb1-LacZ-iCre mouse founders were obtained and crossed with wild-type (WT) mice to study spatiotemporally expression patterns driven by neCtnnb1. 5-bromo-4-chloro-3-indolyl β-D-galactoside (X-Gal) staining of whole-mount and sectioned embryos of multiple lines consistently showed that neCtnnb1 drove LacZ expression predominantly in dorsal forebrains at the peak of Ncx neurogenesis (Fig. 1c; Supplementary Fig. S2a–e), which corroborates with spatiotemporal distributions of epigenetic marks on neCtnnb1. Close-up observations found that at embryonic (E) day 15.5, the LacZ/β-Gal expression driven by neCtnnb1 were mostly concentrated in the SVZ region of developing neocortices (Fig. 1d), with 50.0% of TBR2 + IPs express β-Gal (Fig. 1e). Particularly, immunofluorescent staining showed that neCtnnb1-driving β-Gal signal were detected in 64.8% of IP-differentiating RGCs that co-expressed PAX6 and TBR2 (Fig. 1d, e). Furthermore, the LacZ expression could be observed in cells co-labeled with TBR2 and NEUROD2 (Supplementary Fig. S2h), a marker for differentiating and mature PNs. Together, transgenic reporter mice showed that the enhancer activity of neCtnnb1 is preferentially in embryonic SVZ, the predominantly neurogenic zone of the developing neocortex (Supplementary Fig. S2f, g).
neCtnnb1 associates with the promoter of Ctnnb1 and maintains Ctnnb1 expression
Enhancers usually have close proximity to promoter(s) of its target genes for transcriptional activation34,35,36. Setting neCtnnb1 as the viewpoint, neCtnnb1 was found to interact with Ctnnb1’s promoter (pCtnnb1) in differentiated NPCs and E14.5 cortical neurons using Hi-C and virtual 4C data (Supplementary Fig. S1e–h). Chromosome conformation capture (3C) experiments indeed revealed association of neCtnnb1 with pCtnnb1 in Neuro-2a neuroblastoma cells (Fig. 1f). We next carried out CRISPR/dCas9-mediated chromatin immunoprecipitation (ChIP) assay27. Data showed that the Flag-tagged dCas9 targeting neCtnnb1 could significantly precipitate more pCtnnb1 than the exon 3 of the Ctnnb1 gene (Ctnnb1-Ex3). Consistently, Flag-tagged dCas9 targeting pCtnnb1 could enrich more neCtnnb1 than control locus (Fig. 1g, h). Moreover, CRISPR/dCas9-mediated activation (CRISPRa) or interference (CRISPRi) of the neCtnnb1 locus greatly enhanced (Fig. 1i, j) or inhibited (Fig. 1k, l) Ctnnb1’s transcription. Thus, the long-range interaction between neCtnnb1 and pCtnnb1 might maintain Ctnnb1’s transcription in neural cells.
neCtnnb1 knock-out compromises the production of upper-layer (UL) PNs and Ctnnb1 transcription
The canonical Wnt signaling mediated by nuclear-translocated β-catenin is essential for multiple aspects of Ncx development, including forebrain patterning37,38, area expansion15,18, self-renewal of radial glial cells (RGCs)39,40, and production of layer-specific neurons16,41. In addition, because β-catenin is the key component of apical adherens junctions, loss of β-catenin causes disorganized neuroepithelium15,42. To ask if neCtnnb1 regulates Ncx development via controlling expression of β-catenin, we generated neCtnnb1 knockout mice (neCtnnb1KO) wherein a 5,369-bp-long genomic region encompassing neCtnnb1 was ablated using CRISPR/Cas9-mediated gene editing (Fig. 2a).
a Generation and genotyping of neCtnnb1KO mice. WT, wild-type; gRNA, guide RNA. b Nissl-staining on adult WT and neCtnnb1KO sagittal brain sections. c Comparison of neocortical thickness of adult WT and neCtnnb1KO mice. n = 3 for WT brains and n = 5 for neCtnnb1KO brains. Each point represents an individual brain. d, e Immunofluorescence (d) and quantification (e, top) of SATB2 + , CUX1 + , FOXP2 + and CTIP2 + neurons on boxed area of sagittal sections (e) of adult WT and neCtnnb1KO Ncx. Each point represents an individual brain. f In situ hybridization (ISH) of Ctnnb1 on E14.5 WT (top) and neCtnnb1KO (bottom) coronal brain sections, with boxed regions magnified on the right. g, h Quantification of normalized ISH signal densities in boxed regions of (f). n = 3 for WT brains and n = 3 for neCtnnb1KO brains. Each point represents an individual brain. i RT-qPCR showing expressions of Ctnnb1 in E15.5 WT and neCtnnb1KO neocortex. n = 4 for WT Ncx and n = 6 for neCtnnb1KO Ncx. Each point represents an individual brain. j β-Gal immunostaining on the E15.5 coronal sections of WT_BAT (left) and neCtnnb1KO_BAT (right) Ncx. Boxed regions are enlarged at the bottom left corners. k Quantification of normalized signal density of β-Gal in boxed regions of (j). n = 3 for WT brains and n = 3 for neCtnnb1KO brains. Each point represents an individual brain. Quantification data are shown as means ± SEM. Statistical significance was determined using an unpaired two-tailed Student’s t-test (c, e, h and i). *P < 0.05, **P < 0.01, and ***P < 0.001. ns, not significant. Scale bars, 500 μm (f), 200 μm (b), 100 μm (d and j), 50 μm (magnified views in f and j). CP, cortical plate; VZ, ventricular zone; SVZ, subventricular zone; Ctx, cortex; LV, lateral ventricular; LGE, lateral ganglionic eminences; Hipp, Hippocampal primordium.
Homozygous neCtnnb1KO mice were born according to Mendelian ratio with no visible defects (Supplementary Fig. S3a). Adult neCtnnb1KO brains have the same Ncx hemisphere area and rostral-caudal length as WT controls (Supplementary Fig. S3b, c). However, the Ncx of adult neCtnnb1KO brains were 10.8% thinner than WT controls (Fig. 2b, c). Immunofluorescent (IF) staining revealed that neCtnnb1KO neocortices contained 14.2% and 17.1% fewer SATB2 + and CUX1 + UL PNs respectively with unaltered CTIP2 + and FOXP2 + deep-layer PNs (Fig. 2d, e), suggesting compromised neurogenesis during mid-late embryonic development.
We next examined whether expressions of Ctnnb1 and transcriptional targets of canonical Wnt signaling were compromised in neCtnnb1KO Ncx, particularly during cortical neurogenesis. In situ hybridization (ISH) of WT E14.5 Ncx showed an apical high to basal low gradient of Ctnnb1 expression (Fig. 2f). Strikingly, the Ctnnb1 expression was greatly compromised throughout the neCtnnb1KO Ncx (Fig. 2g, h). Reverse transcription-quantitative PCR (RT-qPCR) studies further confirmed that expressions of Ctnnb1 were greatly reduced in E15.5 neCtnnb1KO Ncx (Fig. 2i). Consistently, the BAT-Gal reporter mice indicated that ablation of neCtnnb1 lead to reduced Wnt-reporter expression, reflected by β-galactosidase IF signals (Fig. 2j, k) and the LacZ staining, throughout E15.5 and postnatal (P) day 8 Ncx (Supplementary Fig. S3d–j).
