Abstract
Patients with teratozoospermia exhibit low phosducin-like protein (Pdcl2) expression. As a member of the phosducin family, chaperonin-related Pdcl2, a germline-specific gene, may be involved in germ cell protein folding. Given that PDCL2 is highly conserved in evolution, it may be indispensable for mammalian spermiogenesis; however, the function of PDCL2 in higher mammalian species remains unknown. To determine the role of PDCL2 in male fertility, we generated Pdcl2 knockout mice using CRISPR/Cas9. Our results revealed that Pdcl2 heterozygous (Pdcl2+/−) male mice were normal, but male Pdcl2-null (Pdcl2−/−) mice were infertile. Accordingly, Pdcl2−/− male mice exhibited lower testis weight, epididymis weight, and sperm number than Pdcl2+/+ mice. Moreover, Pdcl2−/− mice displayed malformed and immotile sperm. Apoptotic cells were significantly enhanced in Pdcl2−/− testes and epididymis when compared with those in wild-type mice. Mechanistically, PDCL2 can interact with the CCT complex, and dysfunction in this complex might lead to infertility in Pdcl2−/− male mice. Collectively, these findings confirm that Pdcl2 knockout leads to male infertility in mice and that PDCL2 may function as a chaperone to promote protein folding during spermiogenesis.
Similar content being viewed by others
Introduction
According to epidemiological investigations, 8–12% of all couples worldwide suffer from infertility, and male factors contribute to approximately 50% of incidences [1]. Sperm malformation is one of the key factors that leads to male infertility in humans. Interestingly, patients with teratozoospermia exhibit low levels of phosducin-like protein 2 (Pdcl2) expression [2]. Chaperonin-related Pdcl2, a member of the phosducin family, is a germline-specific gene [3, 4]. The phosducin family members, except phosducin, play a functional role in assisting protein folding [5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20]. Pdcl2 is highly conserved in evolution [3, 21, 22]. Overexpression of mouse PDCL2 in a deficient yeast strain can completely rescue the meiotic defect phenotype [3], revealing a conserved function from yeast to mammalian cells. However, the role of PDCL2 in higher eukaryotes remains unclear.
Some of the phosducin-like proteins are required for cytosolic chaperonin complex (CCT/TRiC)-mediated protein folding [5, 11, 13,14,15, 17]. CCT belongs to the group II chaperonins, with its two-stack barrel-like structure, which provides sequestered space for protein folding in an ATP-dependent manner [23, 24]. Although the simultaneous presence of phosducin-like proteins and the CCT complex dramatically enhances the protein folding ability when compared with CCT alone, the underlying regulatory mechanism remains elusive.
Considering the conserved sequence of PDCL2 among species, and the fact that the orthologue of PDCL2 plays a role in protein folding [14,15,16], PDCL2 may be involved in protein folding in the germline. In the present study, we generated knockout mouse models to investigate the role of PDCL2 in spermatogenesis.
Results
Pdcl2 is predominantly expressed in Testis with a stage-specific pattern
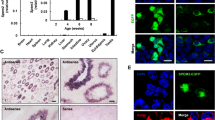
Pdcl2, or named MgcPhLP, refers to mouse germ cell-specific phosducin-like protein [3]. Herein, we examined adult mouse tissues to confirm the specific expression of Pdcl2 in germ cells. Detection of Pdcl2 mRNA and PDCL2 protein in a series of tissues showed its expression in the testis exclusively (Fig. 1A, B), as determined by microarrays in humans (Fig. S3A) and mice (Fig. S3B).
A, B Expression analysis in various tissues from wild-type 11-week-old C57BL/6 mice. A RT-PCR results. The ddH2O group was used as PCR control. The β-actin group was used as loading control. B Western blot analysis. GAPDH was used as loading control. C, D Immunofluorescence staining of different germ cells extracted from testes and mature sperm isolated from cauda epididymis of wild-type 11-week-old C57BL/6 mice. Green indicates different markers of the cells. PLZF is a marker of spermatogonial stem cell. SYCP3 is a synaptonemal complex component. SP56 is a constituent of the acrosomal matrix. PDCL2 was labeled in magenta. Hoechst 33342 labeled cell nuclear was in blue. Bar, 10 µm.
We performed immunofluorescence staining of germ cells from wild-type mouse testis and observed stage-specific expression patterns of PDCL2 during spermatogenesis. Different biomarkers of germ cells have been used to identify distinct cell types. PLZF-positive cells are undifferentiated, differentiating into spermatogonia in the testis [25, 26]. GATA4-positive cells are somatic cells in testis [27, 28]. SYCP3 is a component of the synaptonemal complex in meiotic cells [29, 30]. SP56 is a component of the acrosomal complex in sperm [31]. PDCL2 were detected in germ cells from meiotic cells to elongated spermatids at all spermatogenesis stages, but not in mitotic cells or spermatozoa isolated from cauda epididymis (Fig. 1C, D). Consistently, Pdcl2 microarrays of different types of germ cells revealed expression in pachytene spermatocytes and round spermatids but not in spermatogonia stem cells (Fig. S3C).