neCtnnb1 knockout hampered the production and transit-amplification of IPs
We then asked whether the compromised neurogenesis of UL PNs in neCtnnb1KO cortices was due to alterations of Ncx progenitor pool and/or their behaviors. E16.5 neCtnnb1KO and control embryos were pulse-labeled with 2 h EdU and subjected to IF analyses (Fig. 3a, b). First, although numbers of PAX6 + RGCs and total EdU+ cells were not significantly altered in E16.5 neCtnnb1KO Ncx (Fig. 3c, e), the number of TBR2+ IPs was reduced by 12.7% (Fig. 3f), indicating compromised production of IPs by RGCs and/or hampered transit-amplification of IPs. Indeed, numbers of PAX6 + TBR2 + double-positive cells and the ratio of PAX6 + TBR2 + among all PAX6 + cells were decreased by 17.4% and 18.5% respectively in neCtnnb1KO Ncx (Fig. 3h–j), pointing to compromised production of IPs by RGCs upon ablation of neCtnnb1. Moreover, although the number of RGCs at the proliferation status (PAX6 + EdU +) were unchanged upon loss of neCtnnb1 (Fig. 3e), the dividing capacity of neCtnnb1KO IPs was greatly compromised, as 32.1% fewer or 30.8% less TBR2 + IPs were co-localized with EdU (Fig. 3g) or 0.5 h bromodeoxyuridine (BrdU) (Supplementary Fig. S4a, b) respectively in neCtnnb1KO neocortices. In addition, the neCtnnb1KO Ncx contained 33.7% fewer PAX6 + TBR2 + EdU+ cells and 29.0% less PAX6 + TBR2 + EdU+ cells among PAX6 + EdU+ cells, further indicating compromised IP production from RGCs upon loss of neCtnnb1 (Fig. 3i–k).
a–k Immunofluorescence (a) and quantification (c–k) of EdU + (red), PAX6 + (blue) and TBR2 + (green) cells on boxed area of coronal sections (b) of E16.5 WT and neCtnnb1KO Ncx. Pregnant mice were injected with EdU 2 h before sacrifice. Each point represents an individual brain. l–t Immunofluorescence (l) and quantification (n–t) of Ki67 + (red), TBR2 + (blue) and BrdU + (green) cells on boxed area of coronal sections (m) of E16.5 WT and neCtnnb1KO Ncx. Pregnant mice were injected with BrdU at E15.5. m–t Quantification of data in l. Each point represents an individual brain. Quantification data are shown as means ± SEM. Statistical significance was determined using an unpaired two-tailed Student’s t-test (c–k, n–t). *P < 0.05, **P < 0.01, and ***P < 0.001. ns not significant. Scale bars, 100 μm (a and l), 50 μm (magnified views in a, l).
Next, embryos were pulse-labeled by BrdU 24 h prior to analyses at E16.5 by triple-labeling of BrdU, Ki67 and TBR2 (Fig. 3l, m). Numbers of cells labeled by 24 h BrdU or Ki67 were unchanged in neCtnnb1KO Ncx (Fig. 3n, o). In contrast, 23.8% fewer TBR2 + cells in neCtnnb1KO Ncx were labeled by 24 h BrdU (Fig. 3p), further supporting diminished production of IPs upon loss of neCtnnb1. Moreover, significantly fewer TBR2 + IPs were actively dividing at E16.5 (27.3% fewer Ki67 + TBR2 + cells and 13.2% less Ki67 + TBR2 + /TBR2 + cells, Fig. 3q, s), and stayed in cell cycle during the 24 h period (32.7% fewer BrdU+Ki67 + TBR2 + cells and 11.1% less BrdU+Ki67 + TBR2 + /BrdU+TBR2 + cells, Fig. 3r, t) in neCtnnb1KO Ncx. Collectively, the production and transit-amplification/divisions of neocortical IPs were greatly compromised upon loss of neCtnnb1.
A previous study demonstrated that transient downregulation of canonical Wnt/β-catenin signaling plays a critical permissive role for proper polarization and radial migration of Ncx PNs43, which prompted us to examine whether deletion of neCtnnb1 would hamper neuronal migration. To this end, constructs that express EGFP were electroporated into E14.5 neocortices and Ncx sections were immunostained with SATB2 at E18.5 (Supplementary Fig. S4c, d). In neCtnnb1KO Ncx, the destination and ratio of SATB2 expression of electroporated cells and their progeny labeled by EGFP were the same pattern as those in the WT Ncx (Supplementary Fig. S4e, f), suggesting deletion of neCtnnb1 has no effect on neuronal migration.
Of note, the ventricle surface of neCtnnb1KO Ncx is as smooth and intact as WT Ncx without ectopic neurogenic foci (Fig. 2b, f), indicating that decreased Ctnnb1 expression throughout Ncx caused by neCtnnb1KO has no effect on cellular architecture of developing Ncx.
Stabilization of β-catenin in neCtnnb1-active cells promotes the production and transit-amplification of IPs
We next explored whether activating the canonical Wnt/β-catenin signaling in neCtnnb1-active cells could promote the production and transit-amplification of IPs, hence enhance Ncx neurogenesis. neCtnnb1-LacZ-iCre mice were crossed with conditional Ctnnb1lox(ex3) mice, with tamoxifen (TAM) injected at E13.5 and embryos collected at E16.5 (Fig. 4a). As expected, the non-degradable constitutively-active β-catenin (C.A. β-catenin) could be detected in E16.5 Ctnnb1lox(ex3);neCtnnb1-LacZ-iCre (WNT-iGOF) Ncx, and products of Wnt/β-catenin target genes including C-Myc and Cyclin D1 (Ccnd1) were also significantly enhanced in the WNT-iGOF Ncx (Fig. 4b). Strikingly, the E16.5 WNT-iGOF Ncx and their cortical plates were thicker than Ctnnb1lox(ex3) controls (Fig. 4c, e; Supplementary Fig. S4g–i), with 42.6% more SATB2 + UL PNs and 26.9% more TBR2 + IPs in the E16.5 WNT-iGOF Ncx (Fig. 4h, l). But the numbers of PAX6 + RGCs and CTIP2 + deep-layer PNs were not changed in the WNT-iGOF Ncx (Fig. 4i, k). The length of dorsal ventricle surfaces was not expanded in the WNT-iGOF Ncx (Fig. 4f; Supplementary Fig. S4j), suggesting that activation of the canonical Wnt/β-catenin signaling by neCtnnb1 did not cause lateral expansion of RGCs and Ncx, which could be otherwise observed in other Wnt/β-catenin gain-of-function studies18,40. Importantly, the number of PAX6 + TBR2 + double-positive cells and ratio of PAX6 + TBR2 + among all PAX6 + cells were increased by 38.7% and 30.8% respectively in E16.5 WNT-iGOF Ncx (Fig. 4j, m, n), indicative of enhanced production of IPs by RGCs. In addition, 25% more TBR2 + IPs were actively cycling and expressed Ki67, and the ratio of IPs that expressed Ki67+ increased by 13.4% in WNT-iGOF Ncx (Fig. 4o–r). Consistently, 37.8% more TBR2 + IPs were labeled with 0.5 h BrdU, and a larger portion of 0.5 h BrdU+ cells were IPs in WNT-iGOF Ncx (Supplementary Fig. S4k–n). Again, the integrity of epithelial structure was not altered in WNT-iGOF Ncx, reinforcing the notion that neCtnnb1 controls Ncx neurogenesis independent of the cell-adhesion function of β-catenin (Fig. 4c; Supplementary Fig. S4g). Together, neCtnnb1-mediated canonical Wnt signaling is required and sufficient for generation and transit-amplification of IPs, hence proper production of UL PNs.
a Schematics show mating of Ctnnb1lox(ex3) mice with neCtnnb1-LacZ-iCre mice. Pregnant mice were injected with tamoxifen (TAM) at E13.5 and sacrificed at E16.5. b Top: immunoblotting of β-catenin, c-MYC, Cyclin D1 and ACTIN using protein lysates of Ctrl - Ctnnb1lox(ex3) and WNT-iGOF - neCtnnb1-LacZ-iCre;Ctnnb1lox(ex3) neocortices. Bottom: histograms show protein levels of c-MYC and Cyclin D1 normalized to ACTIN levels. Values of Ctrl neocortices were set as ‘1’. C.A. β-catenin, the constitutively active (non-degradable) β-catenin caused by deletion of Ctnnb1’s exon 3; Endo. β-catenin, endogenous β-catenin. c Nissl-staining on E16.5 Ctrl and WNT-iGOF coronal brain sections. d Comparison of neocortical thickness of E16.5 Ctrl and WNT-iGOF mice. n = 16 for Ctrl brains and n = 12 for WNT-iGOF brains. Each point represents an individual brain. e Comparison of cortical plate thickness of E16.5 Ctrl and WNT-iGOF mice. n = 10 for Ctrl brains and n = 7 for WNT-iGOF brains. Each point represents an individual brain. f Comparison of dorsal ventricle surface length of E16.5 Ctrl and WNT-iGOF mice. n = 10 for Ctrl brains and n = 7 for WNT-iGOF brains. Each point represents an individual brain. g–i Immunofluorescence (g) and quantification (h, i) of SATB2 + and CTIP2 + of coronal sections of E16.5 Ctrl and WNT-iGOF Ncx. j–n Immunofluorescence (j) and quantification (k–n) of of TBR2 + (red) and PAX6 + (green) cells on boxed area of E16.5 coronal sections (p) of Ctrl and WNT-iGOF Ncx. n = 6 for Ctrl brains and n = 6 for WNT-iGOF brains. Each point represents an individual brain. o–r Immunofluorescence (o) and quantification (p–r) of TBR2 + (red) and Ki67 + (green) cells on boxed area of coronal sections (p) of E16.5 Ctrl and WNT-iGOF Ncx. n = 5 for Ctrl brains and n = 5 for WNT-iGOF brains. Each point represents an individual brain. Quantification data are shown as means ± SEM. Statistical significance was determined using an unpaired two-tailed Student’s t-test (d, e, f, h, i, k–n and p–r). *P < 0.05, **P < 0.01, and ***P < 0.001. ns not significant. Scale bars, 500 μm (c), 100 μm (g, j, o and magnified views in c). 50 μm (magnified views in j and o).
ASH2L associates with neCtnnb1 and pCtnnb1 to regulate Ctnnb1’s transcription in mouse and human Ncx progenitors
We next addressed which trans-acting factor(s) can associate with neCtnnb1 to maintain Ctnnb1’s transcription. By analyzing ChIP-seq data of developing Ncx, several transcription factors and epigenetic modifiers were found to be enriched in neCtnnb1 and/or pCtnnb1, including FOXP1, NEUROD2, NEUROG2, TBR2, SATB2, SMAD3, SOX2, ASH2L and EP300 (Fig. 5a). Literatures and ISH data deposited in the Allan Brain Atlas showed most of them are expressed in developing Ncx (Supplementary Fig. S5a). Among them, SOX2, SATB2, NEUROD2 and ASH2L were found to be enriched in both neCtnnb1 and pCtnnb1 and involved in Ncx neurogenesis. Particularly, a recent report found ASH2L, the COMPASS (complex of proteins associated with Set1) family histone methyltransferase co-factor, positively regulates Ctnnb1’s transcription and Ncx neurogenesis44. Sing-cell sequence data showed that, similar to Tbr2/Eomes, the transcript of Ash2l is highly enriched in IPs and immature PNs during mid-late neurogenesis (Supplementary Fig. S5b, c). In silico analyses showed ASH2L strongly associates with neCtnnb1 and pCtnnb1 in developing mouse Ncx but not in non-neural cells (Fig. 5b). Moreover, in neCtnnb1KO cortical NPCs, the association of ASH2L with pCtnnb1 was significantly compromised (Fig. 5c). Depletion of ASH2L could greatly downregulate Ctnnb1’s expression in Neuro-2a cells (Fig. 5e) or in WT Ncx NPCs (Fig. 5d), but not in neCtnnb1KO NPCs (Fig. 5d). In comparison, knocking down Sox2, Satb2, or NeuroD2 elicited no effect on Ctnnb1 expression in Neuro-2a cells (Supplementary Fig. S5d–f), WT or neCtnnb1KO NPCs (Supplementary Fig. S5h–j). However, knocking down Yy1, the gene encoding a transcription factor known to be a positive regulator of Ctnnb145, could decrease Ctnnb1 expression in Neuro-2a cells (Supplementary Fig. S5g), WT and neCtnnb1KO NPCs (Supplementary Fig. S5k), demonstrating that YY1 maintains Ctnnb1’s transcription independent of neCtnnb1. Together, ASH2L associates with pCtnnb1 and maintains Ctnnb1’s expression in an neCtnnb1-dependent manner.
a ChIP-seq tracks for enrichment of indicated trans-acting factors in neCtnnb1 in developing Ncx. neCtnnb1 is highlighted. b ChIP-seq tracks for ASH2L in indicated murine cells. neCtnnb1 is highlighted. c ChIP-qPCR of E15.5 WT and neCtnnb1KO Ncx using an anti-ASH2L antibody. n = 3 for WT Ncx and n = 3 for neCtnnb1KO Ncx. Each point represents an individual brain. d RNA levels of Ash2l (left) and Ctnnb1 (right) in WT and neCtnnb1KO neocortical NPCs transfected with indicated vectors for two days. NPCs were derived from E12.5 Ncx. n = 3 for WT brains and n = 3 for neCtnnb1KO brains. Each point represents an individual brain. e RNA levels of Ash2l (left) and Ctnnb1 (right) in Neuro-2a cells transfected with indicated vectors for two days. Each point represents an independent experiment. f Schematic representation the location of neCtnnb1 (pink shading) and conserved region (orange shading) marked by enrichment of H3K27ac, DNase I hypersensitivity and ASH2L in E14.5 brains. Data were obtained from ENCODE. Locations of sgRNA target sites and ChIP primer were indicated. g, h RNA levels of Ctnnb1 in Neuro-2a cells transfected with indicated CRISPRa (g) and CRISPRi (h) vectors for two days. Each point represents an independent experiment. i Luciferase reporter assay in Neuro-2a cells transfected with indicated vectors for two days. Each point represents an independent experiment. Quantification data are shown as means ± SEM. Statistical significance was determined using two-way ANOVA followed by Sidak’s multiple comparisons test. (c, d, e and i); or using one-way ANOVA analysis (g and h). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. ns, not significant.
We then investigated the precise and minimal region of neCtnnb1 that sufficiently controls Ctnnb1’s transcription. Ten single-guide RNAs (sgRNAs) targeting the 5.19-kb-long region encompassing neCtnnb1 (Fig. 5f) were individually used for CRISPRa and CRISPRi experiments in Neuro-2a cells. Interestingly, sgRNA#3 and #4, the two sgRNAs that target the conserved genomic region of neCtnnb1 (neCtnnb1-CR), displayed the most prominent effects on inducing (Fig. 5g) or inhibiting (Fig. 5h) Ctnnb1’s transcription; and sgRNAs locating away from neCtnnb1-CR showed minor effects on regulating Ctnnb1’s expression. Multiple alignment of genomic sequences of the core conserved region in various species confirmed the high sequence homology across amniotes (Supplementary Fig. S6a, b). We thus cloned the 696-bp-long neCtnnb1-CR into the pGL3-pro vector. Compared to empty vector, pGL3-pro-CR displayed significantly higher luciferase activities in Neuro-2a cells. Moreover, overexpression ASH2L further enhanced the luciferase activities driven by neCtnnb1-CR (Fig. 5i).
In particular, neCTNNB1, the orthologous human genomic region of neCtnnb1, also bears essential enhancer features, such as chromatin accessibility determined by the transposase-accessible chromatin using sequencing (ATAC-seq), peaks for H3K27ac and H3K4me1, in developing human Ncx (Fig. 6a; Supplementary Fig. S7a). Hi-C data and virtual 4C analyses of developing human Ncx also indicated spatial proximity between neCTNNB1 and pCTNNB1 (Fig. 6b, c; Supplementary Fig. S7b, c). ChIP-qPCR experiments showed ASH2L strongly binds to neCTNNB1 and pCTNNB1 in human Ncx NPCs (Fig. 6d). Finally, depletion of ASH2L also significantly decreased expressions of CTNNB1 (Fig. 6e).
a Schematic representation of human CTNNB1 gene and the location of putative neCTNNB1 enhancer (blue shading) marked by H3K27ac and H3K4me1 peaks and ATAC signals. b Hi-C data of H1-derived human neural progenitor cells (hNPC) were obtained from the 3D genome browser. Boundaries of TADs and locations of neCTNNB1 and CTNNB1 (red bars) are indicated below. b' Regions magnified from (b). neCTNNB1 interacts with CTNNB1 in H1-derived hNPC. c Virtual 4C of H1-derived hNPCs revealing interactions between neCTNNB1 with pCTNNB1. d ChIP-qPCR of hNPC using an anti-ASH2L antibody. e RNA levels of ASH2L (left) and CTNNB1 (right) in hNPC transfected with indicated virus for two days. Each point represents an independent experiment (d, e). f The working model. neCtnnb1, the upstream enhancer of Ctnnb1, controls production and transit-amplification of IPs, hence proper productions of UL PNs. ASH2L associates with neCtnnb1 and positively regulates transcription of Ctnnb1 in the neCtnnb1-dependent manner. In e, quantification data are shown as means ± SD. statistical significance was determined using two-way ANOVA followed by Sidak’s multiple comparisons test (e) *P < 0.05, **P < 0.01, and ***P < 0.001. ns not significant. PFC dorsal lateral prefrontal cortex.