Overall, these results confirmed that Pdcl2 is predominantly expressed in male germ cells at a specific stage, indicating that Pdcl2 might have a potential function in spermatogenesis.
Generation of Pdcl2 Knock-out Mice
As PDCL2 protein exists in cell types ranging from meiotic cells to elongated spermatids, we disrupted the Pdcl2 gene to examine its role in germ cell development in vivo. We constructed Pdcl2 knockout mice using CRISPR/Cas9 system. Two gRNAs targeting exon 1 and exon 2 of Pdcl2 (Fig. 2A) were designed using http://crispr.mit.edu. Plasmids expressing Cas9 and gRNAs were transfected into mouse V6.5 embryonic stem cells (ESCs). The genome editing efficiency was determined using the T7E1 assay. Both gRNAs showed notable cleavage of the target site in mouse ESCs (Fig. 2B). Cas9 mRNA and gRNAs were injected into the cytoplasm of mouse zygotes to produce mutant mice. First-generation mutant mice were mated with wild-type C57BL/6 mice for three generations to eliminate off-target effects of the CRISPR/Cas9 system. Fourth-generation of Pdcl2+/− mice were mated to generate Pdcl2+/+, Pdcl2+/−, and Pdcl2−/− mice.
A Schematic representation of the gRNA target sites of the Pdcl2 gene. Exon 1 is the noncoding region. Exon 2 is the beginning of the coding region. B Efficacy of CRISPR/Cas9 with the two designated gRNA. V6.5 mouse embryonic stem cells were transfected with different gRNA-containing pX330 or GFP plasmid as a negative control. Targeted genomic region was PCR amplified and then digested by T7 Endonuclease I. M, molecular weight markers. NC, negative control. C Genotype of the Pdcl2 knock-out mice. Both gRNAs were used simultaneously, generating indels in both gRNA target sites in the Pdcl2 knock-out mice. Indels were generated as highlighted in blue. Protospacer adjacent motifs (PAM) were labeled in red. Minus signs were used to fill in the blanks of the alignment. Apostrophes replaced the omitted sequences due to the demonstration clarity purpose. D, E Confirmation of PDCL2 deficiency in Pdcl2−/− mice. D Representative western blot analysis of testes lysate. GAPDH was used as loading control. E Representative immunofluorescent analysis of testes sections. Mouse testes were paraffin-embedded and sectioned in 7 µm. PDCL2 was labeled in red. Hoechst 33342 labeled cell nuclear was in blue. Bar, 50 µm.
To verify Pdcl2 mutations in mice, the offspring genotypes were detected using Sanger sequencing. Frame-shift mutations were generated at both gRNA target sites (Fig. 2C). We performed western blot analysis of testis lysates with PDCL2 polyclonal antibody, which revealed PDCL2 expression in Pdcl2+/+ and Pdcl2+/− mice, but not in Pdcl2−/−mice (Fig. 2D). As expected, PDCL2 expression was not detected in the seminiferous tubules of Pdcl2−/− mice, as shown by immunostaining (Fig. 2E). These results indicated the successful knockout of Pdcl2 by CRISPR/Cas9.
Male Pdcl2 −/− Mice Were Infertile with Abnormal Spermatogenesis
Pdcl2−/− mice were normal in appearance and presented no obvious phenotypic differences in growth when compared with Pdcl2+/+ and Pdcl2+/− mice (Fig. 3A). However, male Pdcl2−/− mice failed to produce offspring after mating with normal wild-type female mice following one month of continuous mating (Fig. 3B). Meanwhile, Pdcl2+/− mice of both sexes and female Pdcl2−/− mice were fertile.
A Body weight of 11-week-old male mice. B Fertility test. Pdcl2+/+, Pdcl2+/−, and Pdcl2−/− males were mated with wild-type C57BL/6 female mice for one month period of continuous mating. Six adult male mice of each genotype were used. Pdcl2−/− males gave birth to no pups. C, F Appearance of testis or epididymis. Bar, 1 cm. D, G Reduced testis and epididymis weight of Pdcl2−/− mice. The tissue to body ratio was used to normalize the data. Each sample dot represents the average parameter of both sides of testis or epididymis from the same mouse. E, H Hematoxylin and eosin stained cross sections of paraffin-embedded mouse testis or cauda epididymis. Bar, 100 µm. E Stage XI seminiferous tubule cross section was shown. Less elongated spermatids could be found in Pdcl2−/− mice. H Round shaped cells instead of elongated spermatids could be observed in the epididymis of Pdcl2−/− mice. I, J Apoptosis analysis of mouse testis or cauda epididymis section. TUNEL positive signals were counted from pictures of the stained sections. The pictures were taken at random places of the specimens. Each picture covered an area of 375,240 µm2. Each dot of the diagram represented a count number of one picture. Different color of dots represented datasets from different specimens. Lines with different color represented means of the datasets. Lines in black represented means of the three means of each group. K Percentage of germ cells with different ploidy type in testes of 11-week-old male mice. More than 104 cells of each sample were gated. Data of this figure were represented as mean ± standard deviation. *, p < 0.05; **, p < 0.01; ***, p < 0.001. n, mouse number.
To determine the underlying mechanims, we dissected the Pdcl2−/− mice for further examination. Pdcl2−/− mice exhibited significantly smaller testes than Pdcl2+/+ and Pdcl2+/− littermates (Fig. 3C). The testes weight of Pdcl2−/− mice was reduced by approximately 30–40% (testis to body weight ratio of Pdcl2−/− mice was 2.52 ± 0.19‰ compared to 3.91 ± 0.31‰ of Pdcl2+/+ mice and 3.77 ± 0.26‰ of Pdcl2+/− mice) at 11 weeks, whereas no difference was observed between testes of Pdcl2+/+ and Pdcl2+/− mice (Fig. 3D). Hematoxylin and eosin staining of the seminiferous epithelium of Pdcl2−/− mice revealed histological abnormalities. Pdcl2−/− mice showed a disruption of spermiogenesis at elongating spermatid stages. We detected markedly few elongated spermatids in stages XI-VIII seminiferous tubules, and most elongated spermatids presented deformities of spermatid nuclei in Pdcl2−/− mice (Fig. 3E). Compared with Pdcl2+/+ mice, Pdcl2+/− mouse testes showed no histological abnormalities.
Likewise, Pdcl2−/− mice displayed abnormalities in the epididymis, which appeared more transparent (Fig. 3F), thereby suggesting less sperm in the epididymis. The epididymis weight of the Pdcl2−/− mice was reduced by approximately 10-15% (epididymis to body weight ratio of Pdcl2-/- mice was 1.40 ± 0.03‰ compared with 1.64 ± 0.13‰ in Pdcl2+/+ mice and 1.61 ± 0.06‰ in Pdcl2+/− mice) (Fig. 3G). Histological analysis of the epididymis showed that Pdcl2−/− mice had fewer spermatozoa in cauda epididymis and most of the spermatozoa were not elongated while with bigger cell size compared to those of the Pdcl2+/+ and Pdcl2+/− mice (Fig. 3H).
To determine the underlying cause of weight loss in Pdcl2−/− mice testes and epididymis, we first examined germ cells apoptosis. Apoptosis plays a key role in spermatogenesis quality control [32]. Dysfunctional spermatogenesis typically relies on apoptosis to eliminate dysfunctional cells. TUNEL assays of Pdcl2+/+ and Pdcl2−/− testes and epididymis showed increased apoptotic cells in Pdcl2−/− mice (Fig. 3I, J; Fig. S4), potentially accounting for the decrease in testes and epididymis weight.
The DNA content of germ cells in the testes was analyzed using flow cytometry. Although Pdcl2+/+ and Pdcl2+/− had a similar proportion of haploid, diploid and tetraploid cells, Pdcl2−/− presented fewer haploid cells, and hence a greater number of diploid and tetraploid cells (Fig. 3K). No significant difference was found in the ratio of diploid and tetraploid cells (1.00 ± 0.16 in Pdcl2+/+, 0.94 ± 0.22 in Pdcl2+/−, and 1.07 ± 0.17 in Pdcl2−/−), suggesting that spermatogenesis before meiosis MI stage may be normal.
Oligo-astheno-teratozoospermia of Pdcl2 −/− Mice
To investigate how PDCL2 deficiency leads to male infertility, we examined sperm squeezed from the cauda epididymis of Pdcl2+/+, Pdcl2+/−, and Pdcl2−/− mice and quantified the obtained sperm samples. Although we detected no difference in the number of Pdcl2+/+ and Pdcl2+/− sperm, Pdcl2−/− sperm were reduced in number by approximately 70% (sperm count of Pdcl2−/− mice was 4.63 ± 1.34 × 106 compared with 14.96 ± 4.43 × 106 of Pdcl2+/+ mice and 14.57 ± 2.55 × 106 of Pdcl2+/− mice) (Fig. 4A). Sperm motility was determined by computer-assisted sperm analysis. All the sperm from Pdcl2−/− mice were immotile (Fig. 4B).
A Sperm number per cauda epididymis of 11-week-old male mice. B Percentage of immotile sperm. C Percentage of malformed sperm. Data of this figure were represented as mean ± standard deviation. *, p < 0.05; **, p < 0.01; ***, p < 0.001. n, mouse number. D Light microscopic analysis of sperm extracted from cauda epididymis. Bar, 20 µm. E Immunofluorescent analysis of the acrosomal structure of sperm extracted from cauda epididymis. SP56 is a constituent of the acrosomal matrix and was labeled in green. Hoechst 33342 labeled cell nuclear was in blue. Bar, 10 µm. F Observation of ultrastructure of sperm with transmission electron microscope. 1, sperm head of wild-type mice. 2, sperm midpiece of wild-type mice. 3–5, sperm of Pdcl−/− mice, with improper condensed nucleus and abnormal mitochondrial distribution. 3, head and tail were twined together. 4, the axoneme was branched. 5, large vacuole. Bar, 1 µm.