Disscussion
Although previous studies showed the canonical Wnt/β-catenin is involved in multiple aspects of neocortical development, its role in production and behaviors of IPs has not been fully understood. It was postulated that the Wnt/β-catenin signaling regulates both proliferation and differentiation of neural progenitors in vivo in a context-dependent manner, likely separated by either progenitor state and/or developmental time16,46,47. The canonical Wnt/β-catenin signaling displays a VZ-high to SVZ/IZ-medium to CP-low gradient in the developing Ncx, coinciding with the differentiation sequences from RGs to IPs, then to PNs43. Moreover, the strength of canonical Wnt/β-catenin signaling decreases over neurogenic time48, reflecting the progressive fate restriction of early to later RGCs and sequential generation of DL and UL PNs46.
Here we characterized an enhancer-driven canonical Wnt signaling predominantly active in Ncx SVZ, which maintains the generation and transit-amplification of IPs hence proper production of UL PNs (Fig. 6f). Boosting the canonical Wnt signaling in neCtnnb1-active cells greatly promotes IPs’ production and proliferation, leading to overproductions of UL PNs. Interestingly, this function exerted by neCtnnb1 is independent of β-catenin’s roles in self-renewal of RGCs and the maintenance of the epithelial structure of the developing Ncx. Therefore, neCtnnb1 spatiotemporally fine-tunes tones of Wnt signaling to control IP abundance and productions of UL PNs.
Most genetic studies supported the idea that the canonical Wnt/β-catenin signaling promotes RGC self-renewal while inhibits IP production and neuronal differentiation. Loss of β-catenin increases the Ncx IP population, and stabilization of β-catenin or expression of a dominant-active form of β-catenin promotes self-renewal of RGC and inhibits IP production40,49. But an in vitro study showed Wnt3a treatment increased the proportion of embryonic neocortex cells expressing TBR2 and decreased the percentage of RGCs50, which aligns with our findings and supports the notion that canonical Wnt/β-catenin signaling promotes IP production by RGCs. However, these and many other studies could not precisely manipulate the Wnt tone specifically in RGCs or IPs of the developing Ncx. For example, either depletion of RGCs or enhanced differentiation of IPs upon loss or gain of the Wnt/β-catenin signaling respectively could lead to the decrease of IP pool16,51. Even more complicated, manipulating the Wnt signaling by controlling expression levels or stability of β-catenin, overexpressing Wnt ligands, or bending the transactivating machinery using genetic or non-genetic means could result in distinct phenotypes, partly due to affected lateral expansion of RGCs and/or the disturbance of the Ncx epithelial structure41,52,53. We reason that although the overall Wnt/β-catenin signaling mediated by neCtnnb1 is significantly compromised in developing Ncx upon loss of neCtnnb1, it reaches the threshold wherein the production and proliferation of IPs are mostly impacted but the self-renewal capacity and the epithelial architecture of RGCs remain uninfluenced.
As far as we know, neCtnnb1 is the first enhancer that has been identified for Ctnnb1, the gene encoding β-catenin. neCtnnb1 bears typical features for active enhancers during mid-late Ncx neurogenesis: neCtnnb1 resides in the same chromatin subregion/compartment - TAD with the Ctnnb1 gene and contacts with pCtnnb1, and is enriched with histone modifications including H3K27ac and H3K4me1. Moreover, the core-sequence of neCtnnb1 is highly conserved across amniotes with Ncx-like structures in their brains (Supplementary Fig. S6b), coinciding with the presence of IPs in these animals8,54,55, which strongly points to neCtnnb1’s function in Ncx development and expansion. It remains to be elucidated whether the neCtnnb1 sequences of higher animals, particularly the human neCTNNB1, display stronger activity than lower animals to boost IP production and division by enhancing the Wnt//β-catenin signaling.
An enhancer is a region of DNA that can be bound by trans-acting factors and associate with the promoter of its target genes to control the rate of transcription in particular cells at certain time points56. ASH2L has been found to be essential for Ncx development by sustaining expressions of multiple components of canonical Wnt signaling including Ctnnb144. Interestingly, ablation of Ash2l in developing Ncx also caused depletion of IPs. Our study further revealed that ASH2L strongly associates with neCtnnb1 and pCtnnb1. More importantly, we showed that the presence of neCtnnb1 is required for ASH2L’s binding to pCtnnb1, hence for the proper expression of Ctnnb1 in Ncx progenitors (Fig. 6f). In contrast, YY1, a reported transcriptional regulator of Ctnnb157, positively controls Ctnnb1’s expression independent of the presence of neCtnnb1 (Supplementary Fig. S5k). Future studies might unveil additional trans-acting regulators, such as epigenetic modifiers and transcription factors, which can cooperate with ASH2L to facilitate Ctnnb1’s transcription by modifying the neCtnnb1-pCtnnb1 locus.
Previous high-throughput and reporter studies characterized numerous enhancers in the developing telencephalon58. Lately, the Encyclopedia of DNA Elements (ENCODE) project has identified more than 500,000 transposase-accessible regions during mouse fetal development with around five putative enhancers for one gene59,60. Nonetheless, only a fraction of these cis-elements with enhancer features have been functionally characterized61. Thus, there could be other enhancers of Ctnnb1 to control its spatiotemporal expression in the context of development and/or pathophysiological scenarios. Enhancer mutations and aberrant activation are associated with developmental abnormalities such as polydactyly62,63, dysplasia of the sternum27, craniofacial malformation64, as well as malignant transformations65. SNPs inside core regions of neCtnnb1 have yet to be associated with disorders related to neural development, but some might alter transcription factor binding (data not shown). The significance of these SNPs in physiological and pathological contexts deserves further investigation. Moreover, the activity and roles of neCtnnb1 in adult brains, particular in lineages of adult neural stem cells, remain to be explored. Altogether, our findings advance the understanding of transcriptional regulation of Ctnnb1, and imply that canonical Wnt signaling controlled by neCtnnb1 might be essential for Ncx expansion during evolution and development.
Materials and methods
Mice and genotyping
All mice studies were performed in accordance with protocols approved by the Animal Care and Ethical Committee at Wuhan University. All mice were in the C57BL/6 J background. The noon of the day when the vaginal plug was found was counted as embryo (E) day 0.5. neCtnnb1KO mice were generated in the Beijing Biocytogen. neCtnnb1-LacZ-iCre mice were generated in the Shanghai Model Organisms Center Inc. BAT-Gal mice were kind gifts from Dr. Junlei Chang (Jackson Lab, stock number 005317). Mice with conditional activation of β-catenin were obtained by crossing Ctnnb1lox(ex3) (a gift from Dr. Junlei Chang, MGI number: 1858008) with neCtnnb1-LacZ-iCre. Tamoxifen (TAM) was dissolved in corn oil as previously described66. To activate canonical Wnt/β-catenin signaling, TAM was injected into pregnant mice at indicated stages with a concentration of 100 mg/kg body weight. The primer set forward 5′-ccctgcccctgcatatagcatttga-3′/reverse 5′-ccccacctgtgatgctttgatgtct-3′/reverse 5′-atgctgtgtgagtgaccctactcct-3′ were used for neCtnnb1KO mice genotyping. The primer set forward 5′-ggtagtggtccctgcccttgacac-3′/reverse 5′-acgtctggcaagttccgcgtcatcc-3′/reverse 5′-ctaagcttggctggacgtaaactc-3′ were used for Ctnnb1lox(ex3) mice genotyping. The primer set forward 5′-atcctctgcatggtcaggtc-3′/reverse 5′-cgtggcctgattcattcc-3′ was used for BAT-Gal mice and neCtnnb1-LacZ-iCre mice genotyping. The band sizes for neCtnnb1KO mice are 588 base pairs (bp) (wild-type allele), 477 bp (neCtnnb1 knockout allele); ~1500 bp (wild-type allele), ~1300 bp (loxP allele) for Ctnnb1lox(ex3) mice, and 315 bp (LacZ allele) for BAT-Gal mice and neCtnnb1-LacZ-iCre mice.