In addition, sperm of Pdcl2−/− mice were malformed without a regular hook-shaped head (Fig. 4C). On examining sperm morphology, we observed that nearly 50% of sperm from Pdcl2−/− mice had round heads with no tail, whereas the remaining had more slender tails when compared with normal sperm (Fig. 4D). To examine the malformed structure of Pdcl2−/− sperm head, we stained the sperm with the antibody of acrosome maker SP56. The result showed that Pdcl2−/− sperm lacked a normal acrosomal structure, i.e., a typical crescent moon shape (Fig. 4E). We then examined the sperm structure using transmission electron microscopy (Fig. 4F). Sperm of Pdcl2−/− mice failed to exhibit a normal mitochondrial distribution pattern. The mitochondria were clustered in a disordered manner. More white dots were observed in Pdcl2−/− sperm heads than in those of Pdcl2+/+, indicating that nuclei were not properly condensed. Heterogeneous abnormalities were detected in Pdcl2−/− sperm, including twined heads and tails, branched axonemes, and large vacuoles (Figs. 4F, 3–5).
Pdcl2−/− mice had reduced sperm, which were immotile and malformed, indicating that these mice were suffering from oligo-astheno-teratozoospermia, and that was the underlying cause of infertility.
PDCL2 Interacts with CCT and Actin
To elucidate the molecular function of PDCL2 in spermiogenesis, we used the PDCL2 antibody to pull down PDCL2 and PDCL2-associated protein(s) from testis protein extracts and identified the proteins by mass spectrometry. The most abundant proteins that interacted with PDCL2 were CCT components (Fig. 5A, Table S1). The GST pull-down assay further confirmed the interaction of CCT components with PDCL2 (Fig. 5B), in line with previous study performed in yeast [14,15,16, 33, 34]. PLP2 has been identified as a co-factor for actin folding by CCT [14,15,16]. We found that PDCL2 could also interact with actin in mammalian cells (Fig. 5C). Considering that the function of PDCL2 is conserved from yeast to mammals and transfection of mouse PDCL2 could rescue defects in PLP2 deficient yeast [3], PDCL2 might play a role in regulating actin protein folding during spermiogenesis in mice.
A Coimmunoprecipitation of cleared extracts from testes of 11-week-old male mice with the PDCL2 antibody. Proteins were identified using mass spectrometry. Components of the CCT complex were the most abundant proteins interact with PDCL2. B, C GST pull down assay. Fusion proteins with N-terminal GST tag or C-terminal S tag-FLAG tag-SBP tag were over-expressed in 293T cells. Solubilized lysates were immobilized on Glutathione Sepharose 4B. Irrelevant proteins were washed away. Retained proteins were detected by immunoblotting with FLAG antibody. GAPDH was indicated as the loading control. CCT8L1 was used as a negative control. B Interaction between CCT components and PDCL2. C Interaction between actins and PDCL2.
Discussion
PDCL2 is a germline-specific protein. As expected, disruption of Pdcl2 leads to reproductive malfunction in mice; however, this malfunction only occurred in male mice and not in females. This finding did not precisely meet our initial hypothesis, given that Pdcl2 was detected in male spermiogenesis stages, as well as the meiotic stages of both sexes (Fig. 1C and Fig. S3C–F) [3]. In addition, PDCL2 has a conserved meiotic function during evolution. Plp2, the yeast orthologue of mouse Pdcl2, is essential for yeast to produce haploid cells, and the PLP2 defected phenotype can be rescued by overexpressing mouse PDCL2 in yeast [3]. Therefore, it was unexpected that successful meiosis would be observed in male or female Pdcl2−/− mice. It can be speculated that PDCL2 no longer has any meiotic function in mice; however, this fails to explain the phenomena that mouse PDCL2 can rescue yeast meiotic defects.
Another possibility is that PDCL3, a paralog of PDCL2, compensates for the PDCL2 meiotic function. Considering that PDCL2 and PDCL3 exhibit a very high sequence similarity (Fig. S5A), and both can interact with CCT (Fig. 5A, B) [14], they may possess a similar function. In some cases of evolution, after gene duplication, the two daughter genes might mutate and change some expression control elements, resulting in distinct temporal and spatial expression patterns. Although they exhibit similar functions, both PDCL2 and PDCL3 are necessary and maintained in the genome [35]. PDCL2 and PDCL3 were separated and initially fixed in reptiles (Fig. S5B). PDCL2 expression is regulated by MYBL1 (A-MYB)-controlled RFX2 [36] and specifically expressed in the germline (Fig. 1 and Fig. S3) [3, 4], whereas PDCL3 was ubiquitously expressed. During spermatogenesis, during the pachytene stage of meiosis, PDCL2 starts to express, and PDCL3 starts to decay (Fig. S3C, D and Fig. S6A, B). It can be postulated that the remaining PDCL3 protein can function during meiosis. Additional information is required to demonstrate the relationship between PDCL2 and PDCL3.
Although the critical role of PDCL2 in spermiogenesis needs to be comprehensively clarified, we found that PDCL2 can interact with CCT and actin (Fig. 5), which strongly supports the role of PDCL2 for functional actin production. In fact, other members of subgroup II phosducin-like proteins (PhLP2) were found to modulate CCT function and assist or interfere with actin folding [14,15,16]. However, the details of this mechanism remain to be elucidated.
Materials and methods
Ethics statement
All experiments were conducted with protocols approved by The Institutional Animal Care and Use Committee at Sun Yat-sen University.
Animals
Mice used in all experiments were C57BL/6. Mice were housed in specific pathogen-free (SPF) animal facility in Sun Yat-sen University. The Institutional Animal Care and Use Committee of Sun Yat-sen University, P.R.China approved all the experimental protocols concerning the handling of mice. Pdcl2 knock-out mice were generated and tested following the previously described protocol [37].
We generated gRNAs expressing vector with gRNAs sequences as follows:
gRNA1: ttgtgcccctcactgggatc (AGG)
gRNA2: ggtgttgcgcttacagc (AGG)
100 ng/μL gRNA1, 100 ng/μL gRNA2, and 200 ng/μL Cas9 mRNAs were mixed and then 10 pL of the mixture was injected into the cytoplasm of each zygote. Mice were genotyped using the same primers as the T7E1 assay primers.
T7E1 assay primers are as follows:
T7E1-E1-F: 5′-GGACTCAGGGGTAAAAGGCG-3′,
T7E1-E1-R: 5′-TGACTAACAAAGAGAGACACTGCTAAAG-3′,
T7E1-E2-F: 5′-CTGAGTGGATGGGCAGTGTT-3′,
T7E1-E2-R: 5′-TGTGTCCTGTGAATCAAAGCA-3′.
Mutant mice of the first generation were mated with wild-type C57BL/6 for three generations trying to eliminate the uttermost off-target effect of the CRISPR/Cas9 system. The fourth generation of Pdcl2+/− mice were mated to generate Pdcl2+/+, Pdcl2+/−, and Pdcl2−/− mice. Mice of the same age from different groups were selected randomly to be analyzed.
Semi-quantitative RT-PCR
RNA of different mouse tissues were extracted using the TRIzol reagent (Invitrogen, 15596–018). RNA samples were reverse transcripted using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, #K1621). Semi-quantitative RT-PCR conditions were as previously described [37]. Primers information were as follows:
β-actin-F: 5′-TTCTTTGCAGCTCCTTCGTTGCCG-3′,
β-actin-R: 5′-TGGATGGCTACGTACATGGCTGGG-3′,
Pdcl2-F: 5′-TGGCACAACTGAAAGAAGCAGA-3′,
Pdcl2-R: 5′-GCTTGAGATTTATCCCTCCACAT-3′.
Generation of rabbit anti-PDCL2 polyclonal antibody
The entire protein coding sequence of mouse Pdcl2 (NM_023508.7 CDS without stop codon) was cloned into pENTR/D-TOPO vector (Invitrogen) and recombined to pDEST15 (Invitrogen) using LR reaction (Invitrogen, 11791–020). The GST-PDCL2 fusion protein was expressed in Escherichia coli BL21 (DE3) and purified using Glutathione Sepharose 4B (GE Healthcare, 17-0756-01) according to the manufacturer’s instructions. Rabbits were immunized with the purified protein (Shanghai Institutes for Biological Sciences, CAS).
The entire protein coding sequence of mouse Pdcl2 (NM_023508.7 CDS without stop codon) was cloned into pGEX-5X vector (GE Healthcare). The GST-PDCL2 fusion protein was expressed and purified as described above. The purified protein was used as an antigen to purify the rabbit serum using an antigen-coupled column generated with AminoLink Plus Coupling Resin (Pierce, 20501). Antibody specificity and sensitivity were tested (Fig. S2).
Western blot analysis
Experiment procedures were described as previously [37]. Usages of primary antibodies were as follows:
rabbit anti-PDCL2 antibody (1: 1000),
mouse anti-GAPDH antibody (1: 8000; Proteintech group, 60004-1-Ig),
mouse anti-GST antibody (1:5000; Abmart, M20007),
rabbit anti-FLAG antibody (1:5000; Sigma-Aldrich, F7425)
Usages of secondary antibodies were as follows:
Goat anti-mouse IgG (1: 10,000; LI-COR, 926–32220),
goat anti-rabbit IgG (1: 10,000; LI-COR, 926–32211).
Histological analysis
Bouin’s solution fixed mouse testes and epididymis were embedded in paraffin. Tissue-paraffin blocks were sectioned in 2 μm. Tissue slices were rehydrated sequentially and stained with hematoxylin and eosin.