Cell lines
HEK293T cells were gifts from Dr. Hongbing Shu (Wuhan University). Neuro-2a cells were purchased from the Cell Bank of Chinese Academy of Sciences, ReNcell CX human neural precursor cells (hNPCs) were purchased from EMD Millipore (Billerica, MA, USA, SCC008). HEK293T cells were maintained in indicated culture media (DMEM, Gbico) containing 10% fetal bovine serum (FBS) (Life Technologies), Neuro-2a cells were maintained in indicated culture media (MEM, Gbico) containing 10% FBS, non-essential amino acid solution (1×, Gbico, 10370021) and 1 mM sodium pyruvate (Gbico, 11360070). ReNcell CX hNPCs were plated onto laminin (Sigma-Aldrich, 11243217001) coated cell culture plate and maintained in culture media (DMEM/F12, Gbico) containing N2 and B27 supplements (1×, Life Technologies), human recombinant FGF2, and EGF (20 ng/mL each; Life Technologies) in a CO2 cell culture incubator67. Mouse neocortical neural progenitor cells (NPCs) were enriched from E12.5 mouse cortex, cultured on ultra-low-attachment plates (Corning, New York, United States) and maintained in indicated culture media (DMEM/F12, Life Technologies) containing N2 and B27 supplements (1×, Life Technologies), 1 mM Na-pyruvate, 1 mM N-acetyl-L-cysteine (NAC), human recombinant FGF2, and EGF (20 ng/mL each; Life Technologies).
Tissue fixation and sectioning
Pentobarbital sodium [0.7% (w/v), 105 mg/kg of body weight] in 0.9% sodium chloride was peritoneally injected into pregnant mice carefully. Embryos were sequentially removed from uteri, and tails removed for genotyping. Brains were dissected out from embryos in cold phosphate-buffered saline (PBS). For immunofluorescent staining and in situ hybridization, brains were immersed in 4% paraformaldehyde (PFA) overnight at 4 °C; for X-Gal staining, brains were immersed in 4% PFA for 10 min (brains) or 30 min (whole body) at room temperature. For adult mice, mice were deeply anesthetized with intraperitoneal injection of 0.7% (w/v) pentobarbital sodium solution followed by perfusion with 4% PFA. Brains were dissected out and immersed in 4% PFA overnight at 4 °C. The next day, 4% PFA was replaced with 20% (w/v) sucrose for embryo brains or 30% (w/v) sucrose for adult brains overnight at 4 °C. For sectioning, brains were embedded in OCT (SAKURA) and cut at 20 μm for adult brains and X-Gal staining brains, and 14 μm for other stages with a cryotome (Leica CM1950) onto coated slides.
Chromosome conformation capture (3C)
The 3C assay was conducted essentially as described previously68,69. 1 × 107 Neuro-2a cells were cross-linked with 2% formaldehyde for 10 min at room temperature. Next, reaction tubes were transferred into ice and added 0.125 M glycine for 5 min to quench the cross-linking reaction. The pellet was lysed in ice-cold 3C lysis buffer (10 mM Tris-HCl, pH 7.5; 0.1 mM EGTA; 10 mM NaCl; 5 mM MgCl2; 1× protease inhibitor) and incubated on ice for 10 min. Nuclei were pelleted by centrifugation at 400× g for 5 min at 4 °C. Pelleted nuclei were taken up to a new tube with 500 μL 1.2× restriction enzyme buffer (NEB) and 7.5 μL 20% (w/v) SDS on rocker for 1 h at 37 °C. 1 h later, 50 μL of 20% (v/v) Triton X-100 was added to tube followed by 1 h incubation at 37 °C with rotation. Pelleted nuclei were digested by BamH I (NEB) overnight at 37 °C with rotation. The next day, 40 μL of 20% (w/v) SDS were added to tube for 25 min at 65 °C with rotation. Digested nuclei were transfer to a new tube followed by adding 6.125 mL of 1.15× ligation buffer (660 mM Tris-HCl, pH 7.5; 50 mM DTT; 10 mM ATP, 50 mM MgCl2) and 375 μL 20% (v/v) Triton X-100 for 1 h at 37 °C with rotation. 700 U T4 DNA ligase (Takara Bio) was added to the tube and incubate for 4 h at 16 °C followed by 30 min at room temperature. Next, 300 μg proteinase K was added to reverse cross-linked chromatin at 65 °C for overnight. The next day, 300 μg RNase A (TARAKA) was added and incubated at 37 °C for 40 min. The fragments of DNA were extracted by phenol-chloroform extraction and analyzed by RT-qPCR. The specificity and efficiency of all 3C primers can be verified by digestion and ligation of BAC DNA containing the target region. The frequency of cross-linking was calculated using the parameters of BAC DNA standard curve. DNA: value = 10(Ct − b/a) (b: intercept; a: slope). Finally, these values were normalized to Gapdh to generate the relative cross-linking frequency.
CRISPR/dCas9-mediated transcription activation (CRISPRa) and interference (CRISPRi) assay
Clustered regularly interspaced short palindromic repeat (CRISPR) interference (CRISPRi), in which a nuclease-null Cas9 (dCas9) is fused to the Krüppel-associated box (KRAB) repressor, can specifically silence target endogenous gene expression. CRISPR activation (CRISPRa), in which a nuclease-null Cas9 (dCas9) is fused to transcriptional activators like VP64 domain, enables efficient increase in target endogenous gene expression. CRISPRi and CRISPRa assays were performed according to published procedures70,71. SgRNAs were designed to target neCtnnb1 by using online tool (https://zlab.bio/guide-design-resources). SgRNAs were cloned into sgRNA (MS2) cloning vector (Addgene, #61424). For CRISPRi assays, vectors of sgRNA and pHR-SFFV-dCas9-BFP-KRAB (Addgene, #46911) were transfected into cells. For CRISPRa assays, vectors of sgRNA, MS2-P65-HSF1_GFP (Addgene, #61423) and dCAS9-VP64_GFP (Addgene, #61422) were transfected into cells. Fourty-eight hours after transfection, cells were collected to extract RNAs and the expression of Ctnnb1 was quantified by RT-qPCR.
RNA isolation, cDNA synthesis and quantitative RT–PCR (qPCR)
Total RNAs were prepared using the RNAiso Plus (TAKARA) according to the manufacturer’s protocols. Tissues or cells were lysed with 1 mL or 500 μL RNAiso Plus in DNase/RNase-free EP tubes on ice, followed by adding 200 μL or 100 μL chloroform to achieve phase separation. After shaking vigorously, tubes were centrifuged at 12,000× rpm for 15 min at 4 °C, and then transfer aqueous phase to new tubes. The aqueous phase was mixed with equal volumes of isopropyl alcohol to obtain RNA. Precipitation was resuspended with an appropriate DNase/RNase-free water. Complementary DNAs (cDNAs) were synthesized by HiScript® II Q RT SuperMix for qPCR kit (Vazyme; R222-01). qPCR primers were designed by PrimerBank72. cDNAs were used to detect different genes with 2× SYBR Green qPCR master mix (Bimake). Amplifications were performed using the CFX Connect Real-Time PCR Detection System (Bio-Rad) with a final volume of 10 μL under the following condition: 5 min at 95 °C and then 40 cycles at 95 °C for 15 s and 60 °C for 20 s. Relative expression levels for target genes were calculated using the 2−ΔΔCt method73, normalized to the Gapdh or ACTIN housekeeping gene.