Immunofluorescent analysis of germ cells
Mouse germ cells were prepared and stained as previously described [37]. Usages of primary antibodies were as follows:
rabbit anti-PDCL2 antibody (1: 5000),
goat anti-PLZF antibody (1:500; R&D Systems, AF2944),
mouse anti-GATA4 antibody (1:100; Santa Cruz Biotechnology, sc-25310),
mouse anti-SYCP3 antibody (1:200; Santa Cruz Biotechnology, sc-74569),
mouse anti-sp56 antibody (1:1000; QED Bioscience, 55101).
Usages of secondary antibodies were as follows:
Alexa Fluor 488-conjugated donkey anti-goat IgG (1: 500, Molecular Probes, A-11055),
Alexa Fluor 488-conjugated donkey anti-mouse IgG (1: 500, Molecular Probes, A-21202),
Alexa Fluor 555-conjugated donkey anti-rabbit IgG (1: 500, Molecular Probes, A-31572).
Immunofluorescent analysis of testes sections
Experiment procedures were as previously described [37] with modifications. Testes were fixed in PFA. Tissue-paraffin blocks were sectioned into 7 μm slices.
Usages of primary antibodies were as follows:
rabbit anti-PDCL2 antibody (1: 5000),
Usages of secondary antibodies were as follows:
Alexa Fluor 555-conjugated donkey anti-rabbit IgG (1: 500, Molecular Probes, A-31572).
Sperm quantity and motility analysis
Sperm from cauda epididymis was analyzed as previously described [37].
Transmission electron microscope analysis of sperm
Sperm samples were collected as previously described [37]. Samples were centrifuged and washed with PBS for three times at 500 × g for 2 min each time. Sperm samples were fixed by adding 1 mL glutaric dialdehyde and stored at 4 °C overnight. Samples were stained with uranyl acetate and lead citrate performed by Research Center of School of Life Sciences, Sun Yat-sen University. Images were captured with JEM1400 (JEOL).
Analysis of apoptotic cells
Optimal cutting temperature compound (Sakura, 4583) embedded mouse testes and epididymis were cryosectioned into 10 μm slices. TUNEL assay was performed according to the manufacturer’s instruction (Roche, 11684795910).
Analysis of germ cell ploidy types
Germ cell suspensions were prepared and stained with propidium iodide as previously described [37]. Flow cytometry analyses were performed using FACS Calibur (BD).
Coimmunoprecipitation coupled with mass spectrometric identification (IP-MS)
Equally weighted mouse testes tissue of each group were homogenized in cold NETN-G buffer (40 mM Tris-HCl pH 8.0, 100 mM NaCl, 0.5% NP40, 1 mM EDTA pH 8.0, 10% glycerol) with 1:500 Cocktail (Sigma, P3840), 10 mM NaF, 0.2 mM PMSF, 1 mM DTT, 2 mM Na4P2O7 and 1 mM Na3VO4. The homogenates were Ultra-centrifuged at 24,000 rpm for 30 min at 4 °C. The sediment and upper lipid layers were discarded.
Preclear the lysate by incubating with 25 µL Rabbit IgG (Sangon Biotech, D110502) at 4 °C for an hour. Followed the incubation, 15 µL solid volume of precleared protein A/G agarose (Pierce, 20422) and 150 µL solid volume of precleared sefinose CL-6B (Sangon Biotech, SF004-L6B) were added to the mixture then incubated at 4 °C for 30 min. Centrifuge the mixture at 3000 rpm for 1 min at 4 °C (Eppendorf 5810R). The sediments were discarded. Centrifuge the supernatant at 20,000 × g for 30 min at 4 °C (Sigma 3–18 K centrifuge).
Binding the protein with PDCL2 antibody by adding 60 µL purified PDCL2 antibody to the supernatant and incubating at 4 °C for 3 h. After that, 15 µL solid volume of precleared protein A/G agarose were added to the mixture and incubated at 4 °C for 1.5 h. Centrifuge the mixture at 3000 rpm for 1 min at 4 °C (Eppendorf 5810R). The supernatant was discarded and the agarose beads were washed for three times with 1 mL NETN-G buffer each time.
Agarose samples were prepared to run SDS-PAGE. After electrophoresis, the gel was stained in coomassie brilliant blue buffer then cut out and sent to the Beijing Proteomics Research Center to perform the mass spectrometric identification.
GST pull down assay
The full-length coding sequences of the target proteins were subcloned into the vector pDEST27 (Invitrogen) or pBabe-CMV-SFB, which generated fusion proteins with N-terminal GST tag or C-terminal S tag-FLAG tag-SBP tag. The vectors were transfected into 293T cells with polyethyleneimine (Sigma-Aldrich, 408727). Cells were lysed after 40 h of transfection. Cells were incubated with NETN-G buffer with 1: 100 cocktail and 10 mM NaF for 30 min at 4 °C. The homogenates were centrifuged at 15,000 rpm for 15 min at 4 °C. Supernatants were incubated with precleared Glutathione Sepharose 4B (GE Healthcare, 17-0756-01) at 4 °C for an hour. After incubation, beads were washed for three times with 0.5 mL NETN-G buffer each time. Samples were then prepared to run western blot analysis.
Sequence analysis
Mouse PDCL2 and PDCL3 sequence (NP 075997.1 and NP 081126.2) were used to align with the online databases using the blastp suite (NCBI, protein-protein BLAST) to find out orthologues in other species. Multiple alignments were performed using MEGA7 and a phylogenetic tree was constructed. The evolutionary history was inferred using the Neighbor-Joining method. The evolutionary distances were computed using the Poisson correction method.
Statistical analysis
All data are presented as mean ± S.D. SigmaPlot version 12.5 was used for analyzing data. The statistical significance of two groups were analyzed by using two-tailed t-test. The statistical significance of three groups were analyzed by using one way ANOVA. Normality test was performed using the Shapiro–Wilk method. Equal variance test was also performed. If there is a significant difference, Holm–Sidak method was used for all pairwise multiple comparisons. The data were considered significant when p < 0.05 (*), 0.01 (**) or 0.001 (***). Otherwise, it would be considered not significant (N.S.).
Data availability
All data are available in the main text or the supplementary materials.
References
Agarwal A, Baskaran S, Parekh N, Cho CL, Henkel R, Vij S, et al. Male infertility. Lancet 2021;397:319–33.
Platts AE, Dix DJ, Chemes HE, Thompson KE, Goodrich R, Rockett JC, et al. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum Mol Genet. 2007;16:763–73.
Lopez P, Yaman R, Lopez-Fernandez LA, Vidal F, Puel D, Clertant P, et al. A novel germ line-specific gene of the phosducin-like protein (PhLP) family. A meiotic function conserved from yeast to mice. J Biol Chem. 2003;278:1751–7.
Zheng YH, Rengaraj D, Park KJ, Lee SI, Han JY. Expression patterns of germ cell-specific Phosducin-like 2 during testicular and ovarian development in chickens. Asian Austral J Anim. 2010;23:1000–6.
McLaughlin JN, Thulin CD, Hart SJ, Resing KA, Ahn NG, Willardson BM. Regulatory interaction of phosducin-like protein with the cytosolic chaperonin complex. Proc Natl Acad Sci USA. 2002;99:7962–7.
Martin-Benito J, Bertrand S, Hu T, Ludtke PJ, McLaughlin JN, Willardson BM, et al. Structure of the complex between the cytosolic chaperonin CCT and phosducin-like protein. Proc Natl Acad Sci USA. 2004;101:17410–5.
Humrich J, Bermel C, Bunemann MB, Harmark LH, Frost RJA, Quitterer U, et al. Phosducin-like protein regulates G-protein beta gamma folding by interaction with tailless complex polypeptide-1 alpha - Dephosphorylation or splicing of PhLP turns the switch toward regulation of G beta gamma folding. J Biol Chem. 2005;280:20042–50.
Lukov GL, Baker CM, Ludtke PJ, Hu T, Carter MD, Hackett RA, et al. Mechanism of assembly of G protein beta gamma subunits by protein kinase CK2-phosphorylated phosducin-like protein and the cytosolic chaperonin complex. J Biol Chem. 2006;281:22261–74.
Wells CA, Dingus J, Hildebrandt JD. Role of the chaperonin CCT/TRiC complex in G protein beta gamma-dimer assembly. J Biol Chem. 2006;281:20221–32.
Willardson BM, Howlett AC. Function of phosducin-like proteins in G protein signaling and chaperone-assisted protein folding. Cell Signal. 2007;19:2417–27.
Posokhova E, Song H, Belcastro M, Higgins L, Bigley LR, Michaud NA, et al. Disruption of the chaperonin containing TCP-1 function affects protein networks essential for rod outer segment morphogenesis and survival. Mol Cell Proteom. 2011;10:M110 000570.
Gao X, Sinha S, Belcastro M, Woodard C, Ramamurthy V, Stoilov P, et al. Splice isoforms of phosducin-like protein control the expression of heterotrimeric G proteins. J Biol Chem. 2013;288:25760–8.
Plimpton RL, Cuellar J, Lai CW, Aoba T, Makaju A, Franklin S, et al. Structures of the Gbeta-CCT and PhLP1-Gbeta-CCT complexes reveal a mechanism for G-protein beta-subunit folding and Gbetagamma dimer assembly. Proc Natl Acad Sci USA. 2015;112:2413–8.
Stirling PC, Srayko M, Takhar KS, Pozniakovsky A, Hyman AA, Leroux MR. Functional interaction between phosducin-like protein 2 and cytosolic chaperonin is essential for cytoskeletal protein function and cell cycle progression. Mol Biol Cell. 2007;18:2336–45.
McCormack EA, Altschuler GM, Dekker C, Filmore H, Willison KR. Yeast phosducin-like protein 2 acts as a stimulatory co-factor for the folding of actin by the chaperonin CCT via a ternary complex. J Mol Biol. 2009;391:192–206.
Stuart SF, Leatherbarrow RJ, Willison KR. A two-step mechanism for the folding of actin by the yeast cytosolic chaperonin. J Biol Chem. 2011;286:178–84.