X-Gal staining
For frozen sections, sections were fixed in fresh cold fixative (0.2% PFA) in buffer L0 (0.1 M PIPES buffer (pH 6.9), 2 mM MgCl2, 5 mM EGTA) for 10 min. Rinse the slides in PBS plus 2 mM MgCl2 on ice, followed by a 10 min wash in the same solution. Place slides in detergent rinse [0.1 M PBS (pH 7.3), 2 mM MgCl2, 0.01% sodium-deoxycholate, 0.02% Nonidet P-40] on ice for 10 min. Slides were then moved to a freshly made and filtered X-Gal staining solution [0.1 M PBS (pH 7.3), 2 mM MgCl2, 0.01% sodium-deoxycholate, 0.02% Nonidet P-40, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6·3H2O and 1 mg/ml X-Gal]. Sections were incubated at 37 °C from a few minutes to overnight in the dark. Sections were rinsed with water to stop the reaction. Sections were dehydrated with gradient ethanol and xylene sequentially, and mounted with neutral balsam. For whole body or brains, tissues were immersed in 4% PFA 10 min (brains) or 30 min (whole body) at room temperature. Rinse fixed tissues in detergent rinse three times at room temperature for 15–30 min. Tissues were then transferred to freshly made and filtered X-Gal staining solution. The staining time depends on the size of the sample and the strength of bacterial β-Gal expression. After staining, embryos or brains were washed three times in PBS and photographed74.
Nissl staining
Brain sections were stained in 0.25% cresyl violet solution (Sigma-Aldrich) for 15 min at 65 °C. Then, sections were decolorized in ethanol, the decolorization time depends on the strength of color. After decolorizing, sections were dehydrated in ethanol for 5 min, followed by a 10 min wash in xylene and mounted in a neutral balsam.
Immunohistochemical staining
Frozen brain sections were heated to 95°C in citrate antigen retrieval solution (pH 6.0) for 25 min to deactivate endogenous peroxidase. When the temperature of the solution drops to room temperature, sections were rinsed three times in PBS and blocked in solution 1 [PBS with 0.5% Triton X-100 and 5% bovine serum albumin (BSA)] for 1 h at room temperature. Sections were then incubated overnight at 4 °C with primary antibodies in solution 1. The next day, sections were incubated with appropriate secondary antibodies for 1 h at room temperature. Then, sections were incubated with the avidin-biotin-peroxidase complex (A:B = 1:50; VECTASTAIN Elite ABC system, Vector Labs). Peroxidase was reacted in solution 2 (tris-HCl (pH 7.2) with 5 mg/ml 3,3'-diaminobenzidine and 0.075% H2O2). The sections were dehydrated, washed in xylene and mounted in a neutral balsam.
Immunofluorescence
Frozen brain sections were dried at temperature and heated to 95 °C in citrate antigen retrieval solution (pH 6.0) for 25 min. For BrdU staining, sections were incubated with 2 N HCl 30 min at room temperature. Sections were blocked in blocking buffer (3% heat-inactivated normal goat serum, 0.1% Triton X-100 and 0.1% BSA in 10 mM Tris-HCl, pH 7.4; 100 mM NaCl) for 1 h at room temperature. Sections were then incubated overnight at 4 °C with primary antibodies [rat anti-BrdU (1:500; Abcam, ab6326), rabbit anti-TBR2 (1:1000; Abcam, ab23345), rabbit anti NEUROD2 (1:500; Abcam, ab104430), chicken anti- beta Galactosidase (1:500; Immunology Consultants Laboratory, CGAL-45A-Z) rat anti-TBR2 (1:1000; Thermo Fisher Scientific, 14-4875-82), mouse anti-FOXP2 (1:250; Sigma-Aldrich, AMAB91361), rat anti-CTIP2 (1:500; Abcam, ab18465), mouse anti-SATB2 (1:500; Abcam, ab51502), rabbit anti-CUX1 (1:100; Santa Cruz Bio- technology, sc-13024) and rabbit anti-PAX6 (1:500; Millipore, AB2237)] in blocking buffer. The next day, sections were incubated with appropriate secondary antibodies (Alexa Fluor 488-conjugated anti-mouse, Alexa Fluor 488-conjugated anti-rat, Alexa Fluor 488-conjugated anti-rabbit, Alexa Fluor 555-conjugated anti-mouse, Alexa Fluor 555-conjugated anti-rabbit and Alexa Fluor 647-conjugated anti-rabbit; Thermo Fisher Scientific; 1:1000) for 1 h at room temperature. Sections were mounted with anti-fade (1×; Thermo Fisher Scientific) solution with 4′,6-diamidino-2-phenylindole (DAPI, 0.1 μg/ml) in PBS. All comparative expression levels of immunofluorescence were obtained at the same collective conditions.
5-Ethynyl-2′-Deoxyuridine (EdU) staining
Proliferation of cells was investigated with BeyoClickTM EdU Cell Proliferation Kit (C0075S, Beyotime, China) according to the manufacturer’s protocols. In brief, frozen brain sections were dried at temperature and permeated with 0.3% Triton X-100 in PBS for 30 min. After that, the sections were incubated with EdU working solution for 1 h at 37 °C in the dark. After incubation, regular immunofluorescence staining can be followed.
In situ hybridization (ISH)
Templates for antisense RNA probes were cloned from mouse cDNA with specific primers for Ctnnb1. Then, templates were cloned into the pGEM-T Easy vector (Promega). Vector with target template was linearized by appropriate restriction enzyme and then transcribed to DIG-labeled probes by using the DIG-RNA Labeling Mix (Roche). Cryosections were dried in a hybridization oven for 15 min at 50 °C, followed by fixing with 4% PFA for 20 min at room temperature. Then, sections were permeabilized with 2 μg/mL proteinase K (Sigma) in PBS for 10 min at room temperature and acetylation in 0.1 M TEA (triethanolamine) for 10 min at room temperature. After permeabilization and acetylation, sections were blocked in hybridization buffer (5× SSC, 5× Denharts; 500 μg/mL herring sperm DNA; and 250 μg/mL yeast RNA; 50% deionized formamide) for 3 h at room temperature followed by incubating with a DIG-labeled probe diluted (0.2 ng/μL) in hybridization buffer overnight at 65 °C in a hybridization oven. Sections were washed with 0.1× SSC for four times (20 min each) in a hybridization oven at 65 °C, followed by treating with ribonuclease A (TAKARA) (20 μg/mL) for 20 min at 37 °C and then blocked with 10% normal sheep serum in Buffer B1 (0.1 M Tris-HCl, pH 7.4; 150 mM NaCl) for 3.5 h at room temperature. Sections were incubated with 1:5000 dilution of anti-DIG antibody (Roche) overnight at 4 °C. The next day, sections were washed with Buffer B3 (0.1 M Tris-HCl; 0.1 M NaCl; 50 mM MgCl2; 0.1% Tween-20, pH 9.5) for three times (10 min each) at room temperature, followed by colorization with BCIP/NBT (bromochloroindolyl phosphate/nitro blue tetrazolium) (Roche) containing B3 solutions at room temperature in dark. The colorization time depends on the strength of target genes expression. Sections were dehydrated with gradient ethanol and xylene sequentially, and mounted with neutral balsam. The probe of Ctnnb1 were forward 5′-cacgactagttcagctgcttgt-3′/reverse 5′-tccacacatgaacatctccttc-3 according to the ALLEN BRAIN MAP (https://portal.brain-map.org/).
In utero electroporation (IUE) of developing neocortices
Pregnant WT and neCtnnb1KO mice with E14.5 embryos were anesthetized by peritoneally injection of pentobarbital sodium (70 mg/kg), and the uteri were exposed through a 2 cm midline abdominal incision. Embryos were carefully pulled out using ring forceps through the incision and placed on sterile gauze wet with 0.9% sodium chloride. Plasmid of pCIG (1 μg/μL, prepared using Endo Free plasmid purification kit, Tiangen) mixed with 0.05% Fast Green (Sigma) was injected through the uterine wall into the telencephalic vesicle. Five electric pulses (36 V, 50 ms duration at 1 s intervals) were generated using CUY21VIVO-SQ (BEX) and delivered across the head of embryos using 5 mm forceps-like electrodes (BEX). The uteri were then carefully put back into the abdominal cavity, and both peritoneum and abdominal skin were sewed with surgical sutures. The whole procedure was completed within 30 min. Mice were warmed on a heating pad until they regained consciousness and were treated with analgesia (ibuprofen in drinking water) until sacrifice at E18.5.
ChIP-qPCR assay
ChIP-qPCR was conducted essentially as described previously75. 100 mg forebrain cortex (gently homogenized to cell dispersion in ice-cold PBS with protease inhibitors) or 1 × 107 cells were cross-linked with 1% formaldehyde for 10 min at room temperature and quenched by adding 0.125 M glycine for 5 min. Cells were then washed with ice-cold PBS twice (10 min each). Next, cells were harvested in 500 μL lysis buffer (50 mM Tris-HCl, pH 8.0, 0.5% SDS, 5 mM EDTA) and 500 μL digestion buffer (50 mM Tris-HCl, pH 7.6; 1 mM CaCl2, 0.2% Triton X-100) plus 1400 U micrococcal nuclease (NEB; M0247S) for 20 min at 37 °C, followed by adding 5 μL 0.5 M EDTA to stop reaction and incubating on ice for 5 min. Sonicate cells in EP tubes with power output 100 W, 3 min (cells) or 7 min (cortex), 0.5 s on, 0.5 s off on ice. The agarose gel ensures that the size of DNA fragments was proper. One percent of the sonicated lysate was taken as the input. The rest of the sonicated lysates was diluted into 0.1% SDS using dilution buffer (20 mM Tris-HCl, pH 8.0; 150 mM NaCl; 2 mM EDTA; 1% Triton X-100) and divided into two parts for IgG and target protein immunoprecipitation. Samples were incubated with 25 μL pre-washed protein G agarose beads and 2 μg anti-ASH2L (Bethyl Laboratories; A300-489A) or 25 μL anti-Flag affinity gel (Bimake; B23101) overnight at 4 °C on rocker. The next day, beads were wash with Wash Buffer I (20 mM Tris-HCl, pH 8.0; 1% Triton X-100; 2 mM EDTA; 150 mM NaCl; 0.1% SDS), Wash Buffer II (20 mM Tris-HCl, pH 8.0; 1% Triton X-100; 2 mM EDTA; 500 mM NaCl; 0.1% SDS), Wash Buffer III (10 mM Tris-HCl, pH 8.0; 1 mM EDTA; 0.25 M LiCl; 1% NP-40; 1% deoxycholate) and TE buffer. Beads were resuspended in elution buffer (0.1 M NaHCO3, 1% SDS, 20 µg/mL proteinase K) to elute the DNA. The elution was incubated at 65 °C for overnight to reverse cross-linked chromatin. DNA fragments were extracted with DNA purification kit (TIANGEN). The purified DNA was quantified by RT-qPCR.
3D genome browser analysis
The Hi-C and virtual 4C data from cells or tissues were obtained from the 3D Genome Browser76 (http://3dgenome.fsm.northwestern.edu/). In addition, TADs were identified to screen for potentially interacting genes in cells or tissues.
Luciferase reporter assay
The luciferase reporter assay was performed as described77. Briefly, the 696-bp conserved region of neCtnnb1 (chr9:120,869,524-120,870,219, CR) was cloned from mouse brain genomic DNA and subcloned into pGL3-promoter vector (Promega). Neuro-2a cells were transfected with pGL3-promoter vector and pGL3-pro-CR vector or pCDH-ASH2L along with pGL3-promoter vector and pGL3-pro-CR vector. Renilla basic vector was cotransfected as a control for normalization of luciferase activity. Luciferase activity was measured 48 h after transfection using a Promega Dual-Glo assay kit, as per the manufacturer’s instructions.
Quantification and statistical analysis
Sections used for quantification were position-matched for wild-type and experimental brains. Images were binned against dorsal-ventral position to quantify the intensity of layer-specific distributed ISH or LacZ signals of cortex. The signal intensity of those regions was generated using ImageJ as described previously78,79. Statistical tests were performed using GraphPad Prism (version 8.0.2). Data are presented as means ± SEM or ± SD. Unpaired two-tailed Student’s t-test were used for analysis between two groups of equal variances by F-tests. When the F-test of equal variance failed, the Welch t-tests were used. Data comparison of three or more groups with control groups were analyzed using one-way ANOVA followed by Dunnett’s multiple comparison test. Data comparison of three or more groups among each other were analyzed using one-way ANOVA followed by Tukey’s multiple comparison test. Statistically significant was considered by P ≤ 0.05 (*P < 0.05, **P < 0.01, and ***P < 0.001).
Data availability
All data are available in the paper and/or the supplementary materials.
References
Rakic, P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 10, 724–735 (2009).
Florio, M. & Huttner, W. B. Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194 (2014).
Lui, J. H., Hansen, D. V. & Kriegstein, A. R. Development and evolution of the human neocortex. Cell 146, 18–36 (2011).
Haubensak, W., Attardo, A., Denk, W. & Huttner, W. B. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl Acad. Sci. USA 101, 3196–3201 (2004).
Kriegstein, A., Noctor, S. & Martinez-Cerdeno, V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 7, 883–890 (2006).
Molyneaux, B. J., Arlotta, P., Menezes, J. R. & Macklis, J. D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427–437 (2007).
Cardenas, A. et al. Evolution of cortical neurogenesis in amniotes controlled by Robo signaling levels. Cell 174, 590–606 (2018).
Cardenas, A. & Borrell, V. Molecular and cellular evolution of corticogenesis in amniotes. Cell Mol. Life Sci. 77, 1435–1460 (2020).
Attardo, A., Calegari, F., Haubensak, W., Wilsch-Brauninger, M. & Huttner, W. B. Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in Apical versus basal progenitor progeny. Plos One 3, e2388 (2008).
Stoet, G. & Snyder, L. H. Single neurons in posterior parietal cortex of monkeys encode cognitive set. Neuron 42, 1003–1012 (2004).
Vasistha, N. A. et al. Cortical and clonal contribution of Tbr2 expressing progenitors in the developing mouse brain. Cereb. Cortex 25, 3290–3302 (2015).
Xing, L. et al. Serotonin receptor 2A activation promotes evolutionarily relevant basal progenitor proliferation in the developing neocortex. Neuron 108, 1113–1129 (2020).
Kalebic, N. et al. Neocortical expansion due to increased proliferation of basal progenitors is linked to changes in their morphology. Cell Stem Cell 24, 535–550 (2019).
Wang, A. et al. An epigenetic circuit controls neurogenic programs during neocortex development. Development 148, dev199772 (2021).
Machon, O., van den Bout, C. J., Backman, M., Kemler, R. & Krauss, S. Role of beta-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience 122, 129–143 (2003).
Munji, R. N., Choe, Y., Li, G., Siegenthaler, J. A. & Pleasure, S. J. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J. Neurosci. 31, 1676–1687 (2011).
Singh, K. K. et al. Common DISC1 polymorphisms disrupt Wnt/GSK3beta signaling and brain development. Neuron 72, 545–558 (2011).
Chenn, A. & Walsh, C. A. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002).
Inestrosa, N. C. & Arenas, E. Emerging roles of Wnts in the adult nervous system. Nat. Rev. Neurosci. 11, 77–86 (2010).
Lie, D. C. et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature 437, 1370–1375 (2005).
Dong, F. et al. Deletion of CTNNB1 in inhibitory circuitry contributes to autism-associated behavioral defects. Hum. Mol. Genet 25, 2738–2751 (2016).
O’Roak, B. J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012).
Huang, S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
Saito-Diaz, K. et al. APC inhibits ligand-independent Wnt signaling by the clathrin endocytic pathway. Dev. Cell 44, 566–581 (2018).
Durak, O. et al. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat. Neurosci. 19, 1477–1488 (2016).
Croft, B. et al. Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9. Nat. Commun. 9, 1–10 (2018).
Mochizuki, Y. et al. Combinatorial CRISPR/Cas9 approach to Elucidate a far-upstream enhancer complex for tissue-specific Sox9 expression. Dev. Cell 46, 794–806 (2018).
Smith, E. & Shilatifard, A. Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 21, 210–219 (2014).
Jindal, G. A. & Farley, E. K. Enhancer grammar in development, evolution, and disease: dependencies and interplay. Dev. Cell 56, 575–587 (2021).
Lettice, L. A. et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 (2003).
Shi, J. et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 27, 2648–2662 (2013).
Schoenfelder, S. & Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet 20, 437–455 (2019).