Stirling PC, Cuellar J, Alfaro GA, El Khadali F, Beh CT, Valpuesta JM, et al. PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J Biol Chem. 2006;281:7012–21.
Bauer PH, Muller S, Puzicha M, Pippig S, Obermaier B, Helmreich EJ, et al. Phosducin is a protein kinase A-regulated G-protein regulator. Nature 1992;358:73–6.
Hawes BE, Touhara K, Kurose H, Lefkowitz RJ, Inglese J. Determination of the G beta gamma-binding domain of phosducin. A regulatable modulator of G beta gamma signaling. J Biol Chem. 1994;269:29825–30.
Flanary PL, DiBello PR, Estrada P, Dohlman HG. Functional analysis of Plp1 and Plp2, two homologues of phosducin in yeast. J Biol Chem. 2000;275:18462–9.
Bregier C, Krzemien-Ojak L, Wloga D, Jerka-Dziadosz M, Joachimiak E, Batko K, et al. PHLP2 is essential and plays a role in ciliogenesis and microtubule assembly in Tetrahymena thermophila. J Cell Physiol. 2013;228:2175–89.
Wilkinson JC, Richter BW, Wilkinson AS, Burstein E, Rumble JM, Balliu B, et al. VIAF, a conserved inhibitor of apoptosis (IAP)-interacting factor that modulates caspase activation. J Biol Chem. 2004;279:51091–9.
Yebenes H, Mesa P, Munoz IG, Montoya G, Valpuesta JM. Chaperonins: Two rings for folding. Trends Biochem Sci. 2011;36:424–32.
Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: Protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604.
Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–52.
Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–9.
Ketola I, Anttonen M, Vaskivuo T, Tapanainen JS, Toppari J, Heikinheimo M. Developmental expression and spermatogenic stage specificity of transcription factors GATA-1 and GATA-4 and their cofactors FOG-1 and FOG-2 in the mouse testis. Eur J Endocrinol / Eur Federation Endocr Societies. 2002;147:397–406.
Ketola I, Rahman N, Toppari J, Bielinska M, Porter-Tinge SB, Tapanainen JS, et al. Expression and regulation of transcription factors GATA-4 and GATA-6 in developing mouse testis. Endocrinology 1999;140:1470–80.
Kolas NK, Yuan L, Hoog C, Heng HH, Marcon E, Moens PB. Male mouse meiotic chromosome cores deficient in structural proteins SYCP3 and SYCP2 align by homology but fail to synapse and have possible impaired specificity of chromatin loop attachment. Cytogenet Genome Res. 2004;105:182–8.
Kouznetsova A, Novak I, Jessberger R, Hoog C. SYCP2 and SYCP3 are required for cohesin core integrity at diplotene but not for centromere cohesion at the first meiotic division. J Cell Sci. 2005;118:2271–8.
Cheng A, Le T, Palacios M, Bookbinder LH, Wassarman PM, Suzuki F, et al. Sperm-egg recognition in the mouse: characterization of sp56, a sperm protein having specific affinity for ZP3. J Cell Biol. 1994;125:867–78.
Shukla KK, Mahdi AA, Rajender S. Apoptosis, spermatogenesis, and male infertility. Front Biosci (Elite Ed). 2012;4:746–54.
Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, et al. Structure-based assembly of protein complexes in yeast. Science 2004;303:2026–9.
Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature 2006;440:631–6.
Zhang J. Evolution by gene duplication: An update. Trends Ecol Evol. 2003;18:292–8.
Horvath GC, Kistler MK, Kistler WS. RFX2 is a candidate downstream amplifier of A-MYB regulation in mouse spermatogenesis. Bmc Dev Biol. 2009;9:63.
Li M, Huang R, Jiang X, Chen Y, Zhang Z, Zhang X, et al. CRISPR/Cas9 promotes functional study of testis specific X-linked gene in vivo. PLoS One. 2015;10:e0143148.
Acknowledgements
We thank the members of Prof. Huang’s lab. This work was supported by National Natural Science Foundation of China (82201769, 32001063), Guangdong Basic and Applied Basic Research Foundation (2021A1515010759), Guangzhou Science and Technology Planning Project (202201020411), the Guangdong Special Support Program (2019BT02Y276) and the grant from MOE Key Laboratory of Gene Function and Regulation.
Author information
Authors and Affiliations
Contributions
XZ and JH conceived the study. ML, XZ, and JH designed the overall experimental scheme. ML and YC executed the experiments. ML, XZ, and JH analyzed the data. ML and XZ wrote the paper. XZ and JO revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Chen, Y., Ou, J. et al. PDCL2 is essential for spermiogenesis and male fertility in mice. Cell Death Discov. 8, 419 (2022). https://doi.org/10.1038/s41420-022-01210-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-022-01210-2
- Springer Nature Limited
This article is cited by
-
A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle
Nature Communications (2024)
-
STYXL1 regulates CCT complex assembly and flagellar tubulin folding in sperm formation
Nature Communications (2024)