Gonen, N. et al. Sex reversal following deletion of a single distal enhancer of Sox9. Science 360, 1469–1473 (2018).
Chen, H. T. et al. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet 50, 1296–1303 (2018).
Boyd, J. L. et al. Human-Chimpanzee differences in a FZD8 enhancer Alter cell-cycle dynamics in the developing Neocortex. Curr. Biol. 25, 772–779 (2015).
Glasgow, S. M. et al. Glia-specific enhancers and chromatin structure regulate NFIA expression and glioma tumorigenesis. Nat. Neurosci. 20, 1520–1528 (2017).
Lee, S. M., Tole, S., Grove, E. & McMahon, A. P. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development 127, 457–467 (2000).
Galceran, J., Miyashita-Lin, E. M., Devaney, E., Rubenstein, J. L. & Grosschedl, R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development 127, 469–482 (2000).
Kalani, M. Y. et al. Wnt-mediated self-renewal of neural stem/progenitor cells. Proc. Natl. Acad. Sci. USA 105, 16970–16975 (2008).
Wrobel, C. N., Mutch, C. A., Swaminathan, S., Taketo, M. M. & Chenn, A. Persistent expression of stabilized beta-catenin delays maturation of radial glial cells into intermediate progenitors. Dev. Biol. 309, 285–297 (2007).
Nakagawa, N. et al. APC sets the Wnt tone necessary for cerebral cortical progenitor development. Genes Dev. 31, 1679–1692 (2017).
Junghans, D., Hack, I., Frotscher, M., Taylor, V. & Kemler, R. beta-catenin-mediated cell-adhesion is vital for embryonic forebrain development. Dev. Dynam 233, 528–539 (2005).
Boitard, M. et al. Wnt signaling regulates multipolar-to-bipolar transition of migrating neurons in the cerebral cortex. Cell Rep. 10, 1349–1361 (2015).
Li, L. et al. The COMPASS family protein ASH2L mediates corticogenesis via transcriptional regulation of Wnt signaling. Cell Rep. 28, 698–711 (2019).
Yang, F. et al. Cis-Acting circ-CTNNB1 promotes beta-catenin signaling and cancer progression via DDX3-mediated transactivation of YY1. Cancer Res. 79, 557–571 (2019).
Oberst, P. et al. Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature 573, 370–374 (2019).
Harrison-Uy, S. J. & Pleasure, S. J. Wnt signaling and forebrain development. Cold Spring Harb. Perspect. Biol. 4, a008094 (2012).
Mutch, C. A., Funatsu, N., Monuki, E. S. & Chenn, A. Beta-catenin signaling levels in progenitors influence the laminar cell fates of projection neurons. J. Neurosci. 29, 13710–13719 (2009).
Mutch, C. A., Schulte, J. D., Olson, E. & Chenn, A. Beta-catenin signaling negatively regulates intermediate progenitor population numbers in the developing cortex. PLoS One 5, e12376 (2010).
Kuwahara, A. et al. Wnt signaling and its downstream target N-myc regulate basal progenitors in the developing neocortex. Development 137, 1035–1044 (2010).
Draganova, K. et al. Wnt/β-catenin signaling regulates sequential fate decisions of murine cortical precursor cells. Stem Cells 33, 170–182 (2015).
Woodhead, G. J., Mutch, C. A., Olson, E. C. & Chenn, A. Cell-autonomous beta-catenin signaling regulates cortical precursor proliferation. J. Neurosci. 26, 12620–12630 (2006).
Machon, O. et al. A dynamic gradient of Wnt signaling controls initiation of neurogenesis in the mammalian cortex and cellular specification in the hippocampus. Dev. Biol. 311, 223–237 (2007).
Clinton, B. K., Cunningham, C. L., Kriegstein, A. R., Noctor, S. C. & Martinez-Cerdeno, V. Radial glia in the proliferative ventricular zone of the embryonic and adult turtle, Trachemys scripta elegans. Neurogenesis (Austin) 1, e970905 (2014).
Martinez-Cerdeno, V. et al. Evolutionary origin of Tbr2-expressing precursor cells and the subventricular zone in the developing cortex. J. Comp. Neurol. 524, 433–447 (2016).
Andersson, R. & Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet 21, 71–87 (2020).
Shyamsunder, P. et al. Identification of a novel enhancer of CEBPE essential for granulocytic differentiation. Blood 133, 2507–2517 (2019).
Visel, A. et al. A high-resolution enhancer atlas of the developing telencephalon. Cell 152, 895–908 (2013).
Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020).
Song, M. et al. Cell-type-specific 3D epigenomes in the developing human cortex. Nature 587, 644–649 (2020).
Dickel, D. E. et al. Ultraconserved enhancers are required for normal development. Cell 172, 491–499 (2018).
Kvon, E. Z. et al. Comprehensive in vivo interrogation reveals phenotypic impact of human enhancer variants. Cell 180, 1262–1271 (2020).
Lettice, L. A. et al. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev. Cell 22, 459–467 (2012).
Long, H. K. et al. Loss of extreme long-range enhancers in human neural crest drives a craniofacial disorder. Cell Stem Cell 27, 765–783 (2020).
Herz, H. M., Hu, D. & Shilatifard, A. Enhancer malfunction in cancer. Mol. Cell 53, 859–866 (2014).
Guo, C. et al. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons,astrocytes, and oligodendrocytes. Neuron 80, 1167–1174 (2013).
Choi, S. H. et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278 (2014).
Hagege, H. et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 (2007).
Tian, K. et al. Transcriptome analysis identifies SenZfp536, a sense LncRNA that suppresses self-renewal of cortical neural progenitors. Neurosci. Bull. 37, 183–200 (2021).
Alerasool, N., Segal, D., Lee, H. & Taipale, M. An efficient KRAB domain for CRISPRi applications in human cells. Nat. Methods 17, 1093–1096 (2020).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Wang, X., Spandidos, A., Wang, H. & Seed, B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40, D1144–D1149 (2012).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Visel, A., Minovitsky, S., Dubchak, I. & Pennacchio, L. A. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–D92 (2007).
Li X. J. et al. Deficiency of histone methyltransferase SETD2 in liver leads to abnormal lipid metabolism and hepatocarcinoma. Hepatology 73, 1797–1815 (2021).
Wang, Y. et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19, 151 (2018).
Li, Y. et al. Paired related homeobox 1 transactivates dopamine D2 receptor to maintain propagation and tumorigenicity of glioma-initiating cells. J. Mol. Cell Biol. 9, 302–314 (2017).
Golonzhka, O. et al. Pbx regulates patterning of the cerebral cortex in progenitors and postmitotic neurons. Neuron 88, 1192–1207 (2015).
Xu L., et al. Abnormal neocortex arealization and Sotos-like syndrome-associated behavior in Setd2 mutant mice. Sci. Adv. 7, eaba1180 (2021).
Acknowledgements
We thank Drs. Junlei Chang and Haitao Wu for providing BAT-Gal and Ctnnb1lox(ex3) mice. We thank the Core Facility and the Animal Facility of Medical Research Institute of Wuhan University for technical support. We thank Dr. Weimin Zhong and all Zhou lab members for critical reading of the manuscript. This work was supported by grants from National Key R&D Program of China (2018YFA0800700), National Natural Science Foundation of China (No. 31970770 and 31970676) and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
Y.Zho. conceived and designed the study; J.W., A.W. and K.T. performed experiments and analyzed data; X.H. contributed to genotyping; W.L. assisted with 3C experiments; L.X., B.Z., Y.Zhe., X.K., J.W., Z.L. and Y.L. were involved in figure construction; Y.L. and Y.Zho. provided the funding. Y.Z. and J.W. wrote the paper. All authors discussed results and commented to the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, J., Wang, A., Tian, K. et al. A Ctnnb1 enhancer regulates neocortical neurogenesis by controlling the abundance of intermediate progenitors. Cell Discov 8, 74 (2022). https://doi.org/10.1038/s41421-022-00421-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41421-022-00421-2
- Springer Nature Singapore Pte Ltd.
This article is cited by
-
KDM2B regulates hippocampal morphogenesis by transcriptionally silencing Wnt signaling in neural progenitors
Nature Communications (2023